Decellularized Porcine Cartilage Scaffold; Validation of Decellularization and Evaluation of Biomarkers of Chondrogenesis
Abstract
:1. Introduction
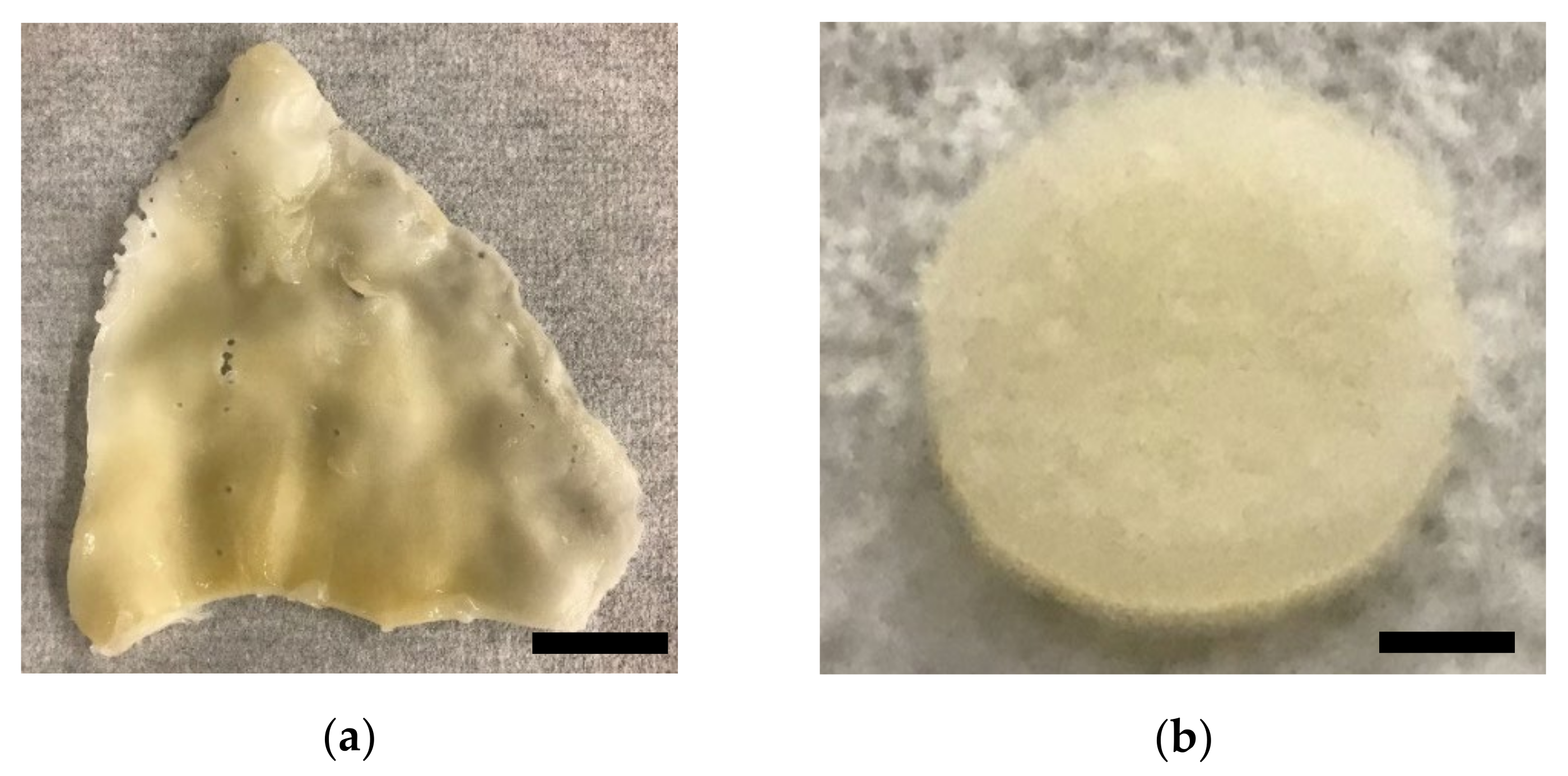
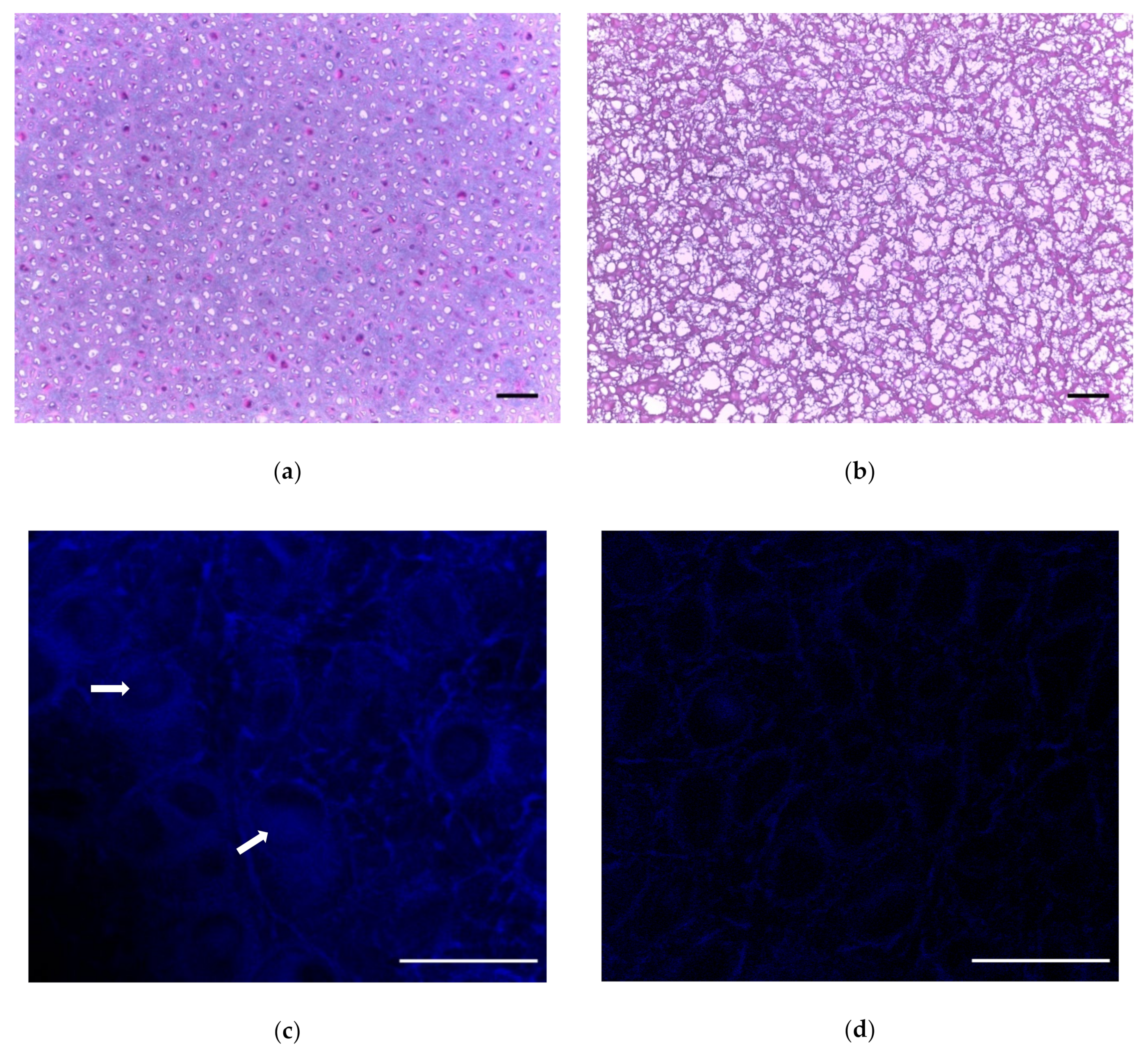
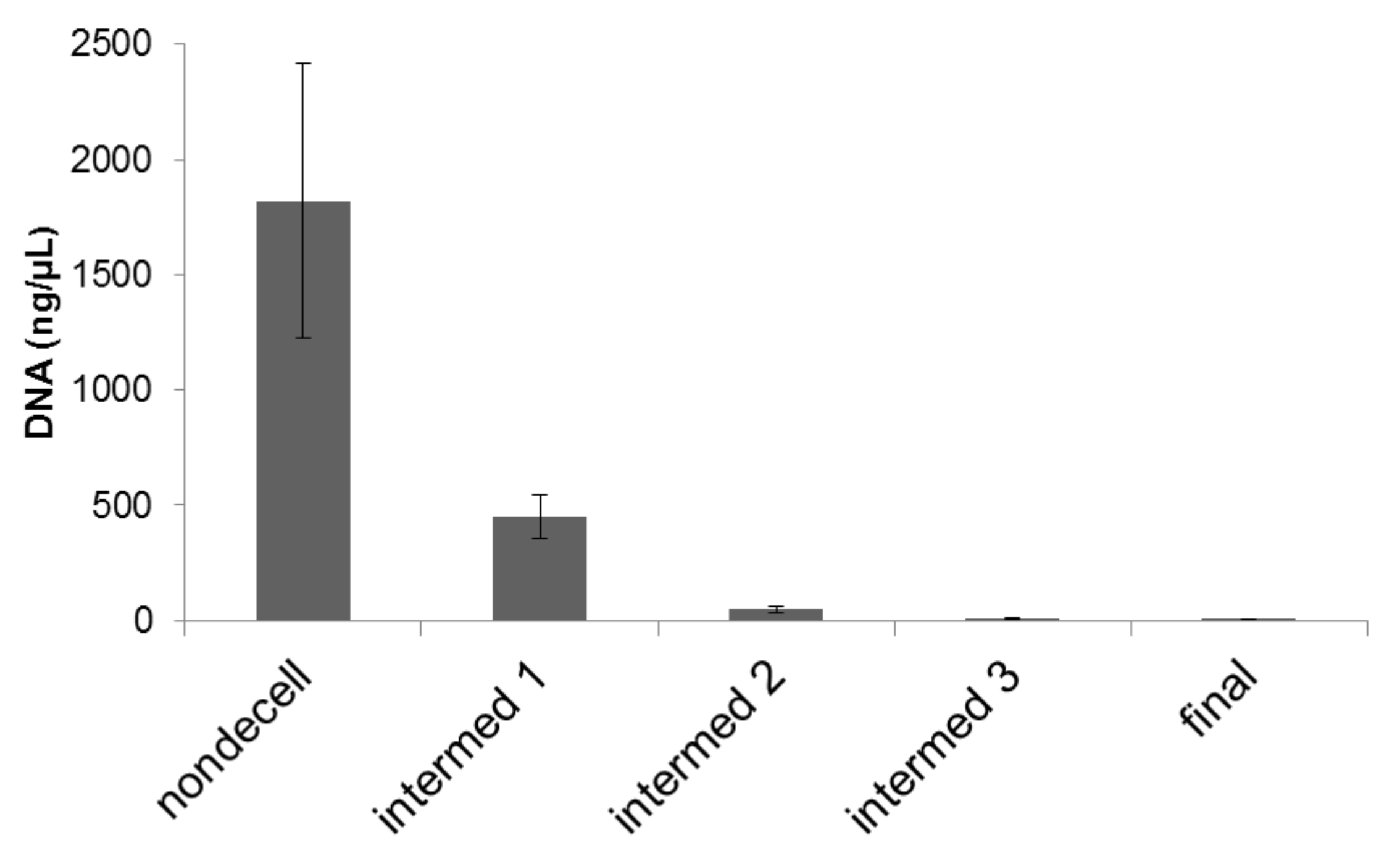
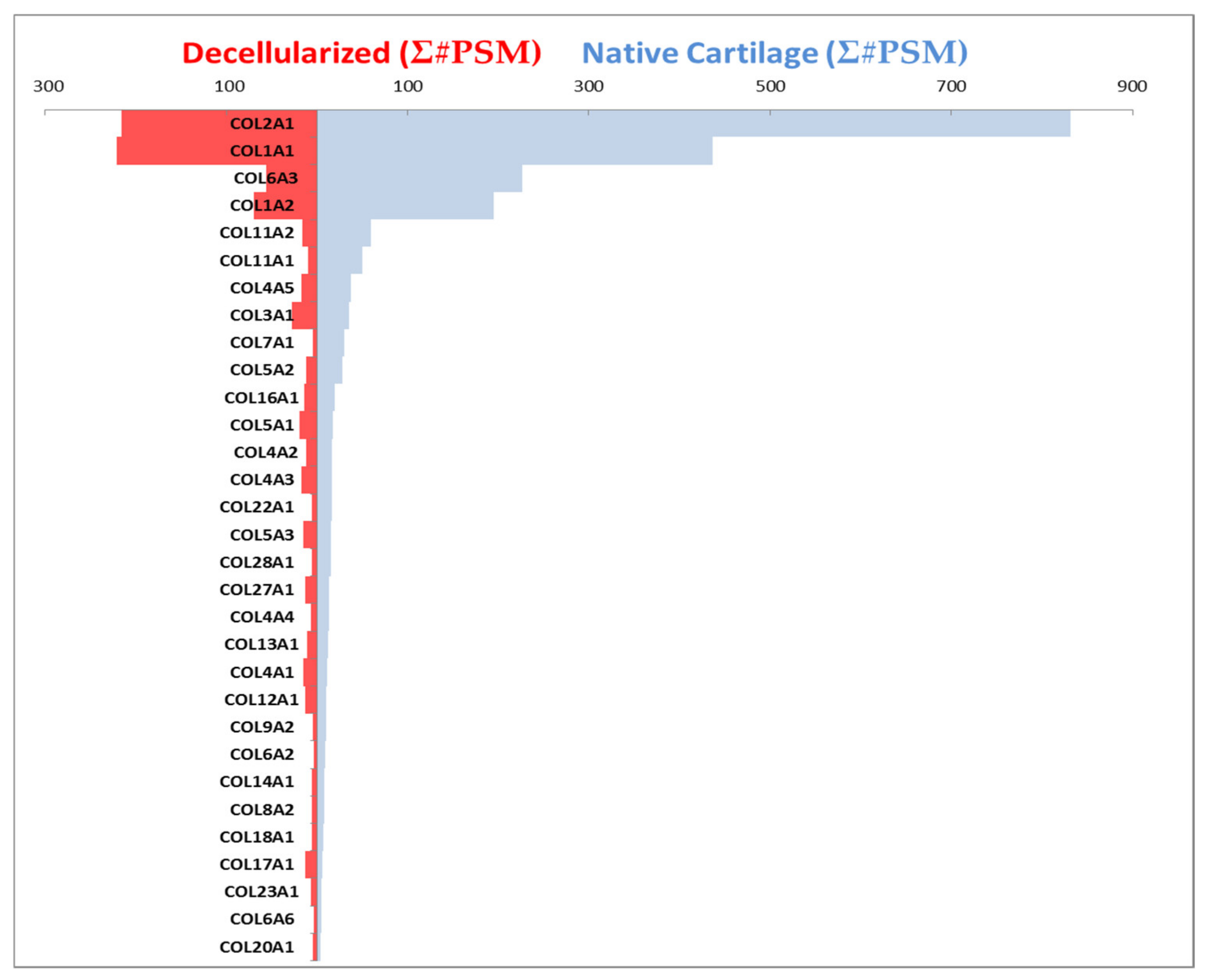
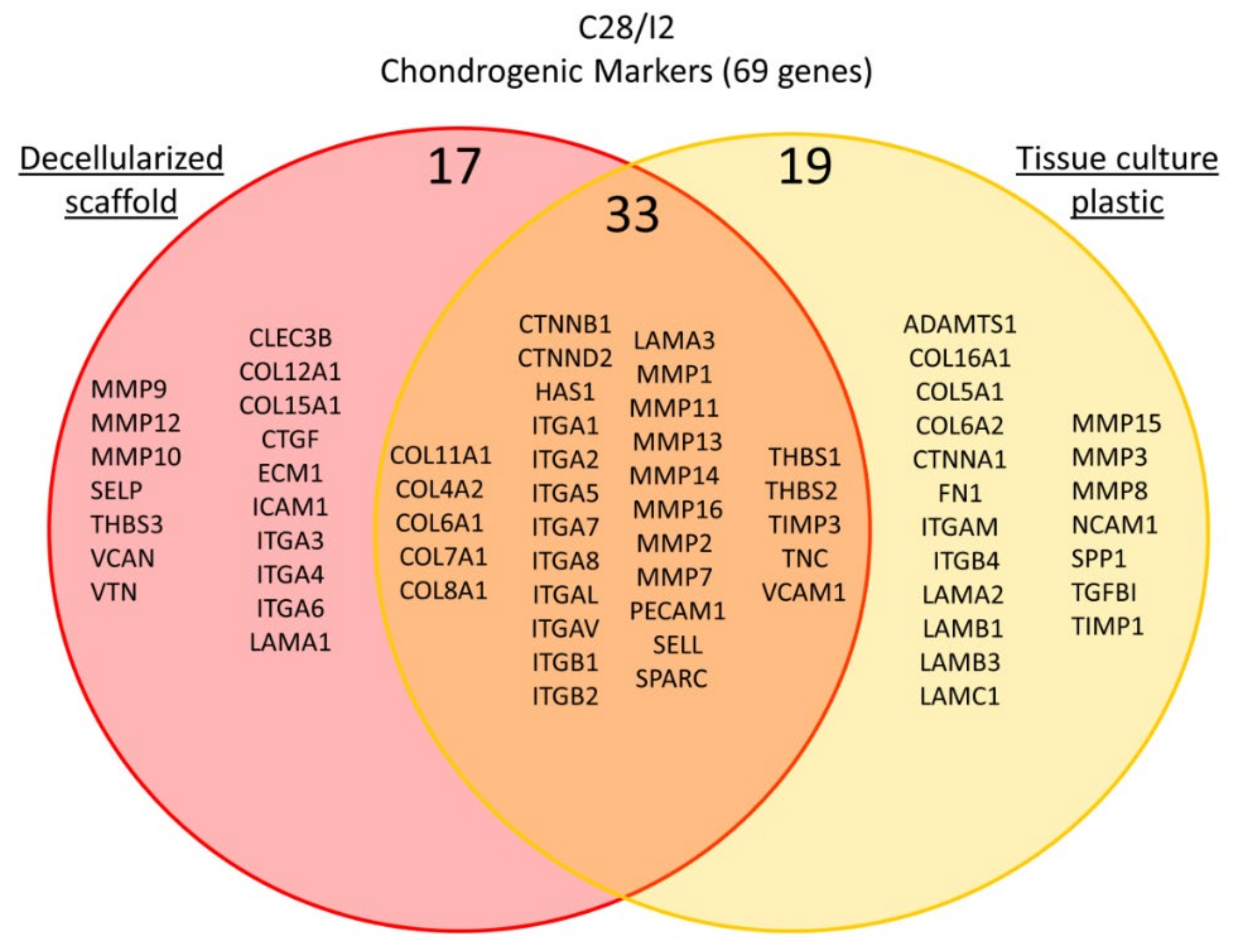
2. Results
3. Discussion and Conclusions
4. Materials and Methods
4.1. Materials
4.2. Methods
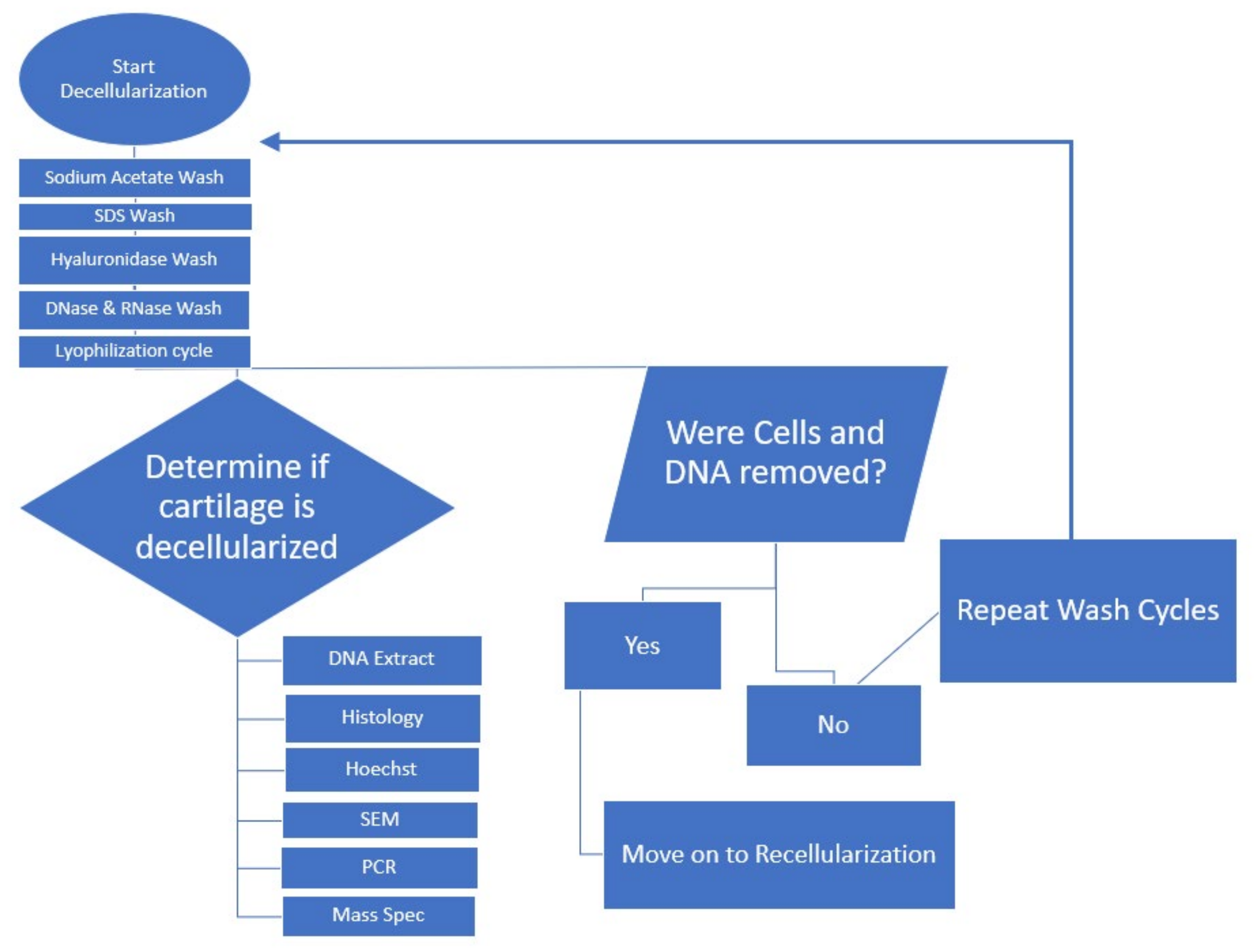
4.2.1. Decellularization
4.2.2. Histology
4.2.3. SEM Preparation and Imaging
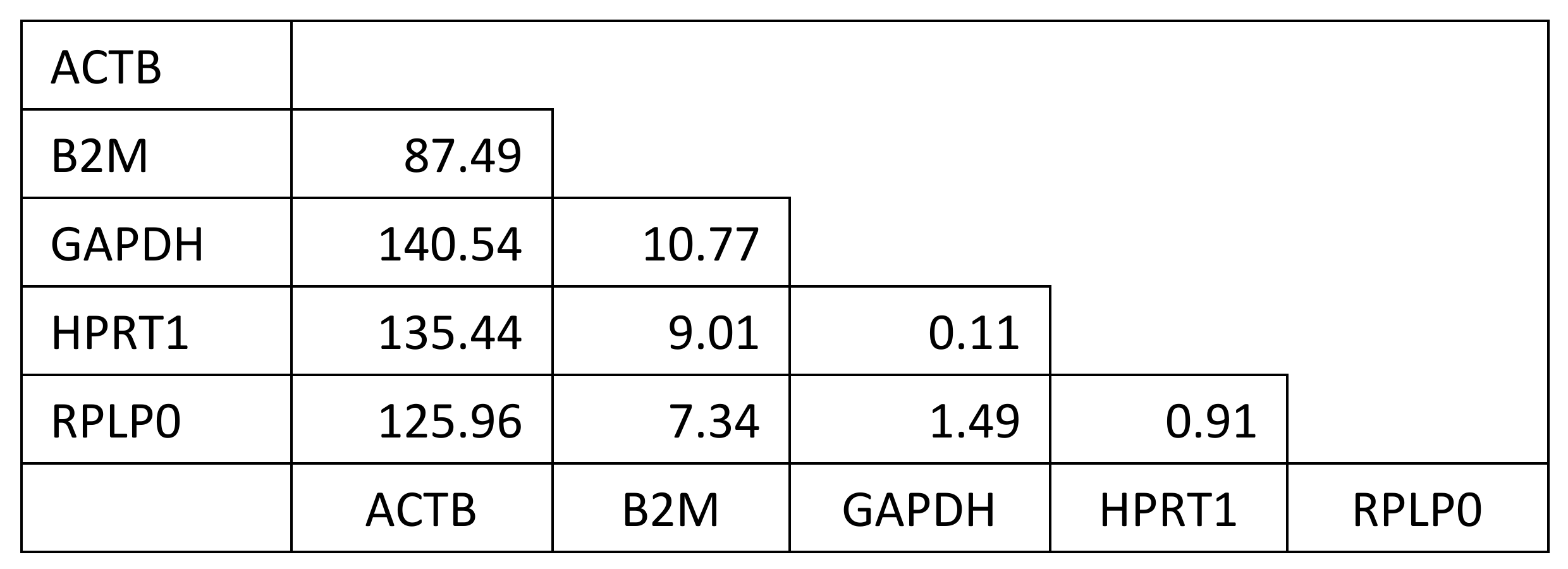
4.2.4. PCR
4.2.5. Protein Concentration Determination
4.2.6. Mass Spectrometry and Proteomics
4.2.7. Recellularization of Decellularized Scaffold
4.2.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Endoplasmic Reticulum | % Depletion | Gene Symbol |
|---|---|---|
| 40S ribosomal protein S13 | 100 | RPS13 |
| 40S ribosomal protein S3 | 100 | RPS3 |
| 5-beta-cholestane-3-alpha,7-alpha-diol 12-alpha-hydroxylase | 100 | CYP8B1 |
| 60S ribosomal protein L14 | 100 | RPL14 |
| 60S ribosomal protein L6 | 100 | RPL6 |
| Beta-crystallin B1 | 100 | CRYBB1 |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2 | 100 | RPN2 |
| Endoplasmic reticulum chaperone BiP | 100 | HSPA5 |
| Glutamate carboxypeptidase 2 | 100 | FOLH1 |
| Heat shock 70 kDa protein 1A | 100 | HSPA1A |
| Heat shock 70 kDa protein 1B | 100 | HSPA1B |
| Heat shock 70 kDa protein 1-like | 100 | HSPA1L |
| Heme oxygenase 1 | 100 | HMOX1 |
| Neutral alpha-glucosidase AB | 100 | GANAB |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 100 | ATP2A2 |
| Transitional endoplasmic reticulum ATPase | 100 | VCP |
| Microsomal triglyceride transfer protein large subunit | 85 | MTTP |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1 | 78 | RPN1 |
| Dual oxidase 1 | 78 | DUOX1 |
| Heat shock protein HSP 90-alpha | 78 | HSP90AA1 |
| Membrane Proteins | % Depletion | Gene Symbol |
|---|---|---|
| ADP-ribosylation factor 6 | 100 | ARF6 |
| Arachidonate 15-lipoxygenase | 100 | ALOX15 |
| Calpain-1 catalytic subunit | 100 | CAPN1 |
| C-X-C chemokine receptor type 4 | 100 | CXCR4 |
| Cysteinyl leukotriene receptor 2 | 100 | CYSLTR2 |
| Desmoglein-1 | 100 | DSG1 |
| Endothelin-1 receptor | 100 | EDNRA |
| Gap junction alpha-1 protein | 100 | GJA1 |
| Geranylgeranyl transferase type-2 subunit alpha | 100 | RABGGTA |
| Glutathione hydrolase 1 proenzyme | 100 | GGT1 |
| Growth hormone receptor | 100 | GHR |
| Guanine nucleotide-binding protein G(q) subunit alpha | 100 | GNAQ |
| Integrin beta-1 | 100 | ITGB1 |
| Interferon alpha/beta receptor 1 | 100 | IFNAR1 |
| Interleukin-4 receptor subunit alpha | 100 | IL4R |
| Interleukin-6 receptor subunit alpha | 100 | IL6R |
| Low-density lipoprotein receptor | 100 | LDLR |
| Mast/stem cell growth factor receptor Kit | 100 | KIT |
| Metalloreductase STEAP1 | 100 | STEAP1 |
| NT-3 growth factor receptor | 100 | NTRK3 |
| Parathyroid hormone/parathyroid hormone-related peptide receptor | 100 | PTH1R |
| PDZ domain-containing protein 11 | 100 | PDZD11 |
| Plasma membrane calcium-transporting ATPase 1 | 100 | ATP2B1 |
| Platelet endothelial cell adhesion molecule | 100 | PECAM1 |
| Protocadherin-11 X-linked | 100 | PCDH11X |
| Receptor activity-modifying protein 1 | 100 | RAMP1 |
| S-Arrestin | 100 | SAG |
| Sialoadhesin | 100 | SIGLEC1 |
| SLA class II histocompatibility antigen, DQ haplotype C alpha chain | 100 | SLA-DQCA |
| SLA class II histocompatibility antigen, DQ haplotype C beta chain | 100 | SLA-DQCB |
| SLA class II histocompatibility antigen, DQ haplotype D beta chain | 100 | SLA-DQDB |
| Small conductance calcium-activated potassium channel protein 3 | 100 | KCNN3 |
| Sodium/glucose cotransporter 1 | 100 | SLC5A1 |
| Solute carrier family 22 member 6 | 100 | SLC22A6 |
| Solute carrier family 22 member 7 | 100 | SLC22A7 |
| Thyroid peroxidase | 100 | TPO |
| Toll-like receptor 9 | 100 | TLR9 |
| Transforming growth factor beta receptor type 3 | 100 | TGFBR3 |
| Beta-1 adrenergic receptor | 94 | ADRB1 |
| Zonadhesin | 94 | ZAN |
| Glutathione S-transferase alpha M14 | 94 | GSTAM14 |
| Activin receptor type-2B | 93 | ACVR2B |
| Solute carrier family 22 member 1 | 91 | SLC22A1 |
| Low-density lipoprotein receptor-related protein 2 | 89 | LRP2 |
| Orexin receptor type 2 | 89 | HCRTR2 |
| Glutamate decarboxylase 2 | 89 | GAD2 |
| Ectonucleotide pyrophosphatase/phosphodiesterase family member 6 | 85 | ENPP6 |
| Tyrosine-protein kinase SYK | 85 | SYK |
| Hormone-sensitive lipase | 85 | LIPE |
| V-type proton ATPase catalytic subunit A | 85 | ATP6V1A |
