Complete Plastid and Mitochondrial Genomes of Aeginetia indica Reveal Intracellular Gene Transfer (IGT), Horizontal Gene Transfer (HGT), and Cytoplasmic Male Sterility (CMS)
Abstract
:1. Introduction
2. Results and Discussion
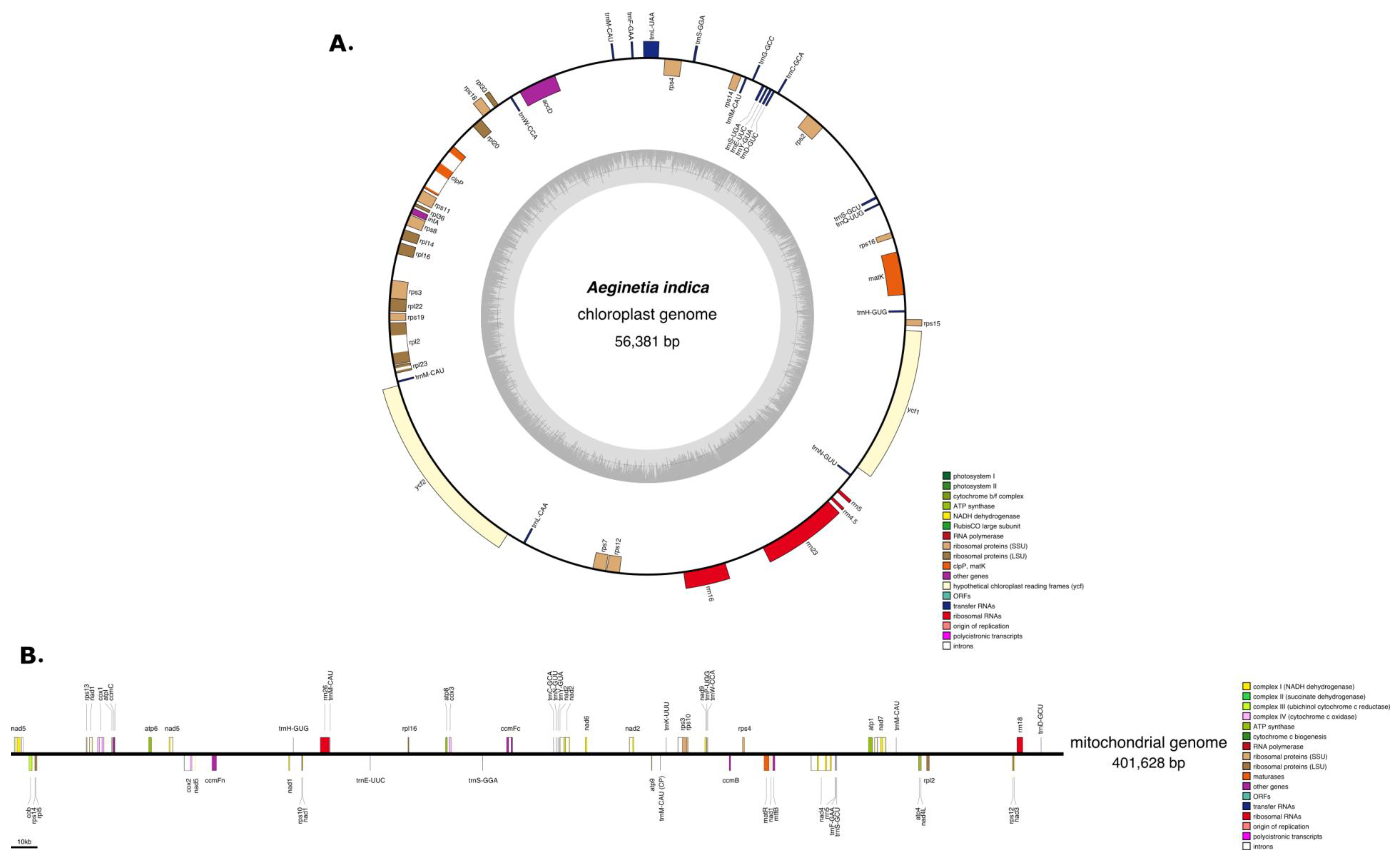
2.1. Characteristics of the A. Indica Plastid Genome
2.2. Characteristics of the A. Indica Mitogenome
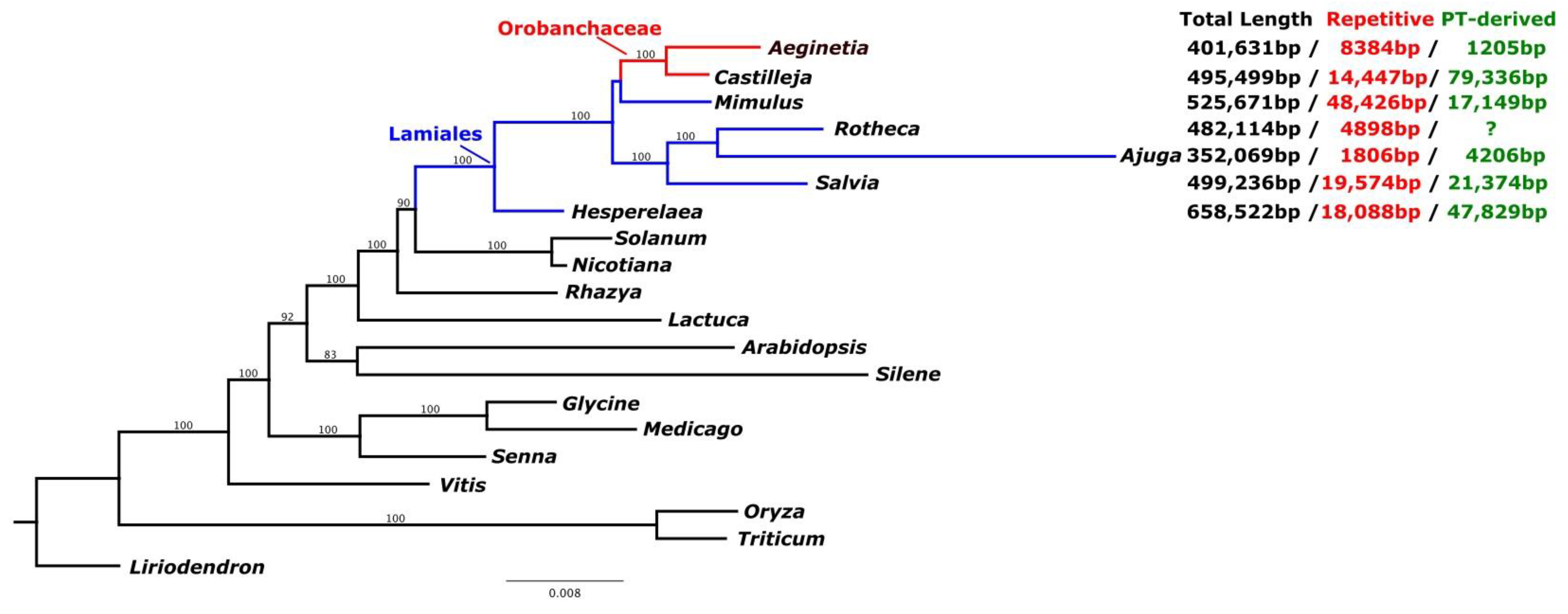
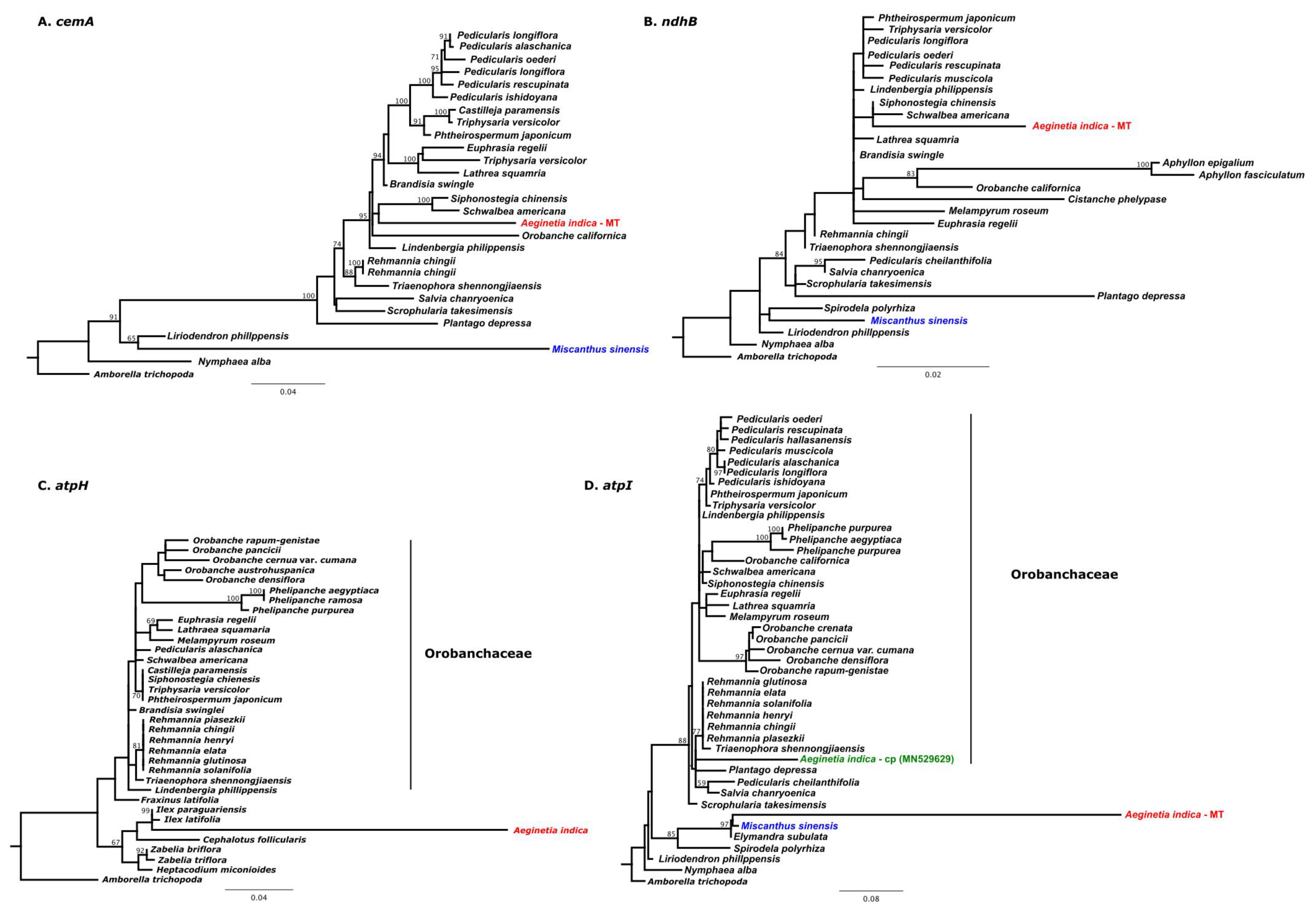
2.3. IGT and HGT of A. Indica Organelle Genomes
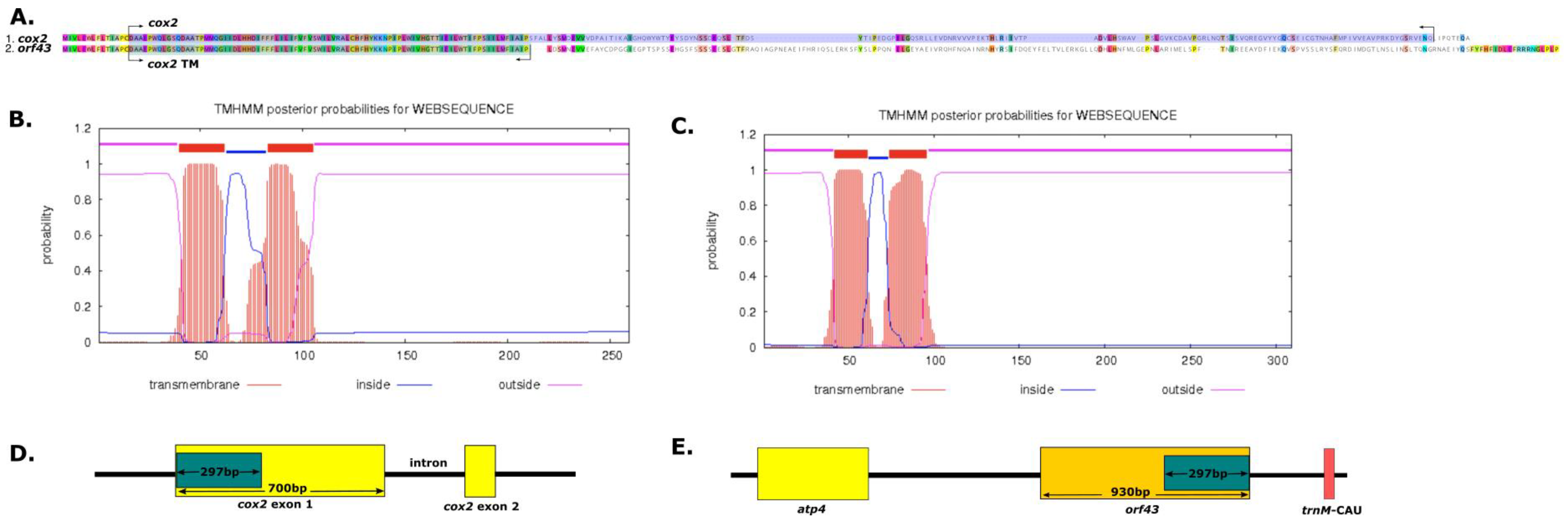
2.4. Cytoplasmic Male Sterility (CMS) of Genes in the A. Indica Mitogenome
3. Materials and Methods
3.1. Plant Samplingand DNA Sequencing
3.2. Organellar Genome Assembly and Annotation
3.3. Plastome and Mitogenome Analyses
3.4. Phylogenetic Analysis of Plastid and Mitochondrial Genes
3.5. Analysis of Intracellular Gene Transfer (IGT), Horizontal Gene Transfer (HGT) and Cytoplasmic Male Sterility (CMS) Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wicke, S.; Schneeweiss, C.W.; Depamphilis, C.W.; Mai, F.M.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [Green Version]
- Kubo, T.; Nishizawa, S.; Sugawara, A.; Otchoda, N.; Estiati, A.; Mikami, T. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulagaris L.) reveals a novel gene for tRNAcys (GCA). Nucleic Acids. Res. 2000, 28, 2571–2576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alverson, A.J.; Wei, X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef] [Green Version]
- Liao, X.; Zhao, Y.; Kong, X.; Khan, A.; Zhou, B.; Liu, D.; Kashif, M.H.; Chen, P.; Wang, H.; Zhou, R. Complete sequence of kenaf (Hibiscus cannabinus) mitochondrial genome and comparative analysis with the mitochondrial genomes of other plants. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kan, S.-L.; Shen, T.-T.; Gong, P.; Ran, J.-H.; Wang, X.-Q. The complete mitochondrial genome of Taxus cuspidata (Taxaceae): Eight protein coding genes have transferred to the nuclear genome. BMC Evol. Biol. 2020, 20, 10. [Google Scholar] [CrossRef] [Green Version]
