Dynamic “Molecular Portraits” of Biomembranes Drawn by Their Lateral Nanoscale Inhomogeneities
Abstract
:1. Introduction
2. Characteristics of Lateral Heterogeneities in Lipid Bilayers
2.1. Direct Experimental Observations of NDs
2.1.1. Model Lipid Membranes
2.1.2. NDs in Cell Membranes
2.1.3. Lessons of Studying NDs in Experiments
2.2. Computer Simulations
- (1)
- Computer simulations of model membranes clearly indicate the existence of NDs on the lipid bilayer surface. Moreover, such objects were first discovered “at the tip of the pen” (i.e., in silico), and only later observed in the direct experiments described above and others. The characteristic sizes and lifetimes of the calculated NDs are >1 nm and >1 ns, respectively, which is perfectly consistent with the results of observations. It is important that the computational data about the mechanisms of NDs formation and the influence of various factors on them (environmental conditions, etc.) are reproduced for the same systems using different calculation technologies: force fields, the level of approximations used (all-atom, united-atom, coarse-grained, etc.), computational protocols of sampling, and other modeling parameters. This indicates that the NDs are not an artifact caused by the choice of computational methods for obtaining data and processing them;
- (2)
- In computer models, it is possible to reproduce well the effect on the DMP of model lipid bilayers of such factors as temperature, the chemical nature of lipids in the membrane, a certain ionic composition, the role of the opposite monolayer, the presence of embedded “alien” objects with different parameters of mobility, etc.
- -
- Analyze in detail all types of interactions in the membrane, both at the level of individual molecules and the entire ensemble, and evaluate their contribution to the DMP characteristics. This allows determination of the balance of forces that cause the formation of the NDs with the observed properties in each particular case;
- -
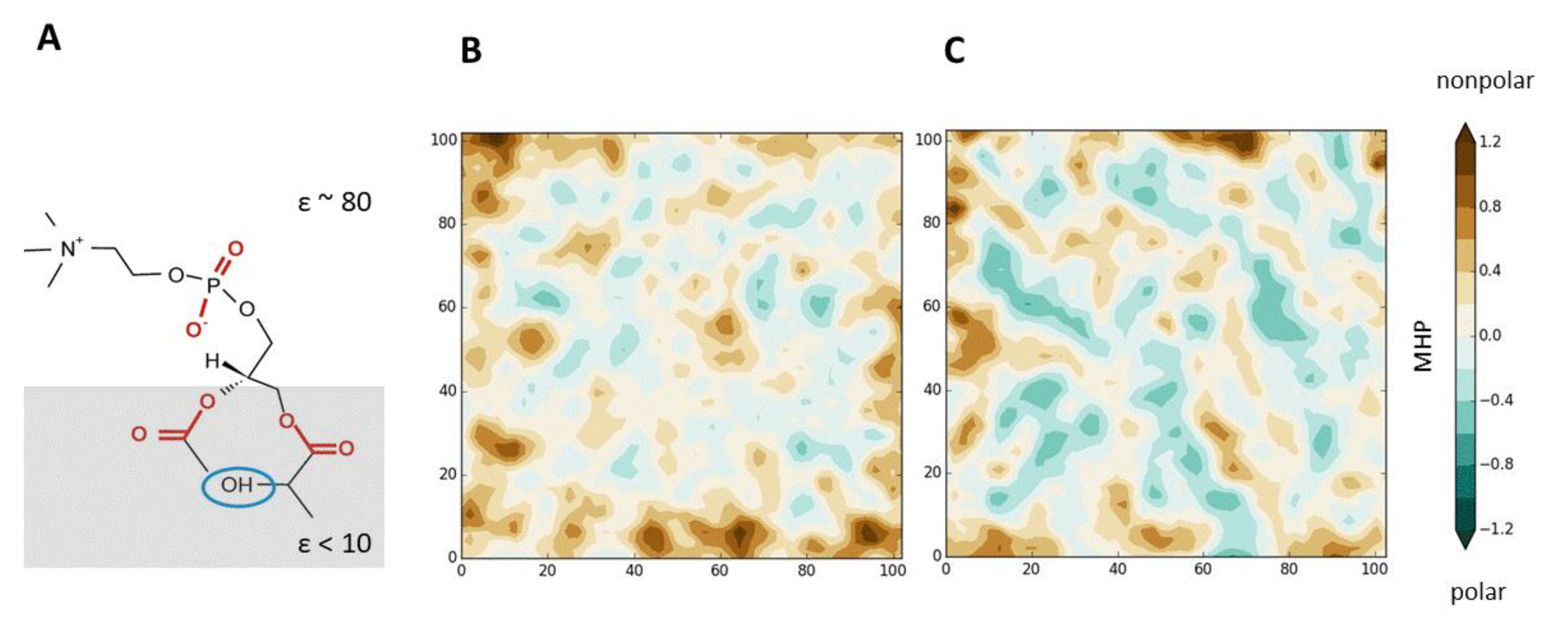
- The possibility of pictorial visualization of the DMPs. They can be presented both in the form of beautiful 3D images, and in the form of much more informative 2D maps of the membrane surface (e.g., [52,53]). In both cases, animation is widely used to represent the dynamics of these complex objects. It is important that the membrane system can be depicted as a whole, or the emphasis can be placed on the behavior of its individual components, for example, NDs, free volume regions, single molecules/groups. In addition, using numerical mapping methods, it is possible to graphically represent a wide variety of physico-chemical