| Potassium-transporting ATPase alpha chain 1 | 82 | ATP4A |
| Electrogenic sodium bicarbonate cotransporter 1 | 81 | SLC4A4 |
| Alpha-2A adrenergic receptor | 78 | ADRA2A |
| Calcium-activated chloride channel regulator 1 | 78 | CLCA1 |
| Gastrin/cholecystokinin type B receptor | 78 | CCKBR |
| Hepatocyte growth factor receptor | 78 | MET |
| Leptin receptor | 78 | LEPR |
| Extracellular calcium-sensing receptor | 70 | CASR |
| Scavenger receptor class B member 1 | 70 | SCARB1 |
| ATP-binding cassette sub-family G member 2 | 66 | ABCG2 |
| Major facilitator superfamily domain-containing protein 6 | 66 | MFSD6 |
| H(+)/Cl(−) exchange transporter 5 | 63 | CLCN5 |
| Prolactin receptor | 55 | PRLR |
| Beta-3 adrenergic receptor | 55 | ADRB3 |
| Lutropin-choriogonadotropic hormone receptor | 55 | LHCGR |
| Cytosolic Proteins | % Depletion | Gene Symbol |
|---|---|---|
| 1-acylglycerol-3-phosphate O-acyltransferase ABHD5 | 100 | ABHD5 |
| 4-hydroxyphenylpyruvate dioxygenase | 100 | HPD |
| Actin, cytoplasmic 1 | 100 | ACTB |
| Alcohol dehydrogenase [NADP(+)] | 100 | AKR1A1 |
| Antileukoproteinase | 100 | SLPI |
| ATP-dependent 6-phosphofructokinase, muscle type | 100 | PFKM |
| Autophagy protein 5 | 100 | ATG5 |
| Bifunctional epoxide hydrolase 2 | 100 | EPHX2 |
| Biogenesis of lysosome-related organelles complex 1 subunit 5 | 100 | BLOC1S5 |
| Calponin-1 OS = Sus scrofa | 100 | CNN1 |
| Calponin-2 | 100 | CNN2 |
| Cas scaffolding protein family member 4 | 100 | CASS4 |
| Coatomer subunit beta | 100 | COPB1 |
| Diacylglycerol kinase alpha | 100 | DGKA |
| Dihydropyrimidine dehydrogenase [NADP(+)] | 100 | DPYD |
| FAST kinase domain-containing protein 4 | 100 | TBRG4 |
| Gastrotropin | 100 | FABP6 |
| Growth factor receptor-bound protein 10 | 100 | Grb10 |
| Integrin beta-1-binding protein 2 | 100 | ITGB1BP2 |
| L-dopachrome tautomerase | 100 | DCT |
| L-lactate dehydrogenase A chain | 100 | LDHA |
| Myosin light chain 4 | 100 | MYL4 |
| Myosin-1 | 100 | MYH1 |
| Myosin-2 | 100 | MYH2 |
| Nucleoside diphosphate kinase B | 100 | NME2 |
| Perilipin-3 | 100 | PLIN3 |
| Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | 100 | PIK3CG |
| Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform | 100 | PPP2R1B |
| Suppressor of cytokine signaling 2 | 100 | SOCS2 |
| Thimet oligopeptidase | 100 | THOP1 |
| Triosephosphate isomerase | 100 | TPI1 |
| Tubulin alpha-1A chain | 100 | TUBA1A |
| Tubulin beta chain | 100 | TUBB |
| Vinculin | 100 | VCL |
| Serine/threonine-protein kinase WNK1 | 95 | WNK1 |
| L-lactate dehydrogenase B chain | 94 | LDHB |
| UTP-glucose-1-phosphate uridylyltransferase | 92 | UGP2 |
| Eukaryotic initiation factor 4A-III | 89 | EIF4A3 |
| Triokinase/FMN cyclase | 85 | TKFC |
| Acylphosphatase-1 | 83 | ACYP1 |
| Glycine N-methyltransferase | 78 | GNMT |
| N-acetylneuraminate lyase | 78 | NPL |
| Phosphatidylinositol 3-kinase catalytic subunit type 3 | 78 | PIK3C3 |
| Serine/threonine-protein phosphatase 1 regulatory subunit 10 | 64 | PPP1R10 |
References
- Cross, M.; Smith, E.; Hoy, D.; Nolte, S.; Ackerman, I.; Fransen, M.; Bridgett, L.; Williams, S.; Guillemin, F.; Hill, C.L.; et al. The global burden of hip and knee osteoarthritis: Estimates from the Global Burden of Disease 2010 study. Ann. Rheum. Dis. 2014, 73, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Escobar Ivirico, J.L.; Bhattacharjee, M.; Kuyinu, E.; Nair, L.S.; Laurencin, C.T. Regenerative Engineering for Knee Osteoarthritis Treatment: Biomaterials and Cell-Based Technologies. Engineering 2017, 3, 16–27. [Google Scholar] [CrossRef]
- Helmick, C.G.; Felson, D.T.; Lawrence, R.C.; Gabriel, S.; Hirsch, R.; Kwoh, C.K.; Liang, M.H.; Maradit Kremers, H.; Mayes, M.D.; Merkel, P.A.; et al. National Arthritis Data Workgroup Estimates of the prevalence of arthritis and other rheumatic conditions in the United States: Part I. Arthritis Rheum. 2008, 58, 15–25. [Google Scholar] [CrossRef]
- Toh, W.S.; Foldager, C.B.; Pei, M.; Hui, J.H.P. Advances in Mesenchymal Stem Cell-based Strategies for Cartilage Repair and Regeneration. Stem Cell Rev. Rep. 2014, 10, 686–696. [Google Scholar] [CrossRef]
- Gupta, P.K.; Das, A.K.; Chullikana, A.; Majumdar, A.S. Mesenchymal stem cells for cartilage repair in osteoarthritis. Stem Cell Res. Ther. 2012, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, A.; Mitchell, K.; Soans, J.; Kim, L.; Zaidi, R. The use of mesenchymal stem cells for cartilage repair and regeneration: A systematic review. J. Orthop. Surg. Res. 2017, 12, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.W.; Solorio, L.D.; Alsberg, E. Decellularized tissue and cell-derived extracellular matrices as scaffolds for orthopaedic tissue engineering. Biotechnol. Adv. 2014, 32, 462–484. [Google Scholar] [CrossRef] [Green Version]