- Pinard, D.; Myburg, A.A.; Mizrachi, E. The plastid and mitochondrial genomes of Eucalyptus grandis. BMC Genom. 2019, 20, 132. [Google Scholar] [CrossRef] [PubMed]
- Raulet, M.E.; Garcia, L.E.; Gandini, C.L.; Sato, H.; Ponce, G.; Sanchez-Puerta, M.V. Multichromosomal structure and foreign tracts in the Ombrophytum subterraneum (Balanophoraceae) mitochondrial genome. Plant Mol. Biol. 2020, 103, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Grewe, F.; Zhu, A.; Ruhlman, T.A.; Sabir, J.; Mower, J.P.; Jansen, R.K. Dynamic evolution of Geranium mitochondrial genomes through multiple horizontal and intracellular gene transfers. New Phytol. 2015, 208, 570–583. [Google Scholar] [CrossRef]
- Gandini, C.L.; Sanchez-Puerta, M.V. Foreign plastid sequences in plant mitochondria are frequently acquired via mitochondrion-to-mitochondrion horizontal transfer. Sci. Rep. 2017, 7, 43402. [Google Scholar] [CrossRef]
- Zhao, N.; Wang, Y.; Hua, J. The roles of mitochondrion in intragenomic gene transfer in plants: A source and a pool. Int. J. Mol. Sci. 2018, 19, 547. [Google Scholar] [CrossRef] [Green Version]
- Dong, S.; Zhao, C.; Chen, F.; Liu, Y.; Zhang, S.; Wu, H.; Zhang, L.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef]
- Hanson, M.R.; Bentolila, S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 2004, 16, S154–S169. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Cui, P.; Zhan, K.; Lin, Q.; Zhuo, G.; Guo, X.; Ding, F.; Yang, W.; Liu, D.; Hu, S.; et al. Comparative analysis of mitochondrial genomes between a wheat K-type cytoplasmic male sterility (CMS) line and its maintainer line. BMC Genom. 2011, 12, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mower, J.P.; Case, A.L.; Floro, E.R.; Willis, J.H. Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol. Evol. 2012, 4, 670–686. [Google Scholar] [CrossRef] [Green Version]
