S100 Calcium-Binding Protein P Secreted from Megakaryocytes Promotes Osteoclast Maturation
Abstract
:1. Introduction
2. Results
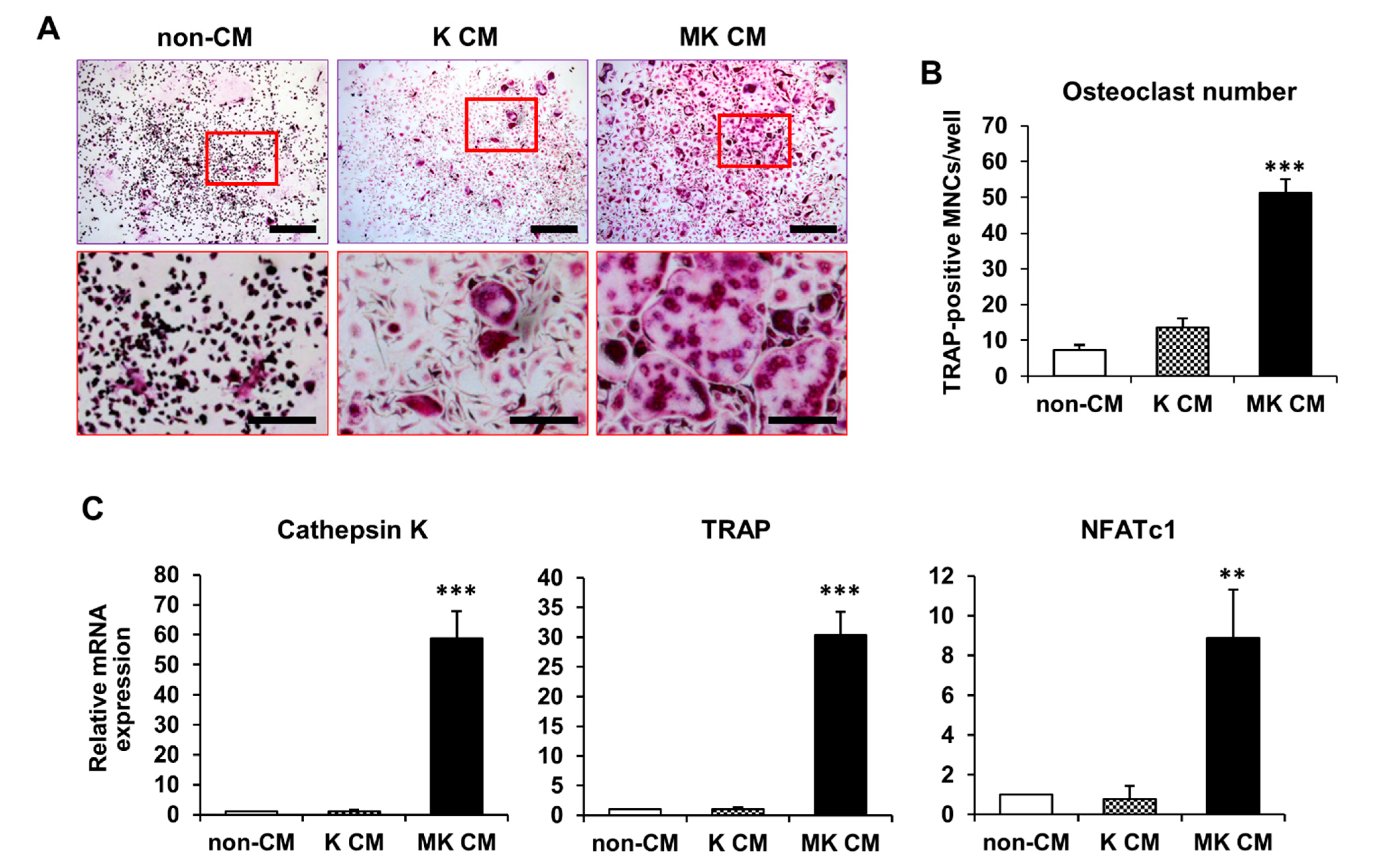
2.1. Osteoclast Differentiation in MK-Cultured CM
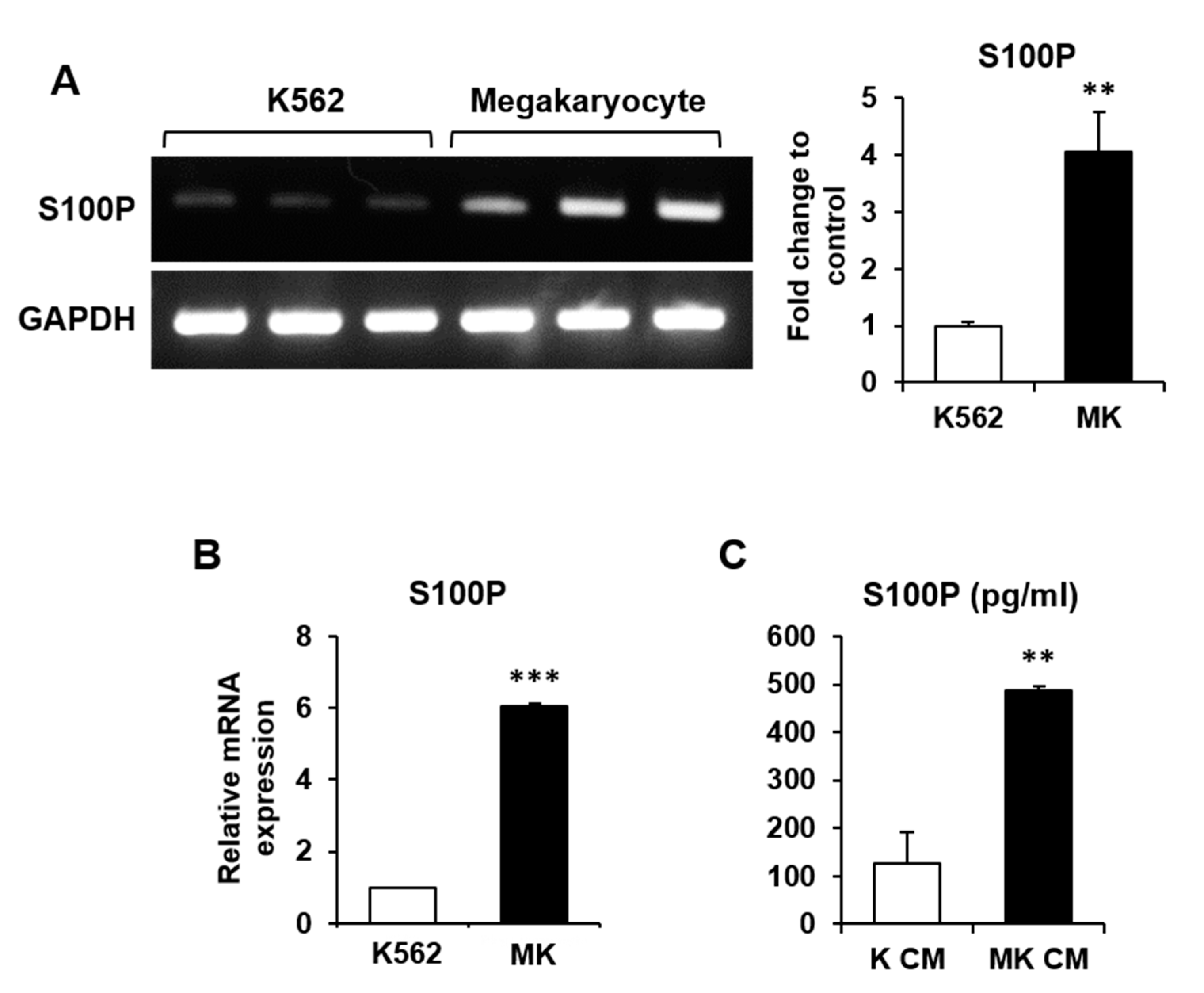
2.2. Differential Gene Expression by Maturation of K562 Cells into MKs
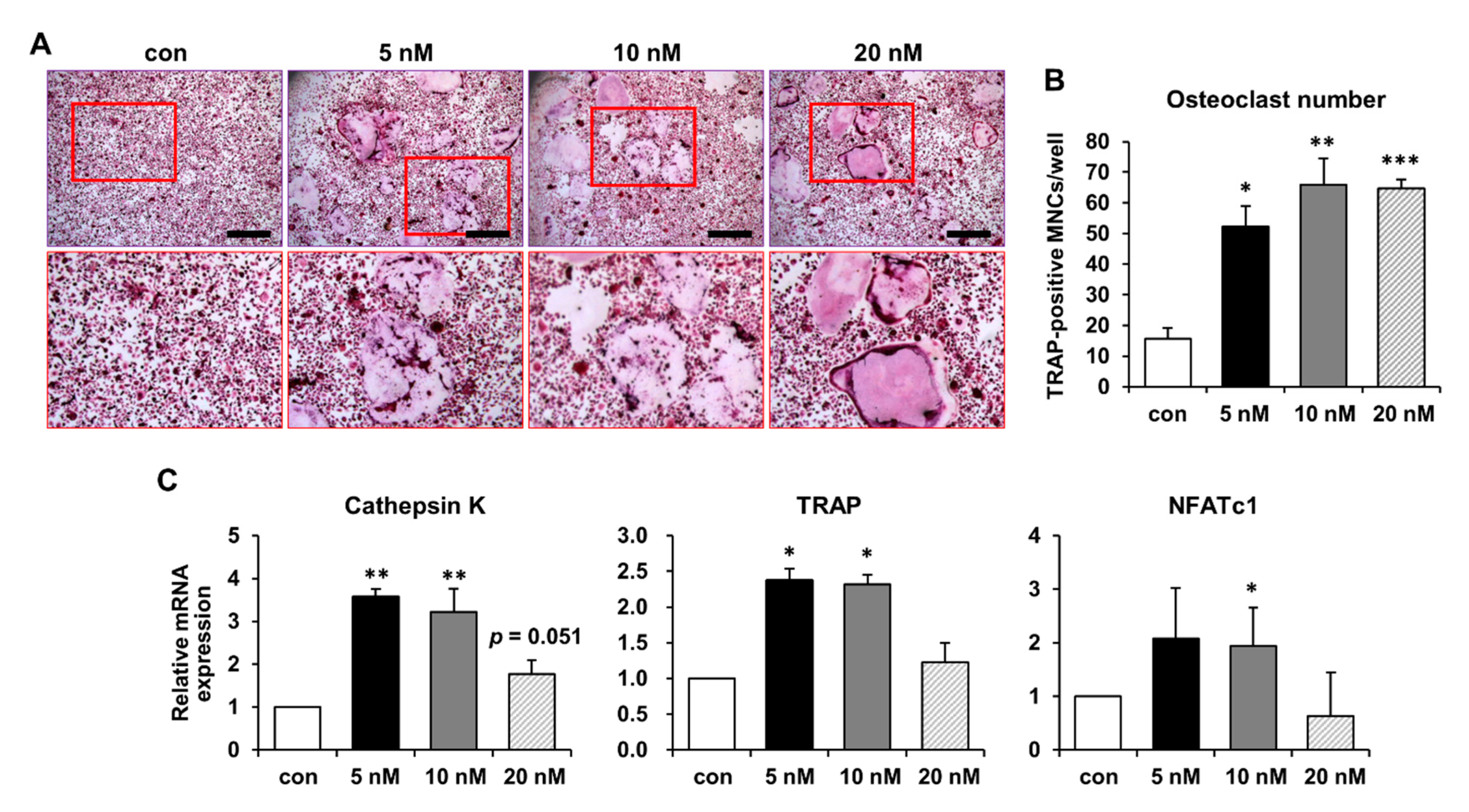
2.3. Increased Osteoclast Differentiation and Bone-Resorbing Activity following S100P Treatment
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Collection of CM
4.2. RNA Extraction, RT-PCR, and qPCR
4.3. Affymetrix cDNA Microarray and Data Analysis
4.4. ELISA
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ishibashi, T.; Ruggeri, Z.M.; Harker, L.A.; Burstein, S.A. Separation of human megakaryocytes by state of differentiation on continuous gradients of Percoll: Size and ploidy analysis of cells identified by monoclonal antibody to glycoprotein IIb/IIIa. Blood 1986, 67, 1286–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, M. Differentiation and proliferation of hematopoietic stem cells. Blood 1993, 81, 2844–2853. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.R.; Hartwig, J.H.; Italiano, J.E., Jr. The biogenesis of platelets from megakaryocyte proplatelets. J. Clin. Investig. 2005, 115, 3348–3354. [Google Scholar] [CrossRef] [Green Version]
- Slayton, W.B.; Wainman, D.A.; Li, X.M.; Hu, Z.; Jotwani, A.; Cogle, C.R.; Walker, D.; Fisher, R.C.; Wingard, J.R.; Scott, E.W.; et al. Developmental differences in megakaryocyte maturation are determined by the microenvironment. Stem Cells 2005, 23, 1400–1408. [Google Scholar] [CrossRef]
- Deutsch, V.R.; Tomer, A. Megakaryocyte development and platelet production. Br. J. Haematol. 2006, 134, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Avecilla, S.T.; Hattori, K.; Heissig, B.; Tejada, R.; Liao, F.; Shido, K.; Jin, D.K.; Dias, S.; Zhang, F.; Hartman, T.E.; et al. Chemokine-mediated interaction of hematopoietic progenitors with the bone marrow vascular niche is required for thrombopoiesis. Nat. Med. 2004, 10, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Galasso, O.; Mariconda, M.; Romano, G.; Capuano, N.; Romano, L.; Iannò, B.; Milano, C. Expandable intramedullary nailing and platelet rich plasma to treat long bone non-unions. J. Orthop. Traumatol. 