Orphan Nuclear Receptor ERRγ Is a Transcriptional Regulator of CB1 Receptor-Mediated TFR2 Gene Expression in Hepatocytes
Abstract
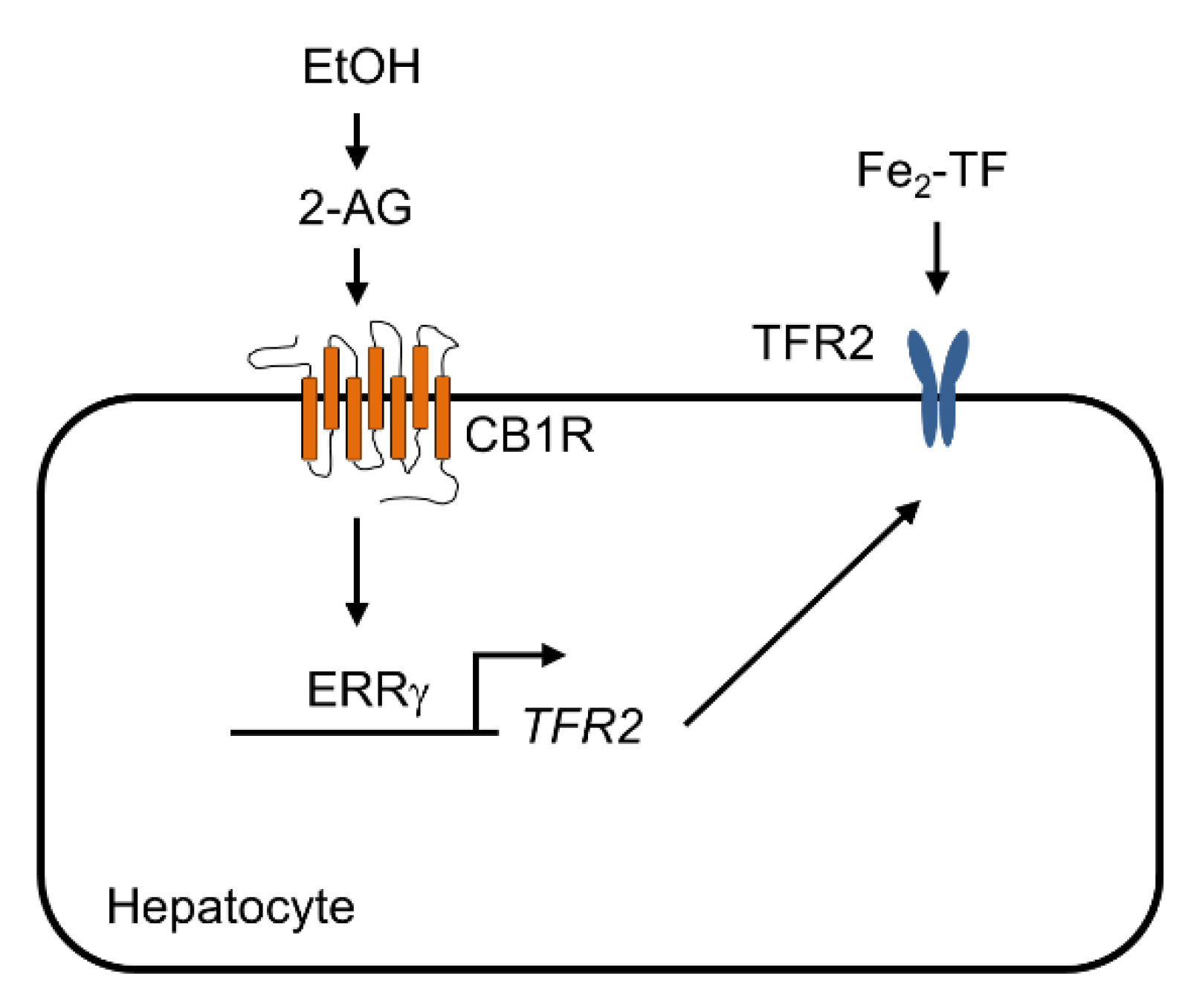
:1. Introduction
2. Results
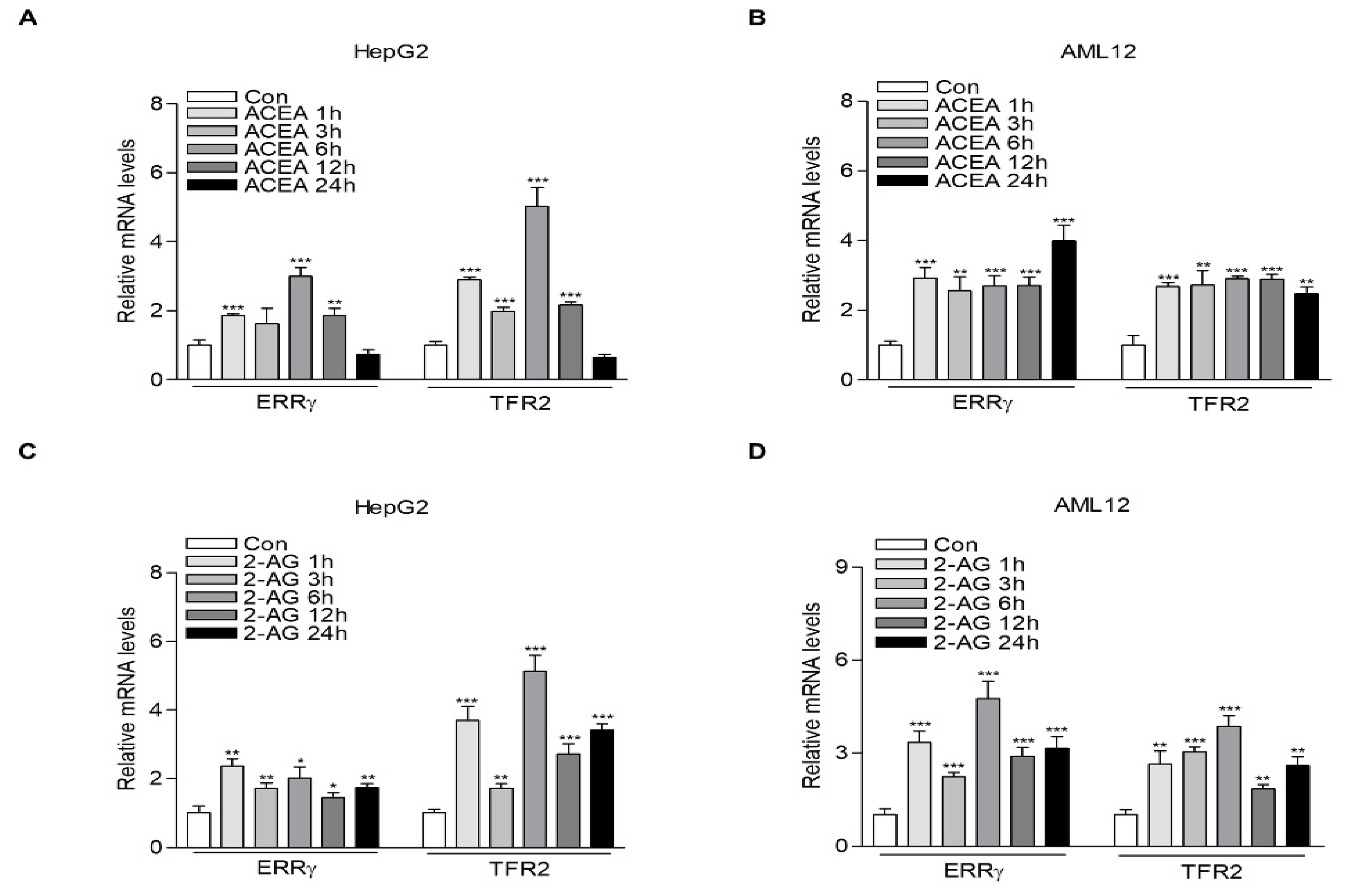
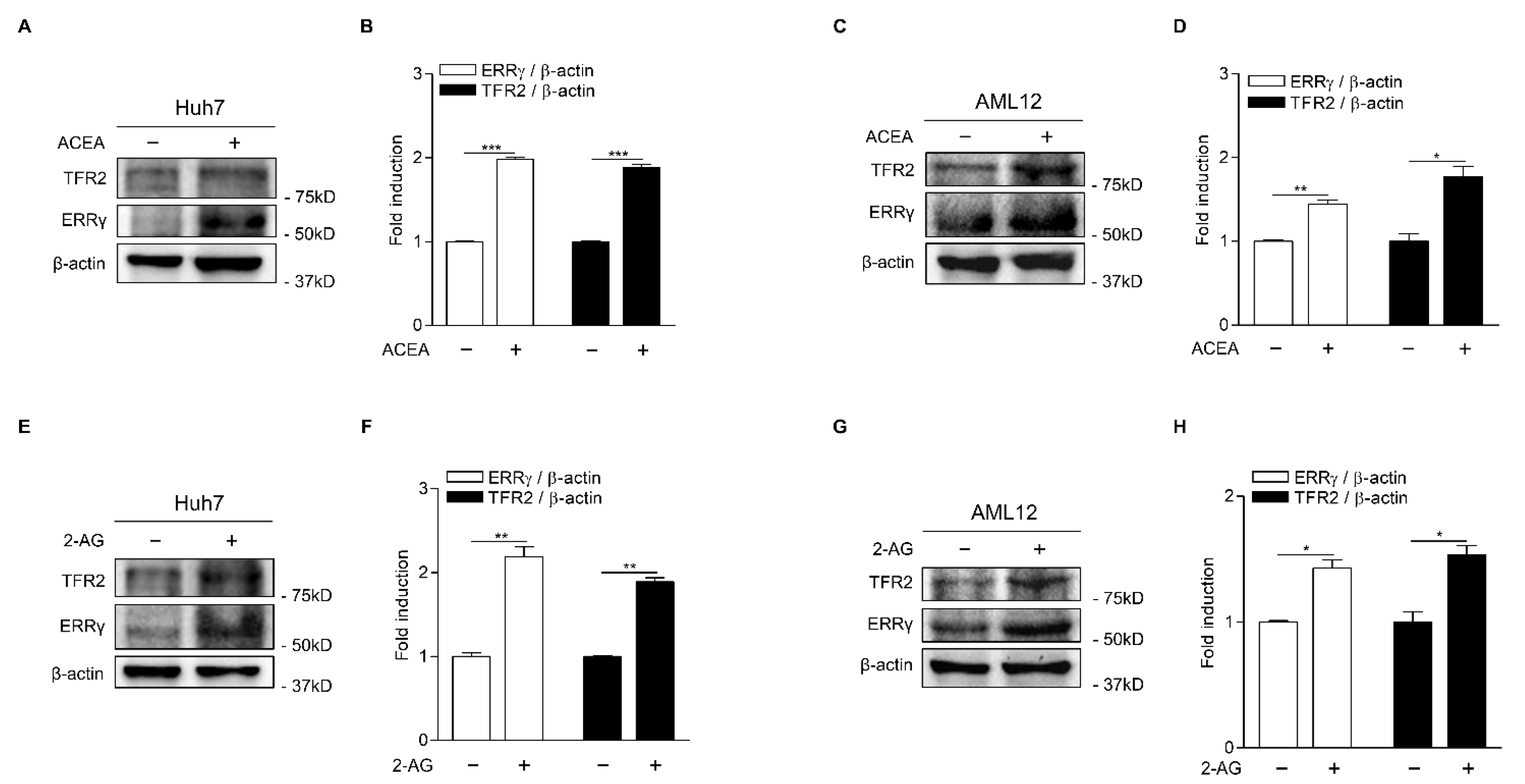
2.1. CB1 Receptor Signaling Increases ERRγ and TFR2 Gene Expression in Hepatocytes
2.2. ERRγ Increases TFR2 Gene Expression
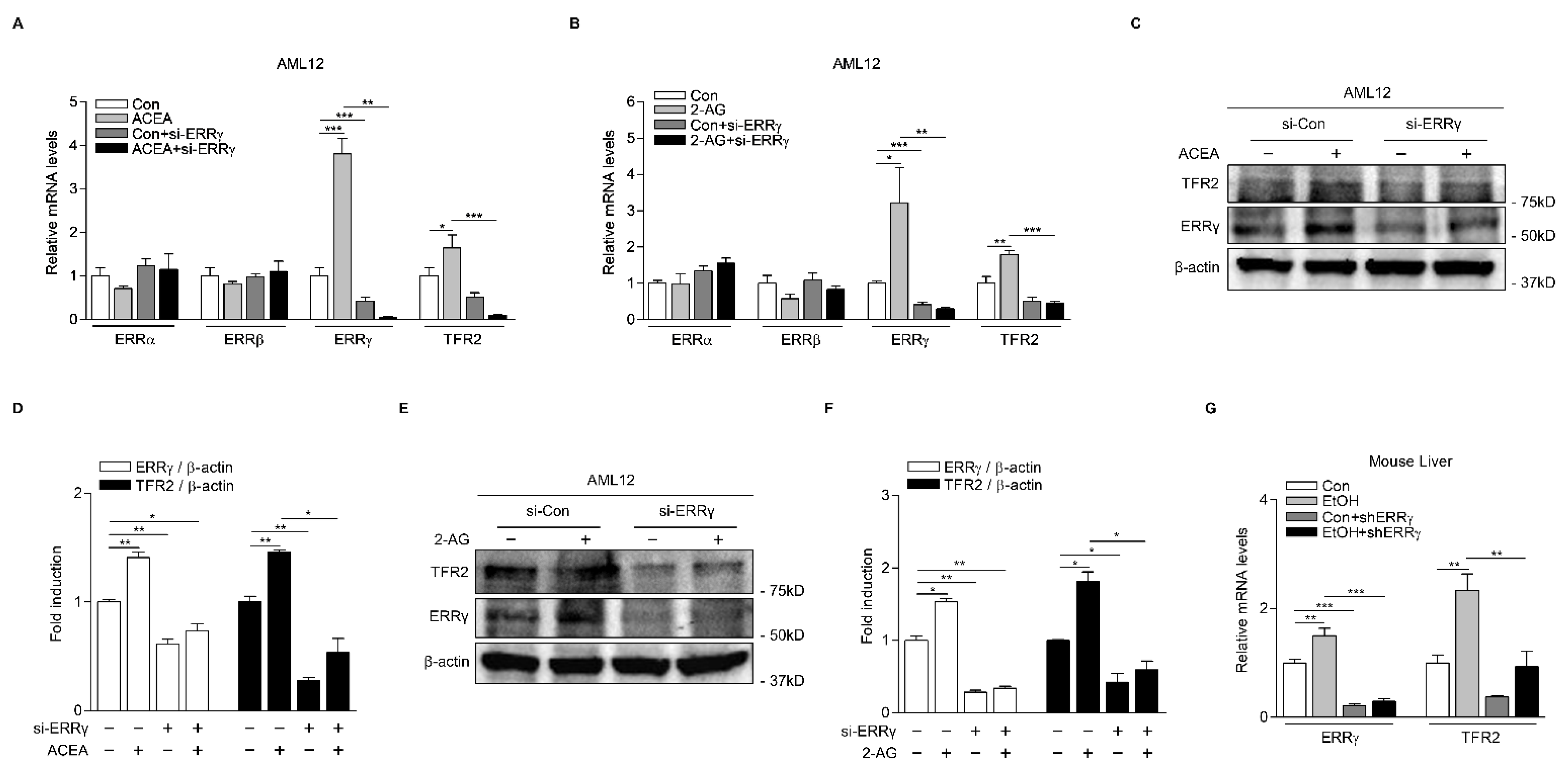
2.3. Knockdown of ERRγ Decreases CB1 Receptor-Mediated TFR2 Expression
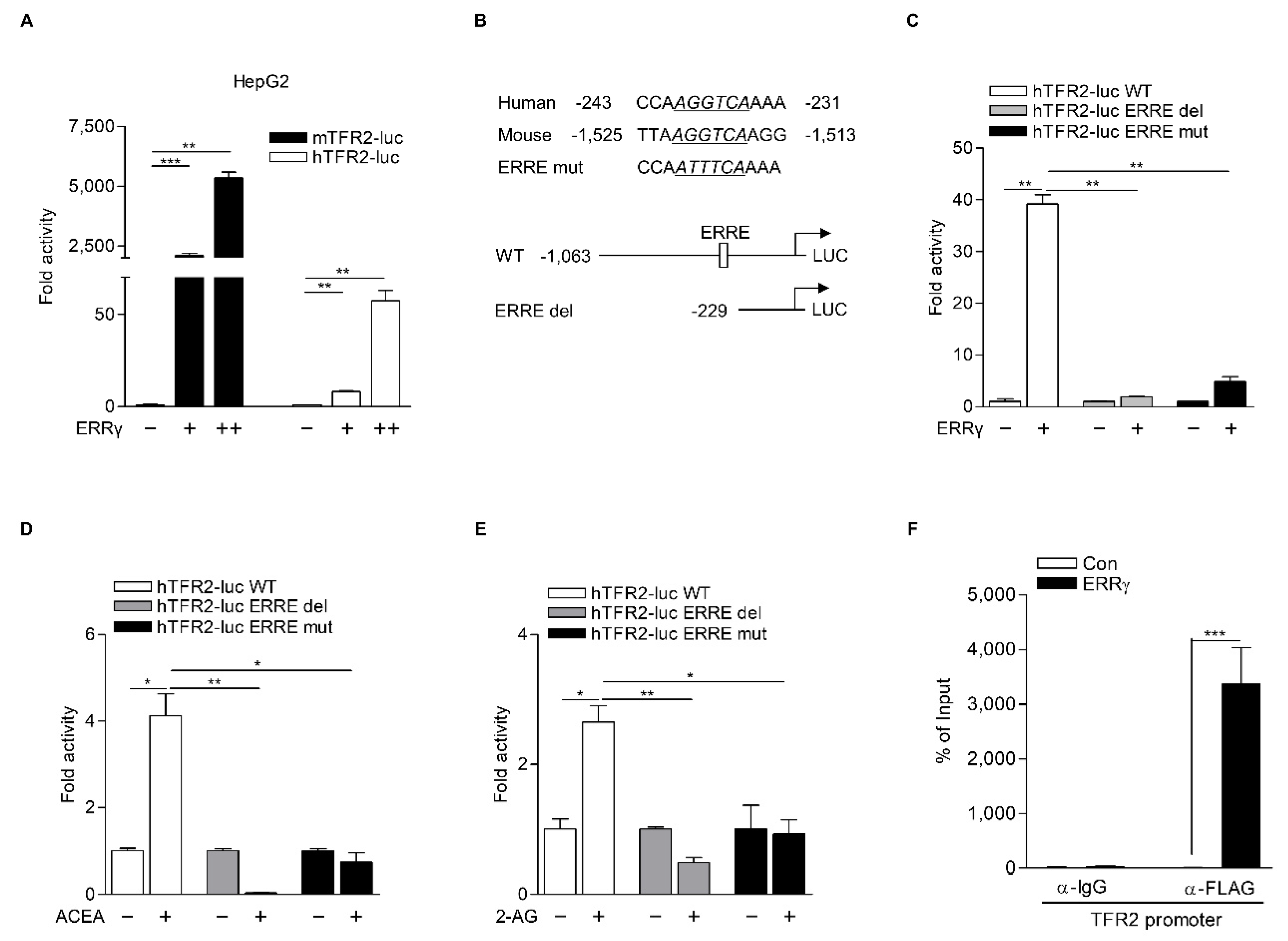
2.4. TFR2 Is a Direct Target of ERRγ
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Plasmid DNAs and Recombinant Adenoviruses
4.3. Cell Culture, Transfection and Luciferase Assay
4.4. Animal Experiment
4.5. Quantitative Real-Time PCR Analysis
4.6. Protein Extraction and Western Blot Analysis
4.7. Chromatin Immunoprecipitation (ChIP) Assay
4.8. RNA Interference
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mallat, A.; Teixeira-Clerc, F.; Deveaux, V.; Manin, S.; Lotersztajn, S. The endocannabinoid system as a key mediator during liver diseases: New insights and therapeutic openings. Br. J. Pharm. 2011, 163, 1432–1440. [Google Scholar] [CrossRef]
- Kunos, G. Interactions between Alcohol and the Endocannabinoid System. Alcoholism 2020, 44, 790–805. [Google Scholar] [CrossRef]
- Bisogno, T.; Howell, F.; Williams, G.; Minassi, A.; Cascio, M.G.; Ligresti, A.; Matias, I.; Schiano-Moriello, A.; Paul, P.; Williams, E.J.; et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J. Cell Biol. 2003, 163, 463–468. [Google Scholar] [CrossRef]