- Štorchová, H.; Stone, J.D.; Sloan, D.B.; Abeyawardana, O.A.J.; Müller, K.; Walterová, J.; Pažoutová, M. Homologous recombination changes the context of Cytochrome b transcription in the mitochondrial genome of Silene vulgaris KRA. BMC Genom. 2018, 19, 874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarenko, M.S.; Usatov, A.V.; Tatarinova, T.V.; Azarin, K.V.; Logacheva, M.D.; Gavrilova, V.A.; Kornienko, I.V.; Horn, R. Organization features of the mitochondrial genome of sunflower (Helianthus annuus L.) with ANN2-type male-sterile cytoplasm. Plants 2019, 8, 439. [Google Scholar]
- Bennett, J.; Mathews, S. Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am. J. Bot. 2006, 93, 1039–1051. [Google Scholar] [CrossRef] [Green Version]
- McNeal, J.R.; Bennett, J.R.; Wolfe, A.D.; Mathews, S. Phylogeny and origins of holoparasitism in Orobanchaceae. Am. J. Bot. 2013, 100, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Shneeweiss, G.M. Phylogenetic relationships and evolution trends in Orobanchaceae. In Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies; Joel, D.M., Gressel, J., Musselman, L.J., Eds.; Springer: Berlin, Germany, 2013; pp. 243–265. [Google Scholar]
- Frailey, D.C.; Chaluvadi, S.R.; Vaughn, J.N.; Coatney, C.G.; Bennetzen, J.L. Gene loss and genome rearrangement in the plastids of five hemiparasites in the family Orobanchaceae. BMC Plant Biol. 2018, 18, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruzdev, E.V.; Kadnikov, V.V.; Beletsky, A.V.; Mardanov, A.V.; Ravin, N.V. Extensive plastome reduction and loss of photosynthesis genes in Diphelypaea coccinea, a holoparasitic plant of the family Orobanchaceae. Peer J. 2019, 7, e7830. [Google Scholar] [CrossRef] [Green Version]