- Djouad, F.; Mrugala, D.; Noël, D.; Jorgensen, C. Engineered mesenchymal stem cells for cartilage repair. Regen. Med. 2006, 1, 529–537. [Google Scholar] [CrossRef]
- Guilak, F.; Nims, R.J.; Dicks, A.; Wu, C.-L.; Meulenbelt, I. Osteoarthritis as a disease of the cartilage pericellular matrix. Matrix Biol. 2010, 71–72, 40–50. [Google Scholar] [CrossRef]
- Sophia Fox, A.J.; Bedi, A.; Rodeo, S.A. The basic science of articular cartilage: Structure, composition, and function. Sports Health 2009, 1, 461–468. [Google Scholar] [CrossRef]
- Martin, J.A.; Buckwalter, J.A. Roles of articular cartilage aging and chondrocyte senescence in the pathogenesis of osteoarthritis. Iowa Orthop. J. 2001, 21, 1–7. [Google Scholar]
- Schulze-Tanzil, G. Intraarticular Ligament Degeneration Is Interrelated with Cartilage and Bone Destruction in Osteoarthritis. Cells 2019, 8, 990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, Y.; Rim, Y.A.; Lee, J.; Ju, J.H. Current therapeutic strategies for stem cell-based cartilage regeneration. Stem Cells Int. 2018, 2018, 8490489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baugé, C.; Boumédiene, K. Use of Adult Stem Cells for Cartilage Tissue Engineering: Current Status and Future Developments. Stem Cells Int. 2015, 2015, 438026. [Google Scholar] [CrossRef] [Green Version]
- Ye, K.; Felimban, R.; Moulton, S.E.; Wallace, G.G.; Di Bella, C.; Traianedes, K.; Choong, P.F.M.; Myers, D.E. Bioengineering of articular cartilage: Past, present and future. Regen. Med. 2013, 8, 333–349. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Xu, H.; Zhang, J. Current Tissue Engineering Approaches for Cartilage Regeneration. In Cartilage Tissue Engineering and Regeneration Techniques; IntechOpen Limited: London, UK, 2019. [Google Scholar] [CrossRef]
- Chang, B.; Cornett, A.; Nourmohammadi, Z.; Law, J.; Weld, B.; Crotts, S.J.; Hollister, S.J.; Lombaert, I.M.A.; Zopf, D.A. Hybrid Three-Dimensional–Printed Ear Tissue Scaffold With Autologous Cartilage Mitigates Soft Tissue Complications. Laryngoscope 2021, 131, 1008–1015. [Google Scholar] [CrossRef]
- Gilpin, A.; Yang, Y. Decellularization Strategies for Regenerative Medicine: From Processing Techniques to Applications. BioMed Res. Int. 2017, 2017, 9831534. [Google Scholar] [CrossRef] [Green Version]
- Heath, D.E. A Review of Decellularized Extracellular Matrix Biomaterials for Regenerative Engineering Applications. Regenerative Eng. Transl. Med. 2019, 5, 155–166. [Google Scholar] [CrossRef]
- Kim, Y.S.; Majid, M.; Melchiorri, A.J.; Mikos, A.G. Applications of decellularized extracellular matrix in bone and cartilage tissue engineering. Bioeng. Transl. Med. 2019, 4, 83–95. [Google Scholar] [CrossRef] [Green Version]
- Goldring, M.B.; Birkhead, J.R.; Suen, L.F.; Yamin, R.; Mizuno, S.; Glowacki, J.; Arbiser, J.L.; Apperley, J.F. Interleukin-1β-modulated gene expression in immortalized human chondrocytes. J. Clin. Investig. 1994, 94, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Eswaramoorthy, R.; Mulhall, K.J.; Kelly, D.J. Decellularization of porcine articular cartilage explants and their subsequent repopulation with human chondroprogenitor cells. J. Mech. Behav. Biomed. Mater. 2016, 55, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, J.; Ritchie, G.R.S.; Roumeliotis, T.I.; Jayasuriya, R.L.; Clark, M.J.; Brooks, R.A.; Binch, A.L.A.; Shah, K.M.; Coyle, R.; Pardo, M.; et al. Integrative epigenomics, transcriptomics and proteomics of patient chondrocytes reveal genes and pathways involved in osteoarthritis. Sci. Rep. 2017, 7, 8935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdes, A.M. Involvement of the CLEC3B gene in osteoarthritis. Osteoarthr. Cartil. 2011, 19, 249. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, C.; Dehne, T.; Lindahl, A.; Brittberg, M.; Pruss, A.; Sittinger, M.; Ringe, J. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthr. Cartil. 2010, 18, 581–592. [Google Scholar] [CrossRef] [Green Version]
- Mazzoni, E.; D’Agostino, A.; Iaquinta, M.R.; Bononi, I.; Trevisiol, L.; Rotondo, J.C.; Patergnani, S.; Giorgi, C.; Gunson, M.J.; Arnett, G.W.; et al. Hydroxylapatite-collagen hybrid scaffold induces human adipose-derived mesenchymal stem cells to osteogenic differentiation in vitro and bone regrowth in patients. Stem Cells Transl. Med. 2020, 9, 377–388. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.; Reynard, L.N.; Loughlin, J. Functional characterisation of the osteoarthritis susceptibility locus at chromosome 6q14.1 marked by the polymorphism rs9350591. BMC Med. Genet. 2015, 16, 81. [Google Scholar] [CrossRef] [Green Version]