- Jeong, W.I.; Osei-Hyiaman, D.; Park, O.; Liu, J.; Bátkai, S.; Mukhopadhyay, P.; Horiguchi, N.; Harvey-White, J.; Marsicano, G.; Lutz, B.; et al. Paracrine activation of hepatic CB1 receptors by stellate cell-derived endocannabinoids mediates alcoholic fatty liver. Cell Metab. 2008, 7, 227–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, Y.S.; Kim, Y.H.; Radhakrishnan, K.; Kim, J.; Kim, D.K.; Lee, J.H.; Oh, H.; Lee, I.K.; Kim, W.; Cho, S.J.; et al. An inverse agonist of estrogen-related receptor γ regulates 2-arachidonoylglycerol synthesis by modulating diacylglycerol lipase expression in alcohol-intoxicated mice. Arch. Toxicol. 2020, 94, 427–438. [Google Scholar] [CrossRef]
- Gonzalez, F.J. Role of cytochromes P450 in chemical toxicity and oxidative stress: Studies with CYP2E1. Mutat. Res. 2005, 569, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Bataller, R. Alcoholic liver disease: Pathogenesis and new therapeutic targets. Gastroenterology 2011, 141, 1572–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.K.; Kim, Y.H.; Jang, H.H.; Park, J.; Kim, J.R.; Koh, M.; Jeong, W.I.; Koo, S.H.; Park, T.S.; Yun, C.H.; et al. Estrogen-related receptor γ controls hepatic CB1 receptor-mediated CYP2E1 expression and oxidative liver injury by alcohol. Gut J. 2013, 62, 1044–1054. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.K.; Kim, Y.H.; Lee, J.H.; Jung, Y.S.; Kim, J.; Feng, R.; Jeon, T.I.; Lee, I.K.; Cho, S.J.; Im, S.S.; et al. Estrogen-related receptor γ controls sterol regulatory element-binding protein-1c expression and alcoholic fatty liver. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 158521. [Google Scholar] [CrossRef] [PubMed]