2008, 9, 129–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghaffarpasand, F.; Shahrezaei, M.; Dehghankhalili, M. Effects of platelet rich plasma on healing rate of long bone non-union fractures: A randomized double-blind placebo controlled clinical trial. Bull. Emerg. Trauma 2016, 4, 134–140. [Google Scholar]
- Sun, Y.; Feng, Y.; Zhang, C.Q.; Chen, S.B.; Cheng, X.G. The regenerative effect of platelet-rich plasma on healing in large osteochondral defects. Int. Orthop. 2010, 34, 589–597. [Google Scholar] [CrossRef] [Green Version]
- Kacena, M.A.; Shivdasani, R.A.; Wilson, K.; Xi, Y.; Troiano, N.; Nazarian, A.; Gundberg, C.M.; Bouxsein, M.L.; Lorenzo, J.A.; Horowitz, M.C. Megakaryocyte-osteoblast interaction revealed in mice deficient in transcription factors GATA-1 and NF-E2. J. Bone Miner. Res. 2004, 19, 652–660. [Google Scholar] [CrossRef]
- Kacena, M.A.; Gundberg, C.M.; Horowitz, M.C. A reciprocal regulatory interaction between megakaryocytes, bone cells, and hematopoietic stem cells. Bone 2006, 39, 978–984. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kwak, M.K.; Moon, S.A.; Choi, Y.J.; Baek, J.E.; Park, S.Y.; Kim, B.J.; Lee, S.H.; Koh, J.M. Regulation of bone metabolism by megakaryocytes in a paracrine manner. Sci. Rep. 2020, 10, 2277. [Google Scholar] [CrossRef] [Green Version]
- Beeton, C.A.; Bord, S.; Ireland, D.; Compston, J.E. Osteoclast formation and bone resorption are inhibited by megakaryocytes. Bone 2006, 39, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Ciovacco, W.A.; Cheng, Y.H.; Horowitz, M.C.; Kacena, M.A. Immature and mature megakaryocytes enhance osteoblast proliferation and inhibit osteoclast formation. J. Cell. Biochem. 2010, 109, 774–781. [Google Scholar] [CrossRef] [Green Version]