characteristics associated with NDs. These include: hydrophobic/hydrophilic and electrical properties (for example, molecular hydrophobicity potential (MHP) or electrostatic potential (EP), charge density, etc.); surface topography and its degree of hydration; mobility of molecules and their individual groups (in particular, calculated during MD); characteristics of various types of interactions involving membrane components (for example, the density of H-bonds, salt bridges, etc.). On such maps, it is useful to plot information about the location of specific atoms and molecules, the area of contact with external agents—for example, binding peptides, proteins, and so on. It is important that the presented surface properties can relate to individual states of the system, as well as to their average values, difference values, etc.;
- -
- Calculate with ultra-high spatiotemporal resolution the motion characteristics of all components of the membrane, up to individual atoms and groups, identify collective movements in the bilayer, quantify their contribution to the integral picture, and predict the influence of specified external factors on them;
- -
- In addition to the lateral NDs, to study such inhomogeneities along the normal to the membrane plane (the first attempts have already been made [54]). Together, the information obtained allows for the creation of an unprecedented detailed 3D dynamic picture of model cell membranes on a nanoscale;
- -
- Consider a wide range of systems in the calculations—from model hydrated bilayers consisting of one or more types of lipids to native-like membranes. Of course, the vast majority of results have so far been obtained for the first case, although it is not so much the less computational complexity of modeling such systems (this is, obviously, an important factor, but it is not critical). The key point is the need to constantly calibrate the results of calculations based on experimental data, and the latter are now available only for such simple systems. In addition, since detailed physical mechanisms for the formation and existence of domains of different scales (including NDs) have not yet been established, modeling of relatively simple systems allows much better control of the contribution of individual factors (for example, lipid composition, the presence of ions, etc.) to the observed phenomena. For the most complex multicomponent membranes, such an analysis is still impossible. The reason is the lack of reliable experimental structural data and other properties for them, on the basis of which it would be possible to carry out parameterization and verification of force fields and simulation protocols.
- -
- The most commonly used is the molecular dynamics (MD) method in its different implementations: classical and Langevin MD, targeted MD, steered MD, and so on. The Monte Carlo (MC) method is still used, although to a lesser extent than before. In addition, there are known examples of the joint application of the MD and MC methods. In relation to the problems of studying membrane DMPs, MD methods seem to be the best choice, since the dynamic aspects of NDs are extremely important —without taking them into account, it is impossible to identify the mechanisms of the phenomenon and make its complete atomistic picture (see below). The availability of several well-developed and constantly supported modeling programs (GROMACS [55], NAMD [56], CHARMM [57], etc.) and the open-source practice used in their implementation also played a major role in the popularity of MD, which allows users to actively implement their developments within these software products;
- -
- Adequate and consistent with both the experiment and the independent modeling studies, the results are obtained using at least several modern force fields: GROMOS96 (with “Berger lipids”) [58], CHARMM36 [59], SLipids [60], MARTINI [61], etc. It is important that these energy functions were specially adapted to the calculations of systems containing all the main components of the cell membrane—lipids, water, proteins, sugars, as well as physiologically significant ions. This allowed us (at least in the main) to get rid of the previously common problems of inconsistent joint application of force field parameters, for example, for lipids and proteins. In contrast to problems that consider only the integral parameters of the membrane, and, therefore, in many cases, are not too sensitive to such details, this question plays an extremely important role (sometimes critical) when it comes to analyzing the interactions of individual molecules, which leads to the formation/decay of NDs, their diffusion, etc.;
- -
- Speaking of force fields, it should also be noted that in the analysis of NDs, different levels of approximations are used: all-/united-atom and coarse-grained (CG) models, as well as, of course, their combinations. In contrast to “conventional” (i.e., non-nanoscale analysis) MD calculations, continuum models are much rarely used now, since individual molecules (water, lipids, and ions) play a key role in the processes under study. In some cases, modeling of complex systems (for example, a protein embedded in a membrane) begins with the use of continuous models of the membrane to find the starting states for further calculations in an explicit solvent;
- -
- Depending on the level of approximations, the duration of MD trajectories, which can currently be achieved on available high-performance clusters (including personal computers with additional GPU cards), reaches tens of microseconds for all-atom models and up to millisecond scale for CG models. In this case, as a rule, several independent MD runs are required for the same system.
3. Molecular Mechanisms of NDs Formation
3.1. Free Energy of Lipid–Lipid Interactions
3.2. Role of H-Bonds in Formation of NDs
3.3. Stochastic Fluctuations
3.4. Effects of Linactants
3.5. Diffusion, Collective Moves, and Free Volume Zones in Membranes
3.6. NDs and Interdigitation of Acyl Chains of Opposite Lipid Monolayers
4. Correspondence between Experimental and Computational Data
5. Representation of NDs via Mapping the Membrane Surface
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| CG | Coarse-grained |
| Chol | Cholesterol |
| DLiPC | Dilineoylphosphatidylcholine |
| DMP | Dynamic molecular portrait |
| DOPC | Dioleoylphosphatidylcholine |
| DOPC-oh | sn-1-β-hydroxy-dioleoylphosphatidylcholine |
| DOPS | Dioleoylphosphatidylserine |
| DPPC | Dipalmitoylphosphatidylcholine |
| DSPC | Distearoylphosphatidylcholine |