- Wicke, S.; Müller, J.F.; de Pmphilis, C.W.; Quandt, D.; Wickett, N.J.; Zhang, Y.; Renner, S.S.; Schneeweiss, G.M. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013, 25, 3711–3725. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Zhu, A.; Kozaczek, M.; Shah, N.; Pabón-Mora, N.; Gonzáles, F.; Mower, J.P. Limited mitogenomic degradation in response to a parasitic lifestyle in Orobanchaceae. Sci. Rep. 2016, 6, 36285. [Google Scholar] [CrossRef]
- Zervas, A.; Petersen, G.; Severg, O. Mitochondrial genome evolution in parasitic plants. BMC Evol. Biol. 2019, 19, 87. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Stefanović, S.; Young, G.J.; Palmer, J.D. Gene transfer from parasitic to host plants. Nature 2004, 432, 165–166. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, T.-C.; Qiao, Q.; Ren, Z.; Zhao, J.; Yonezawa, T.; Hasegawa, M.; Crabbe, M.J.C.; Li, J.; Zhong, Y. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS ONE 2013, 8, e58747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwolek, D.; Denysenko-Bennett, M.; Góralski, G.; Cygan, M.; Mizia, P.; Piwowarczyk, R.; Szklarczyk, M.; Joachimiak, A.K. The first evidence of a host-to-parasite mitochondrial gene transfer in Orobanchaceae. Acta Biol. Cracoviensia Bot. 2017, 59, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Gandini, C.L.; Garcia, L.E.; Abbona, C.C.; Sanchez-Puerta, M.V. The complete organelle genomes of Physochlaina orientalis: Insights into short sequence repeats across seed plant mitochondrial genomes. Mol. Phylognet. Evol. 2019, 137, 274–284. [Google Scholar] [CrossRef]
- Kato, Y.; Inoue, T.; Onishi, Y. In vitro culture of a root parasite, Aeginetia indica L. II. The plane of cell division in the tendril. Plant Cell Physiol. 1984, 25, 981–987. [Google Scholar]