- Kacena, M.A.; Ciovacco, W.A. Megakaryocyte-bone cell interactions. Adv. Exp. Med. Biol. 2010, 658, 31–41. [Google Scholar] [PubMed]
- Kacena, M.A.; Nelson, T.; Clough, M.E.; Lee, S.K.; Lorenzo, J.A.; Gundberg, C.M.; Horowitz, M.C. Megakaryocyte-mediated inhibition of osteoclast development. Bone 2006, 39, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, G.; Ding, Y.; Wang, Z.; Barraclough, R.; Rudland, P.S.; Fernig, D.G.; Rao, Z. The crystal structure at 2Å resolution of the Ca2+-binding protein S100P. J. Mol. Biol. 2003, 325, 785–794. [Google Scholar] [CrossRef]
- Becker, T.; Gerke, V.; Kube, E.; Weber, K. S100P, a novel Ca2+-binding protein from human placenta. cDNA cloning, recombinant protein expression and Ca2+ binding properties. Eur. J. Biochem. 1992, 207, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Emoto, Y.; Kobayashi, R.; Akatsuka, H.; Hidaka, H. Purification and characterization of a new member of the S-100 protein family from human placenta. Biochem. Biophys. Res. Commun. 1992, 182, 1246–1253. [Google Scholar] [CrossRef]
- Carneiro, P.; Moreira, A.M.; Figueiredo, J.; Barros, R.; Oliveira, P.; Fernandes, M.S.; Ferro, A.; Almeida, R.; Oliveira, C.; Carneiro, F.; et al. S100P is a molecular determinant of E-cadherin function in gastric cancer. Cell Commun. Signal. 2019, 17, 155. [Google Scholar] [CrossRef] [Green Version]
- Parkkila, S.; Pan, P.W.; Ward, A.; Gibadulinova, A.; Oveckova, I.; Pastorekova, S.; Pastorek, J.; Martinez, A.R.; Helin, H.O.; Isola, J. The calcium-binding protein S100P in normal and malignant human tissues. BMC Clin. Pathol. 2008, 8, 2. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, T.; Simeone, D.M.; Van Golen, K.; Logsdon, C.D. S100P promotes pancreatic cancer growth, survival, and invasion. Clin. Cancer Res. 2005, 11, 5356–5364. [Google Scholar] [CrossRef] [Green Version]
- Cong, Y.; Cui, Y.; Wang, S.; Jiang, L.; Cao, J.; Zhu, S.; Birkin, E.; Lane, J.; Ruge, F.; Jiang, W.G.; et al. Calcium-binding protein S100P promotes tumor progression but enhances chemosensitivity in breast cancer. Front. Oncol. 2020, 10, 566302. [Google Scholar] [CrossRef]