- Ponka, P.; Lok, C.N. The transferrin receptor: Role in health and disease. Int. J. Biochem. Cell Biol. 1999, 31, 1111–1137. [Google Scholar] [CrossRef]
- Richardson, D.R.; Ponka, P. The molecular mechanisms of the metabolism and transport of iron in normal and neoplastic cells. Biochim. Biophys. Acta Rev. Biomembr. 1997, 1331, 1–40. [Google Scholar] [CrossRef]
- Herbison, C.E.; Thorstensen, K.; Chua, A.C.; Graham, R.M.; Leedman, P.; Olynyk, J.K.; Trinder, D. The role of transferrin receptor 1 and 2 in transferrin-bound iron uptake in human hepatoma cells. Am. J. Physiol. Cell Physiol. 2009, 297, C1567–C1575. [Google Scholar] [CrossRef] [Green Version]
- Kawabata, H.; Germain, R.S.; Ikezoe, T.; Tong, X.; Green, E.M.; Gombart, A.F.; Koeffler, H.P. Regulation of expression of murine transferrin receptor 2. Blood 2001, 98, 1949–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuo, S.; Ogawa, M.; Muckenthaler, M.U.; Mizui, Y.; Sasaki, S.; Fujimura, T.; Takizawa, M.; Ariga, N.; Ozaki, H.; Sakaguchi, M.; et al. Hepatocyte Nuclear Factor 4α Controls Iron Metabolism and Regulates Transferrin Receptor 2 in Mouse Liver. J. Biol. Chem. 2015, 290, 30855–30865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, D.F.; Summerville, L.; Subramaniam, V.N. Targeted disruption of the hepatic transferrin receptor 2 gene in mice leads to iron overload. Gastroenterology 2007, 132, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Roetto, A.; Di Cunto, F.; Pellegrino, R.M.; Hirsch, E.; Azzolino, O.; Bondi, A.; Defilippi, I.; Carturan, S.; Miniscalco, B.; Riondato, F.; et al. Comparison of 3 Tfr2-deficient murine models suggests distinct functions for Tfr2-α and Tfr2-β isoforms in different tissues. Blood 2010, 115, 3382–3389. [Google Scholar] [CrossRef]
- Wallace, D.F.; Summerville, L.; Crampton, E.M.; Frazer, D.M.; Anderson, G.J.; Subramaniam, V.N. Combined deletion of Hfe and transferrin receptor 2 in mice leads to marked dysregulation of hepcidin and iron overload. Hepatology 2009, 50, 1992–2000. [Google Scholar] [CrossRef]
- West, A.P., Jr.; Bennett, M.J.; Sellers, V.M.; Andrews, N.C.; Enns, C.A.; Bjorkman, P.J. Comparison of the interactions of transferrin receptor and transferrin receptor 2 with transferrin and the hereditary hemochromatosis protein HFE. J. Biol. Chem. 2000, 275, 38135–38138. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Chen, J.; Kramer, M.; Tsukamoto, H.; Zhang, A.S.; Enns, C.A. Interaction of the hereditary hemochromatosis protein HFE with transferrin receptor 2 is required for transferrin-induced hepcidin expression. Cell Metab. 2009, 9, 217–227. [Google Scholar] [CrossRef] [Green Version]
- Nemeth, E.; Tuttle, M.S.; Powelson, J.; Vaughn, M.B.; Donovan, A.; Ward, D.M.; Ganz, T.; Kaplan, J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004, 306, 2090–2093. [Google Scholar] [CrossRef] [Green Version]
- Powell, L.W. Normal human iron storage and its relation to ethanol consumption. Australas. Ann. Med. 1966, 15, 110–115. [Google Scholar] [CrossRef]