| ent-SSM | Enantiomer of stearoyl-sphingomyelin |
| EP | Electrostatic potential |
| ESR | Electron spin resonance |
| FRET | Förster resonance energy transfer |
| Ld | Liquid-disordered phase in lipid bilayer |
| Lo | Liquid-ordered phase in lipid bilayer |
| MC | Monte Carlo method |
| MD | Molecular dynamics |
| MHP | Molecular hydrophobicity potential |
| ND | Nanodomain |
| NMR | Nuclear magnetic resonance |
| PC | Phosphatidylcholine |
| POPC | Palmitoyloleoylphosphatidylcholine |
| SM | Sphingomyelin |
| SSM | Stearoyl-sphingomyelin |
References
- Gennis, R.B. Biomembranes: Molecular Structure and Function; Springer: Berlin/Heidelberg, Germany, 1988; p. 533. [Google Scholar]
- Jorgensen, K.; Mouritsen, O.G. Phase separation dynamics and lateral organization of two-component lipid membranes. Biophys. J. 1995, 95, 942–954. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.A.; London, E. Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 1998, 164, 103–114. [Google Scholar] [CrossRef]
- Lingwood, D.H.-J.; Kaiser, I.L.; Simons, K. Lipid rafts as functional heterogeneity in cell membranes. Biochem. Soc. Trans. 2009, 37, 955–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freire, E.; Snyder, B. Estimation of the lateral distribution of molecules in two-component lipid bilayers. Biochemistry 1980, 19, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Curatolo, W.; Sears, B.; Neuringer, L.J. A calorimetry and deuterium NMR study of mixed model membranes of 1-palmitoyl-2-oleylphosphatidylcholine and saturated phosphatidylcholines. Biochim. Biophys. Acta 1985, 817, 261–270. [Google Scholar] [CrossRef]
- Koromyslova, A.D.; Chugunov, A.O.; Efremov, R.G. Deciphering fine molecular details of proteins’ structure and function with a protein surface topography (PST) method. J. Chem. Inf. Mod. 2014, 54, 1189–1199. [Google Scholar] [CrossRef]
- Efremov, R.G.; Gulyaev, D.I.; Vergoten, G.; Modyanov, N.N. Application of 3D molecular hydrophobicity potential to the analysis of spatial organization of membrane domains in proteins: I. Hydrophobic properties of transmembrane segments of Na+, K+-ATPase. J. Protein Chem. 1992, 11, 665–675. [Google Scholar] [CrossRef]
- Engelman, D.M. Membranes are more mosaic than fluid. Nature 2005, 438, 578–580. [Google Scholar] [CrossRef]
- Cebecauer, M.; Amaro, M.; Jurkiewicz, P.; Sarmento, M.J.; Šachl, R.; Cwiklik, L.; Hof, M. Membrane lipid nanodomains. Chem. Rev. 2018, 118, 11259. [Google Scholar] [CrossRef]
- Enkavi, G.; Javanainen, M.; Kulig, W.; Róg, T.; Vattulainen, I. Multiscale simulations of biological membranes: The challenge to understand biological phenomena in a living substance. Chem. Rev. 2019, 119, 5607–5774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinnun, J.J.; Bolmatov, D.; Lavrentovich, M.O.; Katsaras, J. Lateral heterogeneity and domain formation in cellular membranes. Chem. Phys. Lipids 2020, 232, 104976. [Google Scholar] [CrossRef]
- Kure, J.L.; Andersen, C.A.; Mortensen, K.I.; Wiseman, P.W.; Arnspang, E.C. Revealing plasma membrane nano-domains with diffusion analysis methods. Membranes 2020, 10, 314. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.; Ursell, T.; Wiggins, P.; Sens, P. Emerging roles for lipids in shaping membrane-protein function. Nature 2009, 459, 379–385. [Google Scholar] [CrossRef] [Green Version]
- Bocharov, E.V.; Mineev, K.S.; Pavlov, K.V.; Akimov, S.A.; Kuznetsov, A.S.; Efremov, R.G.; Arseniev, A.S. Helix-helix interactions in membrane domains of bitopic proteins: Specificity and role of lipid environment. Biochim. Biophys. Acta 2017, 1859, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Vanni, S.; Hirose, H.; Barelli, H.; Antonny, B.; Gautier, R. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat. Commun. 2014, 5, 4916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Lindau, M. T-Snare transmembrane domain clustering modulates lipid organization and membrane curvature. J. Am. Chem. Soc. 2017, 139, 18440–18443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, F. Physical mechanisms of micro- and nanodomain formation in multicomponent lipid membranes. Biochim. Biophys. Acta 2017, 1859, 509–528. [Google Scholar] [CrossRef]
- Almeida, P.F.F. Thermodynamics of lipid interactions in complex bilayers. Biochim. Biophys. Acta 2009, 1788, 72–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spaar, A.; Saldittm, T. Short range order of hydrocarbon chains in fluid phospholipid bilayers studied by X-ray diffraction from highly oriented membranes. Biophys. J. 2003, 85, 1576–1584. [Google Scholar] [CrossRef] [Green Version]
- De Joannis, J.; Jiang, Y.; Yin, F.; Kindt, J.T. Equilibrium distributions of dipalmitoyl phosphatidylcholine and dilauroyl phosphatidylcholine in a mixed lipid bilayer: Atomistic semigrand canonical ensemble simulations. J. Phys. Chem. B 2006, 110, 25875–25882. [Google Scholar] [CrossRef] [PubMed]
- Dewa, T.; Vigmond, S.J.; Regen, S.L. Lateral heterogeneity in fluid bilayers composed of saturated and unsaturated phospholipids. J. Am. Chem. Soc. 1996, 118, 3435–3440. [Google Scholar] [CrossRef]
- Risselada, H.J.; Marrink, S.J. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA 2008, 105, 17367–17372. [Google Scholar] [CrossRef] [Green Version]
- Ubbelohde, A.R. Melting and Crystal Structure; Oxford University Press: Oxford, UK, 1965; p. 325. [Google Scholar]
- Davies, D.B.; Matheson, A.J. Influence of molecular rotation on some physical properties of liquids. Discuss. Faraday Soc. 1967, 43, 216–222. [Google Scholar] [CrossRef]
- Levine, Y.K. Physical studies of membrane structure. Progr. Biophys. Membr. Struct. 1972, 24, 1–74. [Google Scholar] [CrossRef]
- Lee, A.G.; Birdsall, N.J.M.; Metcalfe, J.C.; Toon, P.A.; Warren, G.B. Clusters in lipid bilayers and the interpretation of thermal effects in biological membranes. Biochemistry 1974, 13, 3699–3705. [Google Scholar] [CrossRef]
- Gordeliy, V.I.; Ivkov, V.G.; Ostanevich, Y.M.; Yaguzhinskij, L.S. Detection of structural defects in phosphatidylcholine membranes by small-angle neutron scattering. The cluster model of a lipid bilayer. Biochim. Biophys. Acta 1991, 1061, 39–48. [Google Scholar] [CrossRef]
- Niemela, P.S.; Ollila, S.; Hyvönen, M.T.; Karttunen, M.; Vattulainen, I. Assessing the nature of lipid raft membranes. PLoS Comput. Biol. 2007, 3, e34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petruzielo, R.S.; Heberle, F.A.; Feigenson, G.W. Phase behavior and domain size in sphingomyelin-containing lipid bilayers. Biochim. Biophys. Acta 2013, 1828, 1302–1313. [Google Scholar] [CrossRef]
- Pathak, P.; London, E. Measurement of lipid nanodomain (raft) formation and size in sphingomyelin/POPC/cholesterol vesicles shows TX-100 and transmembrane helices increase domain size by coalescing preexisting nanodomains but do not induce domain formation. Biophys. J. 2011, 101, 2417–2425. [Google Scholar] [CrossRef] [Green Version]
- DeWit, G.; Danial, J.S.H.; Wallace, M.I. Dynamic label-free imaging of lipid nanodomains. Proc. Natl. Acad. Sci. USA 2015, 112, 12299–12303. [Google Scholar] [CrossRef] [Green Version]
- Honigmann, A.; Mueller, V.; Eggeling, C. STED microscopy detects and quantifies liquid phase separation in lipid membranes using a new far-red emitting fluorescent phosphoglycerolipid analogue. Faraday Discuss. 2013, 161, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Yano, Y.; Hanashima, S.; Hiroshi, T.; Slotte, J.P.; London, E.; Murata, M. Sphingomyelins and ent-sphingomyelins form homophilic nano-subdomains within liquid ordered domains. Biophys. J. 2020, 119, 539–552. [Google Scholar] [CrossRef]
- Winkler, P.M.; Regmi, R.; García-Parajo, M.F. Transient nanoscopic phase separation in biological lipid membranes resolved by planar plasmonic antennas. ACS Nano 2017, 11, 7241–7250. [Google Scholar] [CrossRef] [PubMed]
- Enoki, T.A.; Heberle, F.A.; Feigenson, G.W. FRET detects the size of nanodomains for coexisting liquid-disordered and liquid-ordered phases. Biophys. J. 2018, 114, 1921–1935. [Google Scholar] [CrossRef] [Green Version]
- Yasuda, T.; Matsumori, N.; Murata, M. Formation of gel-like nanodomains in cholesterol-containing sphingomyelin or phosphatidylcholine binary membrane as examined by fluorescence lifetimes and 2H NMR spectra. Langmuir 2015, 31, 13783–13792. [Google Scholar] [CrossRef]
- Ando, J.; Kinoshita, M.; Sodeoka, M. Sphingomyelin distribution in lipid rafts of artificial monolayer membranes visualized by Raman microscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 4558–4563. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.M.; Lin, Y.H.; Hsieh, C.L. Nanoscopic substructures of raft-mimetic liquid-ordered membrane domains revealed by highspeed single-particle tracking. Sci. Rep. 2016, 6, 20542. [Google Scholar] [CrossRef] [Green Version]
- Pathak, P.; London, E. The effect of membrane lipid composition on the formation of lipid ultrananodomains. Biophys. J. 2015, 109, 1630–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eggeling, C.; Ringemann, C.; Medda, R.; Schwarzmann, G.; Sandhoff, K.; Polyakova, S.; Belov, V.N.; Hein, B.; von Middendorff, C.; Schönle, A.; et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 2009, 457, 1159–1162. [Google Scholar] [CrossRef]
- Regmi, R.; Winkler, P.M.; Flauraud, V.; Borgman, K.J.E.; Manzo, C.; Brugger, J.; Rigneault, H.; Wenger, J.; García-Parajo, M.F. Planar optical nanoantennas resolve cholesterol-dependent nanoscale heterogeneities in the plasma membrane of living cells. Nano Lett. 2017, 17, 6295–6302. [Google Scholar] [CrossRef] [Green Version]
- Gladstein, S.; Almassalha, L.M.; Cherkezyan, L.; Chandler, J.E.; Eshein, A.; Eid, A.; Zhang, D.; Wu, W.; Bauer, G.M.; Stephens, A.D.; et al. Multimodal interference-based imaging of nanoscale structure and macromolecular motion uncovers UV induced cellular paroxysm. Nat. Commun. 2019, 10, 1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nickels, J.D.; Chatterjee, S.; Stanley, C.B.; Qian, S.; Cheng, X.; Myles, D.A.A.; Standaert, R.F.; Elkins, J.G.; Katsaras, J. The in vivo structure of biological membranes and evidence for lipid domains. PLoS Biol. 2017, 15, e2002214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouritsen, O.G.; Boothroyd, A.; Harris, R.; Jan, N.; Lookman, T.; MacDonald, L.; Pink, D.A.; Zuckermann, M.J. Computer simulation of the main gel-fluid phase transition of lipid bilayers. J. Chem. Phys. 1983, 79, 2027–2041. [Google Scholar] [CrossRef]
- Jørgensen, K.; Sperotto, M.M.; Mouritsen, O.G.; Ipsen, J.H.; Zuckermann, M.J. Phase equilibria and local structure in binary lipid bilayers. Biochim. Biophys. Acta 1993, 1152, 135–145. [Google Scholar] [CrossRef]
- Sugár, I.P.; Thompson, T.E.; Biltonen, R.L. Monte Carlo simulation of two-component bilayers: DMPC/DSPC mixtures. Biophys. J. 1999, 76, 2099–2110. [Google Scholar] [CrossRef] [Green Version]
- Pandit, S.A.; Jakobsson, E.; Scott, H.L. Simulation of the early stages of nano-domain formation in mixed bilayers of sphingomyelin, cholesterol, and dioleylphosphatidylcholine. Biophys. J. 2004, 87, 3312–3322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polyansky, A.A.; Volynsky, P.E.; Arseniev, A.S.; Efremov, R.G. Adaptation of a membrane active peptide to heterogeneous environment: II. The role of mosaic nature of the membrane surface. J. Phys. Chem. B 2009, 113, 1120–1126. [Google Scholar] [CrossRef]
- Bennett, W.F.D.; Tieleman, D.P. Computer simulations of lipid membrane domains. Biochim. Biophys. Acta 2013, 1828, 1765–1776. [Google Scholar] [CrossRef] [Green Version]
- Marrink, S.J.; Corradi, V.; Souza, P.C.T.; Ingólfsson, H.I.; Tieleman, D.P.; Sansom, M.S.P. Computational modeling of realistic cell membranes. Chem. Rev. 2019, 119, 6184–6226. [Google Scholar] [CrossRef] [Green Version]
- Efremov, R.G.; Chugunov, A.O.; Pyrkov, T.V.; Priestle, J.P.; Arseniev, A.S.; Jacoby, E. Molecular lipophilicity in protein modeling and drug design. Curr. Med. Chem. 2007, 14, 393–415. [Google Scholar] [CrossRef]
- Gapsys, V.; de Groot, B.L.; Briones, R. Computational analysis of local membrane properties. J. Comput. Aid. Mol. Des. 2013, 27, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Pyrkova, D.V.; Tarasova, N.K.; Pyrkov, T.V.; Krylov, N.A.; Efremov, R.G. Atomic-scale lateral heterogeneity and dynamics of two-component lipid bilayers composed of saturated and unsaturated phosphatidylcholines. Soft Matter 2011, 7, 2569–2579. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]
- Brooks, B.R.; Brooks, C.L., III; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1615. [Google Scholar] [CrossRef] [PubMed]
- Berger, O.; Edholm, O.; Jähnig, F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997, 72, 2002–2013. [Google Scholar] [CrossRef] [Green Version]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [Green Version]
- Jämbeck, J.P.M.; Lyubartsev, A.P. Another piece of the membrane puzzle: Extending slipids further. J. Chem. Theory Comput. 2013, 9, 774–784. [Google Scholar] [CrossRef]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI forcefield: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef] [Green Version]
- Von Dreele, P.H. Estimation of lateral species separation from phase transitions in nonideal two-dimensional lipid mixtures. Biochemistry 1978, 17, 3939–3943. [Google Scholar] [CrossRef] [PubMed]
- Yano, Y.; Hanashima, S.; Murata, M. Sphingomyelin stereoisomers reveal that homophilic interactions cause nanodomain formation. Biophys. J. 2018, 115, 1530–1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estep, T.N.; Freire, E.; Thompson, T.E. Thermal behavior of stearoylsphingomyelin-cholesterol dispersions. Biochemistry 1981, 20, 7115–7118. [Google Scholar] [CrossRef]
- Boggs, J.M. Intermolecular hydrogen bonding between lipids: Influence on organization and function of lipids in membranes. Can. J. Biochem. 1986, 58, 755–770. [Google Scholar] [CrossRef]
- Nymeyer, H.; Zhou, H.-X. A method to determine dielectric constants in nonhomogeneous systems: Application to biological membranes. Biophys. J. 2008, 94, 1185–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mombelli, E.; Morris, R.; Taylor, W.; Fraternali, F. Hydrogen-bonding propensities of sphingomyelin in solution and in a bilayer assembly: A molecular dynamics study. Biophys. J. 2003, 84, 1507–1517. [Google Scholar] [CrossRef] [Green Version]
- Efremov, R.G. Dielectric-dependent strength of interlipid H-bonding in biomembranes: Model case study. J. Chem. Inf. Mod. 2019, 59, 2765–2775. [Google Scholar] [CrossRef]
- Pyrkova, D.V.; Tarasova, N.K.; Krylov, N.A.; Nolde, D.E.; Efremov, R.G. Lateral clustering of lipids in hydrated bilayers composed of dioleoylphosphatidylcholine and dipalmitoylphosphatidylcholine. Biochemistry 2011, 5, 278–285. [Google Scholar] [CrossRef]
- Matsumori, N.; Yamaguchi, T.; Murata, M. Orientation and order of the amide group of sphingomyelin in bilayers determined by solid-state NMR. Biophys. J. 2015, 108, 2816–2824. [Google Scholar] [CrossRef] [Green Version]
- Yasuda, T.; Kinoshita, M.; Matsumori, N. Detailed comparison of deuterium quadrupole profiles between sphingomyelin and phosphatidylcholine bilayers. Biophys. J. 2014, 106, 631–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyrkova, D.V.; Tarasova, N.K.; Krylov, N.A.; Nolde, D.E.; Pentkovsky, V.M.; Efremov, R.G. Dynamic clustering of lipids in hydrated two-component membranes: Results of computer modeling and putative biological impact. J. Biomol. Struct. Dyn. 2013, 31, 87–95. [Google Scholar] [CrossRef]
- Björkbom, A.; Róg, T.; Kankaanpää, P.; Lindroos, D.; Kaszuba, K.; Kurita, M.; Yamaguchi, S.; Yamamoto, T.; Jaikishan, S.; Paavolainen, L.; et al. N- and O-methylation of sphingomyelin markedly affects its membrane properties and interactions with cholesterol. Biochim. Biophys. Acta 2011, 1808, 1179–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efremov, R.G.; Pyrkova, D.V.; Krylov, N.A. Fine tuning of microscopic properties in two-component zwitterionic-anionic lipid bilayers: Determinant role of H-bonding. Biophys. J. 2018, 114, 601A. [Google Scholar] [CrossRef]
- Honerkamp-Smith, A.R.; Veatch, S.L.; Keller, S.L. An introduction to critical points for biophysicists; Observations of compositional heterogeneity in lipid membranes. Biochim. Biophys. Acta 2009, 1788, 53–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veatch, S.L.; Cicuta, P.; Sengupta, P.; Honerkamp-Smith, A.; Holowka, D.; Baird, B. Critical fluctuations in plasma membrane vesicles. ACS Chem. Biol. 2008, 3, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Veatch, S.L.; Soubias, O.; Keller, S.L.; Gawrisch, K. Critical fluctuations in domain-forming lipid mixtures. Proc. Natl. Acad. Sci. USA 2007, 104, 17650–17655. [Google Scholar] [CrossRef] [Green Version]
- Fischer, T.; Risselada, H.J.; Vink, R.L.C. Membrane lateral structure: The influence of immobilized particles on domain size. Phys. Chem. Chem. Phys. 2012, 14, 14500. [Google Scholar] [CrossRef] [Green Version]
- Palmieri, B.; Grant, M.; Safran, S.A. Prediction of the dependence of the line tension on the composition of linactants and the temperature in phase separated membranes. Langmuir 2014, 30, 11734–11745. [Google Scholar] [CrossRef]
- Heberle, F.A.; Doktorova, M.; Goh, S.L.; Standaert, R.F.; Katsaras, J.; Feigenson, G.W. Hybrid and nonhybrid lipids exert common effects on membrane raft size and morphology. J. Am. Chem. Soc. 2013, 135, 14932–14935. [Google Scholar] [CrossRef] [PubMed]
- Falck, E.; Róg, T.; Karttunen, M.; Vattulainen, I. Lateral diffusion in lipid membranes through collective flows. J. Am. Chem. Soc. 2008, 130, 44–45. [Google Scholar] [CrossRef]
- Apajalahti, T.; Niemelä, P.; Govindan, P.N.; Miettinen, M.S.; Salonen, E.; Marrink, S.-J.; Vattulainen, I. Concerted diffusion of lipids in raft-like membranes. Faraday Discuss. 2010, 144, 411–430. [Google Scholar] [CrossRef]
- Bolmatov, D.; Cai, Y.O.; Zav’yalov, D.; Zhernenkov, M. Crossover from picosecond collective to single particle dynamics defines the mechanism of lateral lipid diffusion. Biochim. Biophys. Acta 2018, 1860, 2446–2455. [Google Scholar] [CrossRef] [PubMed]
- Metzler, R.; Jeon, J.-H.; Cherstvy, A.G. Non-Brownian diffusion in lipid membranes: Experiments and simulations. Biochim. Biophys. Acta 2016, 1858, 2451–2467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubovskii, P.V.; Efremov, R.G. The role of hydrophobic/hydrophilic balance in the activity of structurally flexible vs. rigid cytolytic polypeptides and analogues developed on their basis. Expert Rev. Proteom. 2018, 15, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Konshina, A.G.; Dubovskii, P.V.; Efremov, R.G. Stepwise insertion of cobra cardiotoxin CT2 into a lipid bilayer occurs as an interplay of protein and membrane “dynamic molecular portraits”. J. Chem. Inf. Mod. 2020, 61, 385–399. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Efremov, R.G. Dynamic “Molecular Portraits” of Biomembranes Drawn by Their Lateral Nanoscale Inhomogeneities. Int. J. Mol. Sci. 2021, 22, 6250. https://doi.org/10.3390/ijms22126250
Efremov RG. Dynamic “Molecular Portraits” of Biomembranes Drawn by Their Lateral Nanoscale Inhomogeneities. International Journal of Molecular Sciences. 2021; 22(12):6250. https://doi.org/10.3390/ijms22126250
Chicago/Turabian StyleEfremov, Roman G. 2021. "Dynamic “Molecular Portraits” of Biomembranes Drawn by Their Lateral Nanoscale Inhomogeneities" International Journal of Molecular Sciences 22, no. 12: 6250. https://doi.org/10.3390/ijms22126250