- Austermann, J.; Nazmi, A.R.; Müller-Tidow, C.; Gerke, V. Characterization of the Ca2+-regulated ezrin-S100P interaction and its role in tumor cell migration. J. Biol. Chem. 2008, 283, 29331–29340. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, T.; Logsdon, C.D. S100P: A novel therapeutic target for cancer. Amino Acids 2011, 41, 893–899. [Google Scholar] [CrossRef]
- Huang, R.; Zhao, L.; Chen, H.; Yin, R.H.; Li, C.Y.; Zhan, Y.Q.; Zhang, J.H.; Ge, C.H.; Yu, M.; Yang, X.M. Megakaryocytic differentiation of K562 cells induced by PMA reduced the activity of respiratory chain complex IV. PLoS ONE 2014, 9, e96246. [Google Scholar] [CrossRef] [Green Version]
- Kanagasabapathy, D.; Blosser, R.J.; Maupin, K.A.; Hong, J.M.; Alvarez, M.; Ghosh, J.; Mohamad, S.F.; Aguilar-Perez, A.; Srour, E.F.; Kacena, M.A.; et al. Megakaryocytes promote osteoclastogenesis in aging. Aging 2020, 12, 15121–15133. [Google Scholar] [CrossRef]
- Jawad, M.D.; Go, R.S.; Reichard, K.K.; Shi, M. Increased multinucleated megakaryocytes as an isolated finding in bone marrow: A rare finding and its clinical significance. Am. J. Clin. Pathol. 2016, 146, 561–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dakhel, S.; Padilla, L.; Adan, J.; Masa, M.; Martinez, J.M.; Roque, L.; Coll, T.; Hervas, R.; Calvis, C.; Messeguer, R.; et al. S100P antibody-mediated therapy as a new promising strategy for the treatment of pancreatic cancer. Oncogenesis 2014, 3, e92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumpler, M.; Würger, T.; Roschger, P.; Zwettler, E.; Sturmlechner, I.; Altmann, P.; Fratzl, P.; Rogers, M.J.; Klaushofer, K. Osteoclasts on bone and dentin in vitro: Mechanism of trail formation and comparison of resorption behavior. Calcif. Tissue Int. 2013, 93, 526–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merrild, D.M.; Pirapaharan, D.C.; Andreasen, C.M.; Kjærsgaard-Andersen, P.; Møller, A.M.; Ding, M.; Delaissé, J.M.; Søe, K. Pit- and trench-forming osteoclasts: A distinction that matters. Bone Res. 2015, 3, 15032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kacena, M.A.; Gundberg, C.M.; Nelson, T.; Horowitz, M.C. Loss of the transcription factor p45 NF-E2 results in a developmental arrest of megakaryocyte differentiation and the onset of a high bone mass phenotype. Bone 2005, 36, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Shivdasani, R.A. Molecular and transcriptional regulation of megakaryocyte differentiation. Stem Cells 2001, 19, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Maupin, K.A.; Himes, E.R.; Plett, A.P.; Chua, H.L.; Singh, P.; Ghosh, J.; Mohamad, S.F.; Abeysekera, I.; Fisher, A.; Sampson, C.; et al. Aging negatively impacts the ability of megakaryocytes to stimulate osteoblast proliferation and bone mass. Bone 2019, 127, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Koh, A.J.; Wang, Z.; Soki, F.N.; Park, S.I.; Pienta, K.J.; McCauley, L.K. Inhibitory effects of megakaryocytic cells in prostate cancer skeletal metastasis. J. Bone Miner. Res. 2011, 26, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Psaila, B.; Lyden, D.; Roberts, I. Megakaryocytes, malignancy and bone marrow vascular niches. J. Thromb. Haemost. 2012, 10, 177–188. [Google Scholar] [CrossRef] [Green Version]
- Maroni, P. Megakaryocytes in Bone Metastasis: Protection or Progression? Cells 2019, 8, 134. [Google Scholar] [CrossRef] [Green Version]
- Nishida, M.; Saegusa, J.; Tanaka, S.; Morinobu, A. S100A12 facilitates osteoclast differentiation from human monocytes. PLoS ONE 2018, 13, e0204140. [Google Scholar] [CrossRef]
- Erlandsson, M.C.; Svensson, M.D.; Jonsson, I.M.; Bian, L.; Ambartsumian, N.; Andersson, S.; Peng, Z.; Vääräniemi, J.; Ohlsson, C.; Andersson, K.M.E.; et al. Expression of metastasin S100A4 is essential for bone resorption and regulates osteoclast function. Biochim. Biophys. Acta 2013, 1833, 2653–2663. [Google Scholar] [CrossRef] [Green Version]
- Grevers, L.C.; de Vries, T.J.; Vogl, T.; Abdollahi-Roodsaz, S.; Sloetjes, A.W.; Leenen, P.J.; Roth, J.; Everts, V.; van den Berg, W.B.; van Lent, P.L. S100A8 enhances osteoclastic bone resorption in vitro through activation of Toll-like receptor 4: Implications for bone destruction in murine antigen-induced arthritis. Arthritis Rheum. 2011, 63, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, T.; Simeone, D.M.; Schmidt, A.M.; Logsdon, C.D. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE). J. Biol. Chem. 2004, 279, 5059–5065. [Google Scholar] [CrossRef] [Green Version]
- Karin, M.; Cao, Y.; Greten, F.R.; Li, Z.W. NF-kappaB in cancer: From innocent bystander to major culprit. Nat. Rev. Cancer 2002, 2, 301–310. [Google Scholar] [CrossRef]
- Boyce, B.F.; Xiu, Y.; Li, J.; Xing, L.; Yao, Z. NF-kappaB-mediated regulation of osteoclastogenesis. Endocrinol. Metab. 2015, 30, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Immel, D.; Xi, C.X.; Bierhaus, A.; Feng, X.; Mei, L.; Nawroth, P.; Stern, D.M.; Xiong, W.C. Regulation of osteoclast function and bone mass by RAGE. J. Exp. Med. 2006, 203, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Héron, E.; Deloukas, P.; van Loon, A.P. The complete exon-intron structure of the 156-kb human gene NFKB1, which encodes the p105 and p50 proteins of transcription factors NF-kappa B and I kappa B-gamma: Implications for NF-kappa B-mediated signal transduction. Genomics 1995, 30, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Kalea, A.Z.; Reiniger, N.; Yang, H.; Arriero, M.; Schmidt, A.M.; Hudson, B.I. Alternative splicing of the murine receptor for advanced glycation end-products (RAGE) gene. FASEB J. 2009, 23, 1766–1774. [Google Scholar] [CrossRef] [Green Version]
- Sims, N.A.; Martin, T.J. Coupling the activities of bone formation and resorption: A multitude of signals within the basic multicellular unit. Bonekey Rep. 2014, 3, 481. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.M.; Lin, C.; Stavre, Z.; Greenblatt, M.B.; Shim, J.H. Osteoblast-osteoclast communication and bone homeostasis. Cells 2020, 9, 2073. [Google Scholar] [CrossRef]
- Kim, S.Y.; Lee, E.H.; Park, S.Y.; Choi, H.; Koh, J.T.; Park, E.K.; Kim, I.S.; Kim, J.E. Ablation of Stabilin-1 enhances bone-resorbing activity in osteoclasts in vitro. Calcif. Tissue Int. 2019, 105, 205–214. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, E.H.; Park, S.Y.; Kim, J.E. Induction of fibrillin-2 and periostin expression in Osterix-knockdown MC3T3-E1 cells. Gene 2017, 596, 123–129. [Google Scholar] [CrossRef]

| Gene Accession | Gene Name (Gene Symbol) | Fold Change | p Value |
|---|---|---|---|
| NM_005139 | annexin A3 (ANXA3) | 4.458 | 0.000 |
| NM_001306129 | fibronectin 1 (FN1) | 3.972 | 0.000 |
| NM_001062 | transcobalamin I (vitamin B12 binding protein, R binder family) (TCN1) | 3.491 | 0.001 |
| NM_000239 | lysozyme (LYZ) | 3.460 | 0.007 |
| NM_003054 | solute carrier family 18 (vesicular monoamine transporter), member 2 (SLC18A2) | 3.364 | 0.000 |
| NM_001024845 | solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (SLC6A9) | 2.715 | 0.005 |
| NM_001243245 | proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) (PRG2) | 2.573 | 0.000 |
| NM_001292045 | neuromedin U (NMU) | 2.469 | 0.000 |
| NM_002031 | fyn-related Src family tyrosine kinase (FRK) | 2.465 | 0.000 |
| NM_000632 | integrin, alpha M (complement component 3 receptor 3 subunit) (ITGAM) | 2.438 | 0.000 |
| NM_001282386 | isocitrate dehydrogenase 1 (NADP+) (IDH1) | 2.377 | 0.000 |
| NM_021109 | thymosin beta 4, X-linked (TMSB4X) | 2.374 | 0.011 |
| NM_000132 | coagulation factor VIII, procoagulant component (F8) | 2.366 | 0.022 |
| NM_000405 | GM2 ganglioside activator (GM2A) | 2.322 | 0.001 |
| NM_002562 | purinergic receptor P2X, ligand gated ion channel, 7 (P2RX7) | 2.313 | 0.003 |
| NM_001303499 | calponin 2 (CNN2) | 2.295 | 0.001 |
| NM_005980 | S100 calcium binding protein P (S100P) | 2.245 | 0.008 |
| NM_001042402 | N-acylethanolamine acid amidase (NAAA) | 2.194 | 0.000 |
| NM_001063 | transferrin (TF) | 2.194 | 0.001 |
| NM_004472 | forkhead box D1 (FOXD1) | 2.130 | 0.000 |
| NM_000189 | hexokinase 2 (HK2) | 2.124 | 0.000 |
| NM_001159629 | solute carrier family 27 (fatty acid transporter), member 2 (SLC27A2) | 2.123 | 0.000 |
| NM_001098503 | protein tyrosine phosphatase, receptor type, J (PTPRJ) | 2.113 | 0.000 |
| NM_001145031 | plasminogen activator, urokinase (PLAU) | 2.102 | 0.002 |
| NM_001025366 | vascular endothelial growth factor A (VEGFA) | 2.065 | 0.000 |
| NM_001078175 | solute carrier family 29 (equilibrative nucleoside transporter), member 1 (SLC29A1) | 2.062 | 0.000 |
| NM_001244984 | sel-1 suppressor of lin-12-like (C. elegans) (SEL1L) | 2.044 | 0.000 |
| NM_004335 | bone marrow stromal cell antigen 2 (BST2) | 2.031 | 0.002 |
| Name | Product | Primer Sequences | |
|---|---|---|---|
| RT-PCR | |||
| S100P | 97 bp | F | 5’-GAG AAG GAG CTA CCA GGC TTC-3’ |
| R | 5’-TCC ACC TGG GCA TCT CCA TT-3’ | ||
| Gapdh | 240 bp | F | 5’-TGA TGA CAT CAA GAA GGT GGT GAA G-3’ |
| R | 5’-TCC TTG GAG GCC ATG TAG GCC AT-3’ | ||
| qPCR | |||
| S100P | 97 bp | F | 5’- GAG AAG GAG CTA CCA GGC TTC-3’ |
| R | 5’- TCC ACC TGG GCA TCT CCA TT-3’ | ||
| CtsK | 178 bp | F | 5’-CAG AAC GGA GGC ATT GAC-3’ |
| R | 5’-CGA TGG ACA CAG AGA TGG-3’ | ||
| Trap | 197 bp | F | 5’-GCA GCC AAG GAG GAC TAC-3’ |
| R | 5’-CCC ACT CAG CAC ATA GCC-3’ | ||
| Nfatc1 | 161 bp | F | 5’-TCT TCC GAG TTC ACA TCC-3’ |
| R | 5’-ACA GCA CCA TCT TCT TCC-3’ | ||
| Gapdh | 126 bp | F | 5’-GCA TCT CCC TCA CAA TTT CCA-3’ |
| R | 5’-GTG CAG CGA ACT TTA TTG ATG G-3’ | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-H.; Ihn, H.J.; Park, E.K.; Kim, J.-E. S100 Calcium-Binding Protein P Secreted from Megakaryocytes Promotes Osteoclast Maturation. Int. J. Mol. Sci. 2021, 22, 6129. https://doi.org/10.3390/ijms22116129
Lee S-H, Ihn HJ, Park EK, Kim J-E. S100 Calcium-Binding Protein P Secreted from Megakaryocytes Promotes Osteoclast Maturation. International Journal of Molecular Sciences. 2021; 22(11):6129. https://doi.org/10.3390/ijms22116129
Chicago/Turabian StyleLee, Seung-Hoon, Hye Jung Ihn, Eui Kyun Park, and Jung-Eun Kim. 2021. "S100 Calcium-Binding Protein P Secreted from Megakaryocytes Promotes Osteoclast Maturation" International Journal of Molecular Sciences 22, no. 11: 6129. https://doi.org/10.3390/ijms22116129