- Suzuki, Y.; Saito, H.; Suzuki, M.; Hosoki, Y.; Sakurai, S.; Fujimoto, Y.; Kohgo, Y. Up-regulation of transferrin receptor expression in hepatocytes by habitual alcohol drinking is implicated in hepatic iron overload in alcoholic liver disease. Alcoholism 2002, 26, S26–S31. [Google Scholar] [CrossRef]
- Misra, J.; Kim, D.K.; Choi, H.S. ERRγ: A Junior Orphan with a Senior Role in Metabolism. Trends Endocrinol. Metab. 2017, 28, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Greschik, H.; Wurtz, J.M.; Sanglier, S.; Bourguet, W.; van Dorsselaer, A.; Moras, D.; Renaud, J.P. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol. Cell 2002, 9, 303–313. [Google Scholar] [CrossRef]
- Hentschke, M.; Süsens, U.; Borgmeyer, U. PGC-1 and PERC, coactivators of the estrogen receptor-related receptor gamma. Biochem. Biophys. Res. Commun. 2002, 299, 872–879. [Google Scholar] [CrossRef]
- Sanyal, S.; Kim, J.Y.; Kim, H.J.; Takeda, J.; Lee, Y.K.; Moore, D.D.; Choi, H.S. Differential regulation of the orphan nuclear receptor small heterodimer partner (SHP) gene promoter by orphan nuclear receptor ERR isoforms. J. Biol. Chem. 2002, 277, 1739–1748. [Google Scholar] [CrossRef] [Green Version]
- Misra, J.; Kim, D.K.; Jung, Y.S.; Kim, H.B.; Kim, Y.H.; Yoo, E.K.; Kim, B.G.; Kim, S.; Lee, I.K.; Harris, R.A.; et al. O-GlcNAcylation of Orphan Nuclear Receptor Estrogen-Related Receptor γ Promotes Hepatic Gluconeogenesis. Diabetes 2016, 65, 2835–2848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.K.; Kim, Y.H.; Hynx, D.; Wang, Y.; Yang, K.J.; Ryu, D.; Kim, K.S.; Yoo, E.K.; Kim, J.S.; Koo, S.H.; et