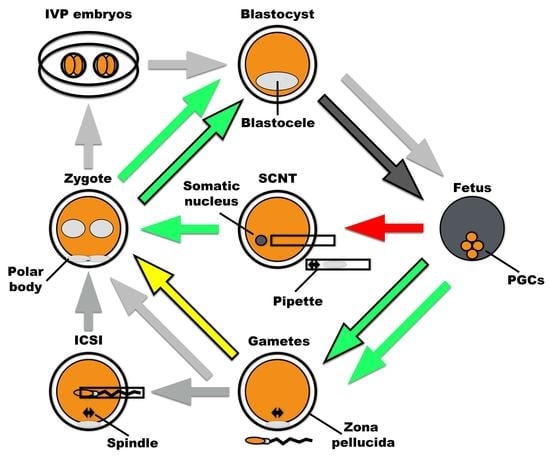
Contextualizing Autophagy during Gametogenesis and Preimplantation Embryonic Development
Abstract
:1. Introduction
2. The Genetic Basis of the Autophagy Pathway
3. Autophagy during Oogenesis
4. Autophagy during Spermatogenesis
5. Autophagy during Fertilization
6. Autophagy during Preimplantation Development
7. Future Directions
8. Concluding Remarks
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Glossary
| 3MA | 3-methyladenine |
| ARTs | assisted reproductive technologies |
| ATG | autophagy-related genes |
| EGA | embryonic genome activation |
| ER | endoplasmic reticulum |
| ICM | inner cell mass |
| ICSI | intracytoplasmic sperm injection |
| IVM | in vitro maturation |
| SCNT | somatic cell nuclear transfer |
| TE | Trophectoderm |
| TFs | transcription factors |
References
- Wassarman, P.M.; Jovine, L.; Litscher, E.S. A profile of fertilization in mammals. Nat. Cell Biol. 2001, 3, E59–E64. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.P.; Florman, H.M. The state of the union: The cell biology of fertilization. Nat. Cell Biol. 2002, 4, S57–S63. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Liu, K. Maternal control of mouse preimplantation development. Results Probl. Cell Differ. 2012, 55, 115–139. [Google Scholar] [CrossRef] [PubMed]
- Frankenberg, S.R.; de Barros, F.R.; Rossant, J.; Renfree, M.B. The mammalian blastocyst. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 210–232. [Google Scholar] [CrossRef] [PubMed]
- Memili, E.; First, N.L. Zygotic and embryonic gene expression in cow: A review of timing and mechanisms of early gene expression as compared with other species. Zygote 2000, 8, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Schier, A.F. The maternal-zygotic transition: Death and birth of RNAs. Science 2007, 316, 406–407. [Google Scholar] [CrossRef] [PubMed]
- Jukam, D.; Shariati, S.A.M.; Skotheim, J.M. Zygotic Genome Activation in Vertebrates. Dev. Cell 2017, 42, 316–332. [Google Scholar] [CrossRef]
- Rossant, J.; Tam, P.P.L. New insights into early human development: Lessons for stem cell derivation and differentiation. Cell Stem Cell 2017, 20, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Madeja, Z.E.; Pawlak, P.; Piliszek, A. Beyond the mouse: Non-rodent animal models for study of early mammalian development and biomedical research. Int. J. Dev. Biol. 2019, 63, 187–201. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.; Tee, W.W. Committing the primordial germ cell: An updated molecular perspective. Wiley Interdiscip. Rev. Syst. Biol. Med. 2019, 11, e1436. [Google Scholar] [CrossRef] [Green Version]
- Lockshin, R.A.; Zakeri, Z. Caspase-independent cell death? Oncogene 2004, 23, 2766–2773. [Google Scholar] [CrossRef] [Green Version]
- Tam, P.P.; Loebel, D.A. Gene function in mouse embryogenesis: Get set for gastrulation. Nat. Rev. Genet. 2007, 8, 368–381. [Google Scholar] [CrossRef]
- Peng, G.; Jing, N. The genome-wide molecular regulation of mouse gastrulation embryo. Sci. China Life Sci. 2017, 60, 363–369. [Google Scholar] [CrossRef]
- Pampfer, S. Apoptosis in rodent peri-implantation embryos: Differential susceptibility of inner cell mass and trophectoderm cell lineages—A review. Placenta 2000, 21 (Suppl. A), S3–S10. [Google Scholar] [CrossRef]
- Qu, X.; Zou, Z.; Sun, Q.; Luby-Phelps, K.; Cheng, P.; Hogan, R.N.; Gilpin, C.; Levine, B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 2007, 128, 931–946. [Google Scholar] [CrossRef] [Green Version]
- Penaloza, C.; Lin, L.; Lockshin, R.A.; Zakeri, Z. Cell death in development: Shaping the embryo. Histochem. Cell Biol. 2006, 126, 149–158. [Google Scholar] [CrossRef]
- Penaloza, C.; Orlanski, S.; Ye, Y.; Entezari-Zaher, T.; Javdan, M.; Zakeri, Z. Cell death in mammalian development. Curr. Pharm. Des. 2008, 14, 184–189. [Google Scholar] [CrossRef]
- Green, D.R.; Llambi, F. Cell Death Signaling. Cold Spring Harb. Perspect. Biol. 2015, 7, a006080. [Google Scholar] [CrossRef]
- Kutscher, L.M.; Shaham, S. Non-apoptotic cell death in animal development. Cell Death Differ. 2017, 24, 1326–1336. [Google Scholar] [CrossRef]
- Harper, J.W.; Elledge, S.J. The DNA damage response: Ten years after. Mol. Cell 2007, 28, 739–745. [Google Scholar] [CrossRef]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chantranupong, L.; Wolfson, R.L.; Sabatini, D.M. Nutrient-sensing mechanisms across evolution. Cell 2015, 161, 67–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efeyan, A.; Comb, W.C.; Sabatini, D.M. Nutrient-sensing mechanisms and pathways. Nature 2015, 517, 302–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef]
- Mizushima, N. Autophagy in protein and organelle turnover. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 397–402. [Google Scholar] [CrossRef] [Green Version]
- Ravanan, P.; Srikumar, I.F.; Talwar, P. Autophagy: The spotlight for cellular stress responses. Life Sci. 2017, 188, 53–67. [Google Scholar] [CrossRef]
- Morishita, H.; Mizushima, N. Diverse Cellular Roles of Autophagy. Annu. Rev. Cell Dev. Biol. 2019, 35, 453–475. [Google Scholar] [CrossRef]
- Zachari, M.; Ganley, I.G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017, 61, 585–596. [Google Scholar] [CrossRef] [Green Version]
- Tooze, S.A.; Yoshimori, T. The origin of the autophagosomal membrane. Nat. Cell Biol. 2010, 12, 831–835. [Google Scholar] [CrossRef]
- Kroemer, G.; Levine, B. Autophagic cell death: The story of a misnomer. Nat. Rev. Mol. Cell Biol. 2008, 9, 1004–1010. [Google Scholar] [CrossRef]
- Kasprowska-Liśkiewicz, D. The cell on the edge of life and death: Crosstalk between autophagy and apoptosis. Postepy Hig. Med. Dosw. 2017, 71, 825–841. [Google Scholar] [CrossRef]
- Okamoto, K. Organellophagy: Eliminating cellular building blocks via selective autophagy. J. Cell Biol. 2014, 205, 435–445. [Google Scholar] [CrossRef]
- Mancias, J.D.; Kimmelman, A.C. Mechanisms of Selective Autophagy in Normal Physiology and Cancer. J. Mol. Biol. 2016, 428, 1659–1680. [Google Scholar] [CrossRef] [Green Version]
- Condello, M.; Pellegrini, E.; Caraglia, M.; Meschini, S. Targeting Autophagy to Overcome Human Diseases. Int. J. Mol. Sci. 2019, 20, 725. [Google Scholar] [CrossRef] [Green Version]
- White, E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer. 2012, 12, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef]