- arcOGEN Consortium; arcOGEN Collaborators; Zeggini, E.; Panoutsopoulou, K.; Southam, L.; Rayner, N.W.; Day-Williams, A.G.; Lopes, M.C.; Boraska, V.; Esko, T.; et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): A genome-wide association study. Lancet 2012, 380, 815–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manon-Jensen, T.; Karsdal, M.A. Type XII Collagen. In Biochemistry of Collagens, Laminins and Elastin: Structure, Function and Biomarkers; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 81–85. [Google Scholar]
- Luo, Y.; Sinkeviciute, D.; He, Y.; Karsdal, M.; Henrotin, Y.; Mobasheri, A.; Önnerfjord, P.; Bay-Jensen, A. The minor collagens in articular cartilage. Protein Cell 2017, 8, 560–572. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, P.; Zwolanek, D.; Keene, D.R.; Schulz, J.-N.; Blumbach, K.; Heinegård, D.; Zaucke, F.; Paulsson, M.; Krieg, T.; Koch, M.; et al. Collagen XII and XIV, new partners of cartilage oligomeric matrix protein in the skin extracellular matrix suprastructure. J. Biol. Chem. 2012, 287, 22549–22559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Sheng, X.; Zhang, Z.; Zhao, K.; Zhu, L.; Guo, G.; Friedenberg, S.G.; Hunter, L.S.; Vandenberg-Foels, W.S.; Hornbuckle, W.E.; et al. Differential genetic regulation of canine hip dysplasia and osteoarthritis. PLoS ONE 2010, 5, e13219. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Muhammad, H.; McLean, C.; Miotla-Zarebska, J.; Fleming, J.; Didangelos, A.; Önnerfjord, P.; Leask, A.; Saklatvala, J.; Vincent, T.L. Connective tissue growth factor contributes to joint homeostasis and osteoarthritis severity by controlling the matrix sequestration and activation of latent TGFβ. Ann. Rheum. Dis. 2018, 77, 1372–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivkovic, S.; Yoon, B.S.; Popoff, S.N.; Safadi, F.F.; Libuda, D.E.; Stephenson, R.C.; Daluiski, A.; Lyons, K.M. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 2003, 130, 2779–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi-Wen, X.; Leask, A.; Abraham, D. Regulation and function of connective tissue growth factor/CCN2 in tissue repair, scarring and fibrosis. Cytokine Growth Factor Rev. 2008, 19, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Lipson, K.E.; Wong, C.; Teng, Y.; Spong, S. CTGF is a central mediator of tissue remodeling and fibrosis and its inhibition can reverse the process of fibrosis. Fibrogenes. Tissue Repair 2012, 5, S24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.; Zhao, Y.P.; Tian, Q.Y.; Feng, J.-Q.; Kobayashi, T.; Merregaert, J.; Liu, C.-J. Extracellular matrix protein 1, a direct targeting molecule of parathyroid hormone-related peptide, negatively regulates chondrogenesis and endochondral ossification via associating with progranulin growth factor. FASEB J. 2016, 30, 2741–2754. [Google Scholar] [CrossRef] [Green Version]
- Mongiat, M.; Fu, J.; Oldershaw, R.; Greenhalgh, R.; Gown, A.M.; Iozzo, R.V. Perlecan protein core interacts with extracellular matrix protein 1 (ECM1), a glycoprotein involved in bone formation and angiogenesis. J. Biol. Chem. 2003, 278, 17491–17499. [Google Scholar] [CrossRef] [Green Version]
- Frahs, S.M.; Reeck, J.C.; Yocham, K.M.; Frederiksen, A.; Fujimoto, K.; Scott, C.M.; Beard, R.S., Jr.; Brown, R.J.; Lujan, T.J.; Solov’yov, I.A.; et al. Prechondrogenic ATDC5 Cell Attachment and Differentiation on Graphene Foam; Modulation by Surface Functionalization with Fibronectin. ACS Appl. Mater. Interfaces 2019, 11, 41906–41924. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Tian, Q.; Guo, F.; Mucignat, M.T.; Perris, R.; Sercu, S.; Merregaert, J.; Di Cesare, P.E.; Liu, C.-J. Interaction between cartilage oligomeric matrix protein and extracellular matrix protein 1 mediates endochondral bone growth. Matrix Biol. 2010, 29, 276–286. [Google Scholar] [CrossRef] [Green Version]
- Yatabe, T.; Mochizuki, S.; Takizawa, M.; Chijiiwa, M.; Okada, A.; Kimura, T.; Fujita, Y.; Matsumoto, H.; Toyama, Y.; Okada, Y. Hyaluronan inhibits expression of ADAMTS4 (aggrecanase-1) in human osteoarthritic chondrocytes. Ann. Rheum. Dis. 2009, 68, 1051–1058. [Google Scholar] [CrossRef]
- Gromova, O.A.; Torshin, I.Y.; Lila, A.M.; Gromov, A.N. Molecular mechanisms of action of glucosamine sulfate in the treatment of degenerative-dystrophic diseases of the joints and spine: Results of proteomic analysis. Nevrol. Neiropsikhiatriya Psikhosomatika 2018, 10, 38–44. [Google Scholar] [CrossRef] [Green Version]
- Rangkasenee, N.; Murani, E.; Schellander, K.; Cinar, M.U.; Ponsuksili, S.; Wimmers, K. Gene expression profiling of articular cartilage reveals functional pathways and networks of candidate genes for osteochondrosis in pigs. Physiol. Genom. 2013, 45, 856–865. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Guo, H.; Yang, X.; Li, Z.; Zhang, D.; Li, B.; Zhang, D.; Li, Q.; Xiong, Y. Potential candidate biomarkers associated with osteoarthritis: Evidence from a comprehensive network and pathway analysis. J. Cell. Physiol. 2019, 234, 17433–17443. [Google Scholar] [CrossRef]
- Djouad, F.; Delorme, B.; Maurice, M.; Bony, C.; Apparailly, F.; Louis-Plence, P.; Canovas, F.; Charbord, P.; Noël, D.; Jorgensen, C. Microenvironmental changes during differentiation of mesenchymal stem cells towards chondrocytes. Arthritis Res. Ther. 2007, 9, R33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weeks, S.; Kulkarni, A.; Smith, H.; Whittall, C.; Yang, Y.; Middleton, J. The effects of chemokine, adhesion and extracellular matrix molecules on binding of mesenchymal stromal cells to poly(l-lactic acid). Cytotherapy 2012, 14, 1080–1088. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Chen, G.; Jiao, W.; Wang, D.; Cao, Y.; Zhang, Q.; Wang, J. Identification of critical genes in nucleus pulposus cells isolated from degenerated intervertebral discs using bioinformatics analysis. Mol. Med. Rep. 2017, 16, 553–564. [Google Scholar] [CrossRef] [Green Version]
- Tu, Y.; Ma, T.; Wen, T.; Yang, T.; Xue, L.; Cai, M.; Wang, F.; Guan, M.; Xue, H. MicroRNA-377–3p alleviates IL-1β-caused chondrocyte apoptosis and cartilage degradation in osteoarthritis in part by downregulating ITGA6. Biochem. Biophys. Res. Commun. 2020, 523, 46–53. [Google Scholar] [CrossRef] [PubMed]
- LaPointe, V.L.S.; Verpoorte, A.; Stevens, M.M. The changing integrin expression and a role for integrin β8 in the chondrogenic differentiation of mesenchymal stem cells. PLoS ONE 2013, 8, e82035. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Chen, M.; Wang, H.; Huang, J.; Bao, Y.; Gan, X.; Liu, B.; Lu, X.; Wang, L. Cell Adhesion-Related Molecules Play a Key Role in Renal Cancer Progression by Multinetwork Analysis. BioMed Res. Int. 2019, 2019, 2325765. [Google Scholar] [CrossRef]
- Soki, F.N.; Yoshida, R.; Paglia, D.N.; Duong, L.T.; Hansen, M.F.; Drissi, H. Articular cartilage protection in Ctsk-/- mice is associated with cellular and molecular changes in subchondral bone and cartilage matrix. J. Cell. Physiol. 2018, 233, 8666–8676. [Google Scholar] [CrossRef] [PubMed]
- Adapala, N.S.; Kim, H.K.W. Comprehensive genome-wide transcriptomic analysis of immature articular cartilage following ischemic osteonecrosis of the femoral head in piglets. PLoS ONE 2016, 11, e0153174. [Google Scholar] [CrossRef] [Green Version]
- Grogan, S.P.; Duffy, S.F.; Pauli, C.; Koziol, J.A.; Su, A.I.; D’Lima, D.D.; Lotz, M.K. Zone-specific gene expression patterns in articular cartilage. Arthritis Rheum. 2013, 65, 418–428. [Google Scholar] [CrossRef] [Green Version]
- Mann, V.; Grimm, D.; Corydon, T.J.; Krüger, M.; Wehland, M.; Riwaldt, S.; Sahana, J.; Kopp, S.; Bauer, J.; Reseland, J.E.; et al. Changes in human foetal osteoblasts exposed to the random positioning machine and bone construct tissue engineering. Int. J. Mol. Sci. 2019, 20, 1357. [Google Scholar] [CrossRef] [Green Version]
- Dehne, T.; Schenk, R.; Perka, C.; Morawietz, L.; Pruss, A.; Sittinger, M.; Kaps, C.; Ringe, J. Gene expression profiling of primary human articular chondrocytes in high-density micromasses reveals patterns of recovery, maintenance, re- and dedifferentiation. Gene 2010, 462, 8–17. [Google Scholar] [CrossRef]
- Göhring, A.R.; Lübke, C.; Andreas, K.; Kaps, C.; Häupl, T.; Pruss, A.; Perka, C.; Sittinger, M.; Ringe, J. Tissue-engineered cartilage of porcine and human origin as in vitro test system in arthritis research. Biotechnol. Prog. 2010, 26, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.J.; Peng, Y.; Lim, F.L.; Sun, Y.; Lv, M.; Zhou, L.; Wang, H.; Zheng, Z.; Cheung, K.; Leung, V. Matrix metalloproteinase 12 is an indicator of intervertebral disc degeneration co-expressed with fibrotic markers. Osteoarthr. Cartil. 2016, 24, 1826–1836. [Google Scholar] [CrossRef] [Green Version]
- Challa, T.D.; Rais, Y.; Ornan, E.M. Effect of adiponectin on ATDC5 proliferation, differentiation and signaling pathways. Mol. Cell. Endocrinol. 2010, 323, 282–291. [Google Scholar] [CrossRef]