al. PKB/Akt phosphorylation of ERRγ contributes to insulin-mediated inhibition of hepatic gluconeogenesis. Diabetologia 2014, 57, 2576–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.K.; Jeong, J.H.; Lee, J.M.; Kim, K.S.; Park, S.H.; Kim, Y.D.; Koh, M.; Shin, M.; Jung, Y.S.; Kim, H.S.; et al. Inverse agonist of estrogen-related receptor γ controls Salmonella typhimurium infection by modulating host iron homeostasis. Nat. Med. 2014, 20, 419–424. [Google Scholar] [CrossRef]
- Zuercher, W.J.; Gaillard, S.; Orband-Miller, L.A.; Chao, E.Y.; Shearer, B.G.; Jones, D.G.; Miller, A.B.; Collins, J.L.; McDonnell, D.P.; Willson, T.M. Identification and structure-activity relationship of phenolic acyl hydrazones as selective agonists for the estrogen-related orphan nuclear receptors ERRβ and ERRγ. J. Med. Chem. 2005, 48, 3107–3109. [Google Scholar] [CrossRef]
- Kim, D.K.; Ryu, D.; Koh, M.; Lee, M.W.; Lim, D.; Kim, M.J.; Kim, Y.H.; Cho, W.J.; Lee, C.H.; Park, S.B.; et al. Orphan nuclear receptor estrogen-related receptor γ (ERRγ) is key regulator of hepatic gluconeogenesis. J. Biol. Chem. 2012, 287, 21628–21639. [Google Scholar] [CrossRef] [Green Version]
- Zakhari, S. Overview: How is alcohol metabolized by the body? Alcohol Res. Health 2006, 29, 245–254. [Google Scholar] [PubMed]
- Kano, M.; Ohno-Shosaku, T.; Hashimotodani, Y.; Uchigashima, M.; Watanabe, M. Endocannabinoid-mediated control of synaptic transmission. Physiol. Rev. 2009, 89, 309–380. [Google Scholar] [CrossRef] [PubMed]
- Valerio, L.G., Jr.; Parks, T.; Petersen, D.R. Alcohol mediates increases in hepatic and serum nonheme iron stores in a rat model for alcohol-induced liver injury. Alcoholism 1996, 20, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, H.; Horne, W.; Kamimura, S.; Niemelä, O.; Parkkila, S.; Ylä-Herttuala, S.; Brittenham, G.M. Experimental liver cirrhosis induced by alcohol and iron. J. Clin. Investig. 1995, 96, 620–630. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.C.; Merlino, G.; Fausto, N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl. Acad. Sci. USA 1994, 91, 674–678. [Google Scholar] [CrossRef] [Green Version]
- Sefried, S.; Häring, H.U.; Weigert, C.; Eckstein, S.S. Suitability of hepatocyte cell lines HepG2, AML12 and THLE-2 for investigation of insulin signalling and hepatokine gene expression. Open Biol. 2018, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, P.A.; Clarkson, R.; Hussain, A.; Weeks, R.J.; Morison, I.M. DNA methylation of hepatic iron sensing genes and the regulation of hepcidin expression. PLoS ONE 2018, 13, e0197863. [Google Scholar] [CrossRef] [Green Version]
- Rapisarda, C.; Puppi, J.; Hughes, R.D.; Dhawan, A.; Farnaud, S.; Evans, R.W.; Sharp, P.A. Transferrin receptor 2 is crucial for iron sensing in human hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G778–G783. [Google Scholar] [CrossRef] [Green Version]
- Giguère, V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 2008, 29, 677–696. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.K.; Gang, G.T.; Ryu, D.; Koh, M.; Kim, Y.N.; Kim, S.S.; Park, J.; Kim, Y.H.; Sim, T.; Lee, I.K.; et al. Inverse agonist of nuclear receptor ERRγ mediates antidiabetic effect through inhibition of hepatic gluconeogenesis. Diabetes 2013, 62, 3093–3102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzog, B.; Cardenas, J.; Hall, R.K.; Villena, J.A.; Budge, P.J.; Giguère, V.; Granner, D.K.; Kralli, A. Estrogen-related receptor alpha is a repressor of phosphoenolpyruvate carboxykinase gene transcription. J. Biol. Chem. 2006, 281, 99–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, B.-E.; Choi, B.; Park, W.-R.; Kim, Y.-J.; Kim, I.-Y.; Jung, Y.S.; Kim, Y.-H.; Lee, C.-H.; Choi, H.-S.; Kim, D.-K. Orphan Nuclear Receptor ERRγ Is a Transcriptional Regulator of CB1 Receptor-Mediated TFR2 Gene Expression in Hepatocytes. Int. J. Mol. Sci. 2021, 22, 6021. https://doi.org/10.3390/ijms22116021
Kim B-E, Choi B, Park W-R, Kim Y-J, Kim I-Y, Jung YS, Kim Y-H, Lee C-H, Choi H-S, Kim D-K. Orphan Nuclear Receptor ERRγ Is a Transcriptional Regulator of CB1 Receptor-Mediated TFR2 Gene Expression in Hepatocytes. International Journal of Molecular Sciences. 2021; 22(11):6021. https://doi.org/10.3390/ijms22116021
Chicago/Turabian StyleKim, Bo-Eun, Byungyoon Choi, Woo-Ram Park, Yu-Ji Kim, In-Young Kim, Yoon Seok Jung, Yong-Hoon Kim, Chul-Ho Lee, Hueng-Sik Choi, and Don-Kyu Kim. 2021. "Orphan Nuclear Receptor ERRγ Is a Transcriptional Regulator of CB1 Receptor-Mediated TFR2 Gene Expression in Hepatocytes" International Journal of Molecular Sciences 22, no. 11: 6021. https://doi.org/10.3390/ijms22116021