- Sakamaki, J.I.; Long, J.S.; New, M.; Van Acker, T.; Tooze, S.A.; Ryan, K.M. Emerging roles of transcriptional programs in autophagy regulation. Transcription 2018, 9, 131–136. [Google Scholar] [CrossRef] [Green Version]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef] [Green Version]
- Kuma, A.; Komatsu, M.; Mizushima, N. Autophagy-monitoring and autophagy-deficient mice. Autophagy 2017, 13, 1619–1628. [Google Scholar] [CrossRef] [Green Version]
- Delorme-Axford, E.; Klionsky, D.J. Transcriptional and post-transcriptional regulation of autophagy in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2018, 293, 5396–5403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, Z.; Jin, S.; Yang, C.; Levine, A.J.; Heintz, N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA 2003, 100, 15077–15082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuma, A.; Hatano, M.; Matsui, M.; Yamamoto, A.; Nakaya, H.; Yoshimori, T.; Ohsumi, Y.; Tokuhisa, T.; Mizushima, N. The role of autophagy during the early neonatal starvation period. Nature 2004, 432, 1032–1036. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Kuma, A.; Murakami, M.; Kishi, C.; Yamamoto, A.; Mizushima, N. Autophagy is essential for preimplantation development of mouse embryos. Science 2008, 321, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.H.; Yu, H.Y.; Wang, P.; Mao, G.K.; Liu, W.X.; Li, M.N.; Wang, H.N.; Shang, Y.L.; Liu, C.; Xu, Z.L.; et al. Germ cell-specific Atg7 knockout results in primary ovarian insufficiency in female mice. Cell Death Dis. 2015, 6, e1589. [Google Scholar] [CrossRef] [Green Version]
- Behrends, C.; Sowa, M.E.; Gygi, S.P.; Harper, J.W. Network organization of the human autophagy system. Nature 2010, 466, 68–76. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, M.M.; Zheng, B.; Lu, T.; Yan, Z.; Py, B.F.; Ng, A.; Xavier, R.J.; Li, C.; Yankner, B.A.; Scherzer, C.R.; et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2010, 107, 14164–14169. [Google Scholar] [CrossRef] [Green Version]
- Jacomin, A.C.; Gul, L.; Sudhakar, P.; Korcsmaros, T.; Nezis, I.P. What we learned from big data for autophagy research. Front. Cell Dev. Biol. 2018, 6, 92. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J.; Kumar, A. A systems biology approach to learning autophagy. Autophagy 2006, 2, 12–23. [Google Scholar] [CrossRef] [Green Version]
- Ng, A.C. Integrative systems biology and networks in autophagy. Semin. Immunopathol. 2010, 32, 355–361. [Google Scholar] [CrossRef]
- Till, A.; Saito, R.; Merkurjev, D.; Liu, J.J.; Syed, G.H.; Kolnik, M.; Siddiqui, A.; Glas, M.; Scheffler, B.; Ideker, T.; et al. Evolutionary trends and functional anatomy of the human expanded autophagy network. Autophagy 2015, 11, 1652–1667. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J. The autophagy connection. Dev. Cell 2010, 19, 11–12. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, M.M. Towards the global understanding of the autophagy regulatory network. Autophagy 2010, 6, 1218–1220. [Google Scholar] [CrossRef] [Green Version]
- Broz, D.K.; Mello, S.S.; Bieging, K.T.; Jiang, D.; Dusek, R.L.; Brady, C.A.; Sidow, A.; Attardi, L.D. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013, 27, 1016–1031. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.H.; Li, Y.; Deng, S.L.; Liu, Y.X.; Lian, Z.X.; Yu, K. Recent research advances in mitosis during mammalian gametogenesis. Cells 2019, 8, 567. [Google Scholar] [CrossRef] [Green Version]