- Li, X.; Feng, T.; Randle, C.; Schneeweiss, G.M. Phylogenetic relationships in Orobanchaceae inferred from low-copy nuclear genes: Consolidation of major clades and identification of a novel position of the non-photosynthetic Orobanche clade sister to all other parasitic Orobanchaceae. Front. Plant Sci. 2019, 10, 902. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Yu, R.; Dai, J.; Liu, Y.; Zhou, R. The loss of photosynthesis pathway and genomic locations of the lost plastid genes in a holoparasitic plant Aeginetia indica. BMC Plant Biol. 2020, 20, 199. [Google Scholar] [CrossRef]
- Sloan, D.B. One ring to rule them all? Genome sequencing provides new insights into the master cirlce model of plant mitochondrial DNA structure. New Phytol. 2013, 200, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Khan, A.R.; Waqas, M.; Kang, S.-M.; Khan, M.A.; Shahzad, R.; Seo, C.-W.; Shin, J.-H.; Lee, I.-J. Mitochondrial genome analysis of wild rice (Oryza minuta) and its comparison with other related species. PLoS ONE 2016, 11, e0152937. [Google Scholar] [CrossRef]
- Adams, K.L.; Qiu, Y.-L.; Stoutemyer, M.; Palmer, J.D. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. USA 2002, 99, 9905–9912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clifton, S.W.; Minx, P.; Fauron, C.M.; Gibson, M.; Allen, J.O.; Sun, H.; Tompson, M.; Barbazuk, W.B.; Kanuganti, S.; Tayloe, C.; et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004, 136, 3486–3503. [Google Scholar] [CrossRef] [Green Version]
- Kubo, T.; Mikamu, T. Organization and variation of angiosperm mitochondrial genome. Physiol. Plant. 2007, 129, 6–13. [Google Scholar] [CrossRef]
- Grewe, F.; Zhu, A.; Mower, J.P. Loss of a trans-splicing and1 intron from Geraniaceae and transfer of the maturase gene matR to the nucleus in Pelargonium. Genome Biol. Evol. 2016, 8, 3193–3201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raman, G.; Park, S.; Lee, E.M.; Park, S. Evidence of mitochondrial DNA in the chloroplast genome of Convalaria keiskei and its subsequent evolution in the Asparagales. Sci. Rep. 2019, 9, 5028. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-C.; Su, Y.-Y.; Wu, C.-H.; Liu, Y.-C.; Huang, C.-H.; Chang, C.-C. Analysis of mitochondrial genomics and transcriptomics reveal abundant RNA edits and differential editing status in moth orchid, Phalaenopsis aphrodite subsp. formosana. Sci. Hortic. 2020, 267, 109304. [Google Scholar] [CrossRef]
- Cusimano, N.; Wicke, S. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. 2016, 210, 680–693. [Google Scholar] [CrossRef] [Green Version]
- Park, J.-M.; Manen, J.-F.; Schneeweissm, G.M. Horizontal gene transfer of a plastid gene in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobancheae). Mol. Phylogenet. Evol. 2007, 43, 974–985. [Google Scholar] [CrossRef]
- Garcia, L.E.; Edera, A.A.; Palmer, J.D.; Sato, H.; Sanchez-Puerta, M.V. Horizontal gene transfers dominate the functional mitochondrial gene space of a holoparastic plant. New Phytol. 2021, 229, 1701–1714. [Google Scholar] [CrossRef] [PubMed]
- Abdelnoor, R.V.; Yule, R.; Elo, A.; Christensen, A.C.; Meyer-Gauen, G.; Mackenzie, S.A. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to Muts. Proc. Natl. Acad. Sci. USA 2003, 100, 5968–5973. [Google Scholar] [CrossRef] [Green Version]
- Jo, Y.D.; Choi, Y.; Kim, D.-H.; Kum, B.-D.; Kang, B.-C. Extensive structural variations between mitochondrial genomes of CMS and normal peppers (Capsicum annuum L.) revealed by complete nucleotide sequencing. BMC Genom. 2014, 15, 561. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, M.P.; Shinada, H.; Onodera, Y.; Komaki, C.; Mikami, T.; Kubo, T. A male sterility-associated mitochondrial protein in wild beets causes pollen disruption in transgenic plants. Plant J. 2008, 54, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Li, P.; Wnag, A.; Ge, X.; Li, Z. A novel cytoplasmic male sterility in Brassica napus (inap CMS) with carpelloid staens via protoplast fusion with Chinese woad. Font. Plant Sci. 2017, 8, 529. [Google Scholar]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-Se: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Category | Group of Gene | Genes | 1 * | 2 * | Category | Group of Gene | Genes | 1 * | 2 * |
|---|---|---|---|---|---|---|---|---|---|
| Photosynthethic | Phothsystem I | psaA | - | - | Self-replication | Large ribosomal subunit | rpl2 | o(x2) | o |
| psaB | - | - | rpl14 | o(x2) | o | ||||
| psac | - | - | rpl16 | o(x2) | o | ||||
| psaI | - | - | rpl20 | o | o | ||||
| psaJ | - | - | rpl22 | o(x2) | o | ||||
| Photosystem II | psbA | - | - | rpl23 | o(x2) | - | |||
| psbB | - | - | rpl32 | - | - | ||||
| psbC | - | - | rpl33 | o | o | ||||
| psbD | - | - | rpl36 | o | o | ||||
| psbE | - | - | Small ribosomal subunit | rps2 | o | o | |||
| psbF | - | - | rps3 | o(x2) | o | ||||
| psbH | - | - | rps4 | o | o | ||||
| psbI | - | - | rps7 | o(x2) | o | ||||
| psbJ | - | - | rps8 | o(x2) | o | ||||
| psbK | - | - | rps11 | o | o | ||||
| psbL | - | - | rps12 | o(x2) | o | ||||
| psbM | - | - | rps14 | o | o | ||||
| psbN | - | - | rps15 | o | o | ||||
| psbT | - | - | rps16 | o | o | ||||
| psbZ | - | - | rps18 | o | o | ||||
| NADH dehydrogenase | nadA | - | - | rps19 | o(x2) | o | |||
| ndhB | ψ | ψ | RNA polymerase | rpoA | - | - | |||
| ndhC | - | - | rpoB | - | - | ||||
| ndhD | - | - | rpoC1 | - | - | ||||
| ndhE | - | - | rpoC2 | - | - | ||||
| ndhF | - | - | ribosomal RNAs | rrn23S | o(x2) | o | |||
| ndhG | - | - | rrn16S | o(x2) | o | ||||
| ndhH | - | - | rrn5S | o(x2) | o | ||||
| ndhI | - | - | rrn4.5S | o(x2) | o | ||||
| ndhK | - | - | Biosynthesis | Maturase | matK | o | o | ||
| Cytochrome b/f complex | petA | - | - | Protease | clpP | o | o | ||
| petB | - | - | Envelope membrane protein | cemA | - | - | |||
| petD | - | - | Acetyl-CoA carboxylase | accD | o | o | |||
| petG | - | - | cytochrome synthesis gene | ccsA | - | - | |||
| petL | - | - | Translation initiation factor | infA | o | o | |||
| petN | - | - | Unknown function | hypothetical chloroplast reading frame | ycf1 | o | o | ||
| atpA | - | - | ycf2 | o | o | ||||
| ATP synthase Rubisco | atpB | - | - | ycf3 | - | - | |||
| atpE | - | - | ycf4 | - | - | ||||
| atpF | - | - | |||||||
| atpH | - | o | |||||||
| atpI | - | - | |||||||
| rbcL | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, K.-S.; Park, S. Complete Plastid and Mitochondrial Genomes of Aeginetia indica Reveal Intracellular Gene Transfer (IGT), Horizontal Gene Transfer (HGT), and Cytoplasmic Male Sterility (CMS). Int. J. Mol. Sci. 2021, 22, 6143. https://doi.org/10.3390/ijms22116143
Choi K-S, Park S. Complete Plastid and Mitochondrial Genomes of Aeginetia indica Reveal Intracellular Gene Transfer (IGT), Horizontal Gene Transfer (HGT), and Cytoplasmic Male Sterility (CMS). International Journal of Molecular Sciences. 2021; 22(11):6143. https://doi.org/10.3390/ijms22116143
Chicago/Turabian StyleChoi, Kyoung-Su, and Seonjoo Park. 2021. "Complete Plastid and Mitochondrial Genomes of Aeginetia indica Reveal Intracellular Gene Transfer (IGT), Horizontal Gene Transfer (HGT), and Cytoplasmic Male Sterility (CMS)" International Journal of Molecular Sciences 22, no. 11: 6143. https://doi.org/10.3390/ijms22116143
APA StyleChoi, K.-S., & Park, S. (2021). Complete Plastid and Mitochondrial Genomes of Aeginetia indica Reveal Intracellular Gene Transfer (IGT), Horizontal Gene Transfer (HGT), and Cytoplasmic Male Sterility (CMS). International Journal of Molecular Sciences, 22(11), 6143. https://doi.org/10.3390/ijms22116143