- Yang, Q.; Attur, M.; Kirsch, T.; Lee, Y.L.; Yakar, S.; Liu, Z.; Abramson, S.B.; Mignatti1, P. Membrane-type 1 matrix metalloproteinase controls osteo-and chondrogenesis by a proteolysis-independent mechanism mediated by its cytoplasmic tail. Osteoarthr. Cartil. 2015, 23, A64. [Google Scholar] [CrossRef] [Green Version]
- Miao, D.; Bai, X.; Panda, D.K.; Karaplis, A.C.; Goltzman, D.; McKee, M.D. Cartilage abnormalities are associated with abnormal Phex expression and with altered matrix protein and MMP-9 localization in Hyp mice. Bone 2004, 34, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Gari, M.A.; AlKaff, M.; Alsehli, H.S.; Dallol, A.; Gari, A.; Abu-Elmagd, M.; Kadam, R.; Abuzinadah, M.F.; Gari, M.; Abuzenadah, A.M.; et al. Identification of novel genetic variations affecting osteoarthritis patients. BMC Med. Genet. 2016, 17, 68. [Google Scholar] [CrossRef] [Green Version]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database J. Biol. Databases Curation 2016, 2016, baw100. [Google Scholar] [CrossRef] [PubMed]
- Bonn, F.; Pantakani, K.; Shoukier, M.; Langer, T.; Mannan, A.U. Functional evaluation of paraplegin mutations by a yeast complementation assay. Hum. Mutat. 2010, 31, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Vos, H.L.; Devarayalu, S.; De Vries, Y.; Bornstein, P. Thrombospondin 3 (Thbs3), a new member of the thrombospondin gene family. J. Biol. Chem. 1992, 267, 12192–12196. [Google Scholar] [CrossRef]
- Adolph, K.W.; Long, G.L.; Winfield, S.; Ginns, E.I.; Bornstein, P. Structure and organization of the human thrombospondin 3 gene (thbs3). Genomics 1995, 27, 329–336. [Google Scholar] [CrossRef]
- Posey, K.L.; Hankenson, K.; Veerisetty, A.C.; Bornstein, P.; Lawler, J.; Hecht, J.T. Skeletal abnormalities in mice lacking extracellular matrix proteins, thrombospondin-1, thrombospondin-3, thrombospondin-5, and type IX collagen. Am. J. Pathol. 2008, 172, 1664–1674. [Google Scholar] [CrossRef] [Green Version]
- Hankenson, K.D.; Hormuzdi, S.G.; Meganck, J.A.; Bornstein, P. Mice with a Disruption of the Thrombospondin 3 Gene Differ in Geometric and Biomechanical Properties of Bone and Have Accelerated Development of the Femoral Head. Mol. Cell. Biol. 2005, 25, 5599–5606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamiya, N.; Watanabe, H.; Habuchi, H.; Takagi, H.; Shinomura, T.; Shimizu, K.; Kimata, K. Versican/PG-M regulates chondrogenesis as an extracellular matrix molecule crucial for mesenchymal condensation. J. Biol. Chem. 2006, 281, 2390–2400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choocheep, K.; Hatano, S.; Takagi, H.; Watanabe, H.; Kimata, K.; Kongtawelert, P.; Watanabe, H. Versican facilitates chondrocyte differentiation and regulates joint morphogenesis. J. Biol. Chem. 2010, 285, 21114–21125. [Google Scholar] [CrossRef] [Green Version]
- Sztrolovics, R.; Grover, J.; Cs-Szabo, G.; Shi, S.L.; Zhang, Y.; Mort, J.S.; Roughley, P.J. The characterization of versican and its message in human articular cartilage and intervertebral disc. J. Orthop. Res. 2002, 20, 257–266. [Google Scholar] [CrossRef]
- Luo, H.; Yao, L.; Zhang, Y.; Li, R. Liquid chromatography–mass spectrometry-based quantitative proteomics analysis reveals chondroprotective effects of astragaloside IV in interleukin-1β-induced SW1353 chondrocyte-like cells. Biomed. Pharmacother. 2017, 91, 796–802. [Google Scholar] [CrossRef]
- Vieira, A.E.; Repeke, C.E.; De Barros Ferreira, S.; Colavite, P.M.; Biguetti, C.C.; Oliveira, R.C.; Assis, G.F.; Taga, R.; Trombone, A.P.; Garlet, G.P. Intramembranous bone healing process subsequent to tooth extraction in mice: Micro-computed tomography, histomorphometric and molecular characterization. PLoS ONE 2015, 10, e0128021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, M.; Zhang, Y.; Li, J.; Chen, D. Antioxidation of decellularized stem cell matrix promotes human synovium-derived stem cell-based chondrogenesis. Stem Cells Dev. 2013, 22, 889–900. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, S.; Koerber, L.; Elsaesser, A.F.; Goldberg-Bockhorn, E.; Seitz, A.M.; Dürselen, L.; Ignatius, A.; Walther, P.; Breiter, R.; Rotter, N. Decellularized cartilage matrix as a novel biomatrix for cartilage tissue-engineering applications. Tissue Eng. Part A 2012, 18, 2195–2209. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Xu, B.; Yang, Q.; Li, X.; Ma, X.; Xia, Q.; Zhang, Y.; Zhang, C.; Wu, Y.; Zhang, Y. Comparison of decellularization protocols for preparing a decellularized porcine annulus fibrosus scaffold. PLoS ONE 2014, 9, e86723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiyotake, E.A.; Beck, E.C.; Detamore, M.S. Cartilage extracellular matrix as a biomaterial for cartilage regeneration. Ann. N. Y. Acad. Sci. 2016, 1383, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Peretti, G.M.; Randolph, M.A.; Caruso, E.M.; Rossetti, F.; Zaleske, D.J. Bonding of cartilage matrices with cultured chondrocytes: An experimental model. J. Orthop. Res. 1998, 16, 89–95. [Google Scholar] [CrossRef]
- Vas, W.J.; Shah, M.; Blacker, T.S.; Duchen, M.R.; Sibbons, P.; Roberts, S.J. Decellularized Cartilage Directs Chondrogenic Differentiation: Creation of a Fracture Callus Mimetic. Tissue Eng. Part A 2018, 24, 1364–1376. [Google Scholar] [CrossRef]
- Li, J.; Pei, M. Cell Senescence: A Challenge in Cartilage Engineering and Regeneration. Tissue Eng. Part. B Rev. 2012, 18, 270–287. [Google Scholar] [CrossRef]
- Gong, Y.Y.; Xue, J.X.; Zhang, W.J.; Zhou, G.D.; Liu, W.; Cao, Y. A sandwich model for engineering cartilage with acellular cartilage sheets and chondrocytes. Biomaterials 2011, 32, 2265–2273. [Google Scholar] [CrossRef]
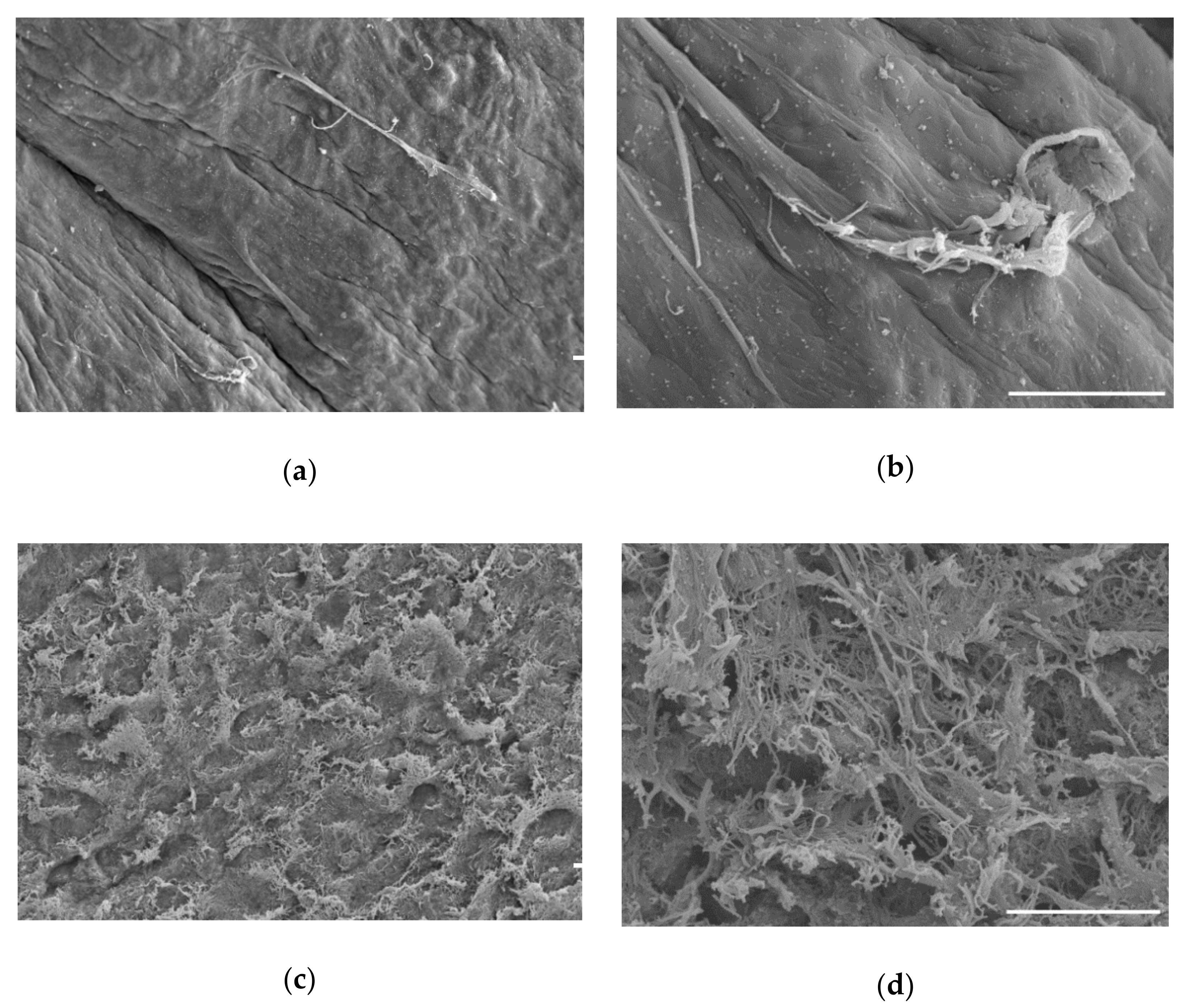


| Nuclear Proteins Depleted by Decellularization Process | % Depletion | Gene Symbol |
|---|---|---|
| Aprataxin | 100 | APTX |
| BRCA1-A complex subunit RAP80 | 100 | UIMC1 |
| Doublesex- and mab-3-related transcription factor 1 | 100 | DMRT1 |
| Histone H3.3 | 100 | H3F3A |
| Interferon-induced GTP-binding protein Mx1 | 100 | MX1 |
| Interferon-stimulated gene 20 kDa protein | 100 | ISG20 |
| Iron-responsive element-binding protein 2 | 100 | IREB2 |
| Myocardin | 100 | MYOCD |
| Nuclear factor of activated T-cells, cytoplasmic 1 | 100 | NFATC1 |
| Nuclear receptor subfamily 0 group B member 1 | 100 | NR0B1 |
| Polypyrimidine tract-binding protein 1 | 100 | PTBP1 |
| POU domain, class 5, transcription factor 1 | 100 | POU5F1 |
| Sorbin and SH3 domain-containing protein 2 | 100 | SORBS2 |
| SRSF protein kinase 3 | 100 | SRPK3 |
| Sterol regulatory element-binding protein 1 | 100 | SREBF1 |
| Histone H4 | 96 | Histone H4 |
| Signal transducer and activator of transcription 5A | 89 | STAT5A |
| Hepatocyte nuclear factor 1-beta | 85 | HNF1B |
| Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 | 85 | DHX16 |
| V(D)J recombination-activating protein 1 | 83 | RAG1 |
| Golgi Proteins | % Depletion | Gene Symbol |
|---|---|---|
| UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 | 100 | B3GALNT1 |
| Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA | 100 | MAN1A1 |
| Galactoside 2-alpha-l-fucosyltransferase 2 | 100 | FUT2 |
| Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C | 100 | MGAT4C |
| Lactosylceramide 1,3-N-acetyl-beta-d-glucosaminyltransferase | 66 | B3GNT5 |
| Mitochondrial Proteins | % Depletion | Gene Symbol |
|---|---|---|
| Aconitate hydratase, mitochondrial | 100 | ACO2 |
| A-kinase anchor protein 10, mitochondrial | 100 | AKAP10 |
| Aspartate aminotransferase, mitochondrial | 100 | GOT2 |
| Carnitine O-palmitoyltransferase 1, muscle isoform | 100 | CPT1B |
| Cholesterol side-chain cleavage enzyme, mitochondrial | 100 | CYP11A1 |
| Cysteine protease ATG4D | 100 | ATG4D |
| Cytochrome b-245 heavy chain | 100 | CYBB |
| Cytochrome c oxidase copper chaperone | 100 | COX17 |
| Cytochrome P450 11B1, mitochondrial | 100 | CYP11B1 |
| Glycerol-3-phosphate acyltransferase 1, mitochondrial | 100 | GPAM |
| Glycine amidinotransferase, mitochondrial | 100 | GATM |
| Methylmalonyl-CoA mutase, mitochondrial | 100 | MUT |
| Mitochondrial Rho GTPase 2 | 100 | RHOT2 |
| Mitochondrial uncoupling protein 2 | 100 | UCP2 |
| Mitochondrial uncoupling protein 3 | 100 | UCP3 |
| NADH-ubiquinone oxidoreductase chain 5 | 100 | MT-ND5 |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 100 | SDHA |
| Succinate-CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | 100 | SUCLG1 |
| Valine-tRNA ligase, mitochondrial | 100 | VARS2 |
| Amine oxidase [flavin-containing] B | 91 | MAOB |
| Mitochondria-eating protein | 91 | SPATA18 |
| Nicotinamide phosphoribosyltransferase | 91 | NAMPT |
| Hexokinase-2 OS=Sus scrofa | 87 | HK2 |
| Kynurenine 3-monooxygenase | 85 | KMO |
| NADP-dependent malic enzyme | 83 | ME1 |
| Cytochrome P450 3A29 | 78 | CYP3A29 |
| Glyceraldehyde-3-phosphate dehydrogenase | 66 | GAPDH |
| Hydroxymethylglutaryl-CoA synthase, mitochondrial | 66 | HMGCS2 |
| Creatine kinase U-type, mitochondrial | 55 | CKMT1 |
| Extracellular Matrix Noncollagenous Proteins | % Depleted | Gene Symbol |
|---|---|---|
| Fibromodulin | 100 | FMOD |
| Dystroglycan | 100 | DAG1 |
| Fibrillin-1 | 100 | FBN1 |
| Aggrecan core protein | 100 | ACAN |
| Decorin | 100 | DCN |
| Lactadherin | 98 | MFGE8 |
| Hyaluronan and proteoglycan link protein 1 | 90 | HAPLN1 |
| Tenascin | 70 | TNC |
| Biglycan | 57 | BGN |
| Gene Symbol | Name | Function | Reference |
|---|---|---|---|
| CLEC3B | C-type lectin domain family 3, member B | Encodes tetranectin. Cellular response to transforming growth factor stimulus. | Steinberg 2017 [24] Valdes 2011 [25] Karlsson 2010 [26] Mazzoni 2020 [27] |
| COL12A1 | Collagen, type XII, alpha 1 | Encodes the alpha chain of type XII collagen. Modifies the interactions between collagen fibrils and the surrounding matrix. A component of cartilage ECM. | Johnson 2015 [28] Zeggini 2012 [29] Manon-Jensen 2016 [30] Luo 2017 [31] Agarwal 2012 [32] |
| COL15A1 | Collagen, type XV, alpha 1 | Encodes the alpha chain of type XV collagen. Strongest expression in basement membrane zones; may function to adhere basement membranes to underlying connective tissue. | Karlsson 2010 [26] Zhou 2010 [33] Valdes 2011 [25] |
| CTGF | Connective tissue growth factor | Modulates signaling pathways leading to cell adhesion and migration, along with ECM deposition and remodeling, which together lead to tissue remodeling. | Tang 2018 [34] Ivkovic 2003 [35] Shi-Wen 2008 [36] Lipson 2012 [37] |
| ECM1 | Extracellular matrix protein 1 | Inhibits chondrocyte hypertrophy, matrix mineralization, and endochondral bone formation. | Kong 2016 [38] Mongiat 2003 [39] Frahs 2019 [40] Kong 2010 [41] |
| ICAM1 | Intercellular adhesion molecule 1 | Encodes cell surface glycoprotein. | Yatabe 2009 [42] Gromova 2018 [43] Rangkasenee 2013 [44] |
| ITGA3 | Integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | Involved in cell adhesion and collagen binding. | Zhang 2019 [45] |
| ITGA4 | Integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | Functions in cell surface adhesion and signaling, ECM receptor interaction. | Djouad 2007 [46] Weeks 2012 [47] Zhu 2017 [48] |
| ITGA6 | Integrin, alpha 6 | Functions in cell surface adhesion and signaling. | Tu 2020 [49] LaPointe 2013 [50] |
| LAMA1 | Laminin, alpha 1 | Major component of the basement membrane. Associated with cell adhesion, differentiation, migration, and signaling. | Zhang 2019 [45] Wang 2019 [51] Soki 2018 [52] Adapala 2016 [53] Grogan 2013 [54] Mann 2019 [55] |
| MMP10 | Matrix metallopeptidase 10 (stromelysin 2) | Involved in the breakdown of extracellular matrix in normal physiological processes, such as tissue remodeling. | Dehne 2010 [56] Gohring 2010 [57] |
| MMP12 | Matrix metallopeptidase 12 | Involved in the breakdown of extracellular matrix in normal physiological processes, such as tissue remodeling. | Dehne 2010 [56] Lv 2016 [58] |
| MMP9 | Matrix metallopeptidase 9 (gelatinase B, 92 kDa gelatinase, 92 kDa type IV collagenase) | Involved in the breakdown of extracellular matrix in normal physiological processes, such as tissue remodeling. | Challa 2010 [59] Yang 2015 [60] Miao 2004 [61] |
| SELP | Selectin P (granule membrane protein 140 kDa, antigen CD62) | This protein redistributes to the plasma membrane during platelet activation and degranulation. | Weeks 2012 [47] Gari 2016 [62] Rouillard 2016 [63] Bonn 2010 [64] |
| THBS3 | Thrombospondin 3 | Mediates cell-to-cell and cell-to-matrix interactions. Found in developing cartilage. | Vos 1992 [65] Adolph 1995 [66] Djouad 2007 [46] Posey 2008 [67] Hankenson 2005 [68] |
| VCAN | Versican | Major component of the ECM; involved in cell adhesion and proliferation during chondrogenesis. | Kamiya 2006 [69] Choocheep 2010 [70] Sztrolovics 2002 [71] |
| VTN | Vitronectin | ECM markers that promote cell adhesion and spreading. | Luo 2017 [72] Vieira 2015 [73] Pei 2013 [74] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stone, R.N.; Frahs, S.M.; Hardy, M.J.; Fujimoto, A.; Pu, X.; Keller-Peck, C.; Oxford, J.T. Decellularized Porcine Cartilage Scaffold; Validation of Decellularization and Evaluation of Biomarkers of Chondrogenesis. Int. J. Mol. Sci. 2021, 22, 6241. https://doi.org/10.3390/ijms22126241
Stone RN, Frahs SM, Hardy MJ, Fujimoto A, Pu X, Keller-Peck C, Oxford JT. Decellularized Porcine Cartilage Scaffold; Validation of Decellularization and Evaluation of Biomarkers of Chondrogenesis. International Journal of Molecular Sciences. 2021; 22(12):6241. https://doi.org/10.3390/ijms22126241
Chicago/Turabian StyleStone, Roxanne N., Stephanie M. Frahs, Makenna J. Hardy, Akina Fujimoto, Xinzhu Pu, Cynthia Keller-Peck, and Julia Thom Oxford. 2021. "Decellularized Porcine Cartilage Scaffold; Validation of Decellularization and Evaluation of Biomarkers of Chondrogenesis" International Journal of Molecular Sciences 22, no. 12: 6241. https://doi.org/10.3390/ijms22126241