- Kierszenbaum, A.L. Cell-cycle regulation and mammalian gametogenesis: A lesson from the unexpected. Mol. Reprod. Dev. 2006, 73, 939–942. [Google Scholar] [CrossRef]
- Larose, H.; Shami, A.N.; Abbott, H.; Manske, G.; Lei, L.; Hammoud, S.S. Gametogenesis: A journey from inception to conception. Curr. Top. Dev. Biol. 2019, 132, 257–310. [Google Scholar] [CrossRef]
- Grive, K.J. Pathways coordinating oocyte attrition and abundance during mammalian ovarian reserve establishment. Mol. Reprod. Dev. 2020, 87, 843–856. [Google Scholar] [CrossRef]
- You, S.Y.; Park, Y.S.; Jeon, H.J.; Cho, D.H.; Jeon, H.B.; Kim, S.H.; Chang, J.W.; Kim, J.S.; Oh, J.S. Beclin-1 knockdown shows abscission failure but not autophagy defect during oocyte meiotic maturation. Cell Cycle 2016, 15, 1611–1619. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.H.; Jin, Y.X.; Liang, S.; Kwon, J.W.; Zhu, J.W.; Lei, L.; Kim, N.H. Autophagy is required for proper meiosis of porcine oocytes maturing in vitro. Sci. Rep. 2018, 8, 12581. [Google Scholar] [CrossRef]
- Latorraca, L.B.; Feitosa, W.B.; Mariano, C.; Moura, M.T.; Fontes, P.K.; Nogueira, M.F.G.; Paula-Lopes, F.F. Autophagy is a pro-survival adaptive response to heat shock in bovine cumulus-oocyte complexes. Sci. Rep. 2020, 10, 13711. [Google Scholar] [CrossRef]
- Lin, F.H.; Zhang, W.L.; Li, H.; Tian, X.D.; Zhang, J.; Li, X.; Li, C.Y.; Tan, J.H. Role of autophagy in modulating post-maturation aging of mouse oocytes. Cell Death Dis. 2018, 9, 308. [Google Scholar] [CrossRef] [Green Version]
- Moura, M.T.; Paula-Lopes, F.F. Thermoprotective molecules to improve oocyte competence under elevated temperature. Theriogenology 2020, 156, 262–271. [Google Scholar] [CrossRef]
- Bang, S.; Shin, H.; Song, H.; Suh, C.S.; Lim, H.J. Autophagic activation in vitrified-warmed mouse oocytes. Reproduction 2014, 148, 11–19. [Google Scholar] [CrossRef]
- Gao, H.H.; Li, J.T.; Liu, J.J.; Yang, Q.A.; Zhang, J.M. Autophagy inhibition of immature oocytes during vitrification-warming and in vitro mature activates apoptosis via caspase-9 and -12 pathway. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017, 217, 89–93. [Google Scholar] [CrossRef]
- Bang, S.; Lee, G.K.; Shin, H.; Suh, C.S.; Lim, H.J. Vitrification, in vitro fertilization, and development of Atg7 deficient mouse oocytes. Clin. Exp. Reprod. Med. 2016, 43, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, G.K.; Shin, H.; Lim, H.J. Rapamycin Influences the Efficiency of In vitro Fertilization and Development in the Mouse: A Role for Autophagic Activation. Asian-Australas. J. Anim. Sci. 2016, 29, 1102–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Yin, Q.; Wei, D.; Yang, Z.; Du, Y.; Ma, Y. Autophagy in male reproduction. Syst. Biol. Reprod. Med. 2019, 65, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wan, H.; Li, X.; Liu, W.; Chen, Q.; Wang, Y.; Yang, L.; Tang, H.; Zhang, X.; Duan, E.; et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014, 24, 852–869. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Wang, H.; Jia, P.; Zhao, H.; Liu, C.; Liu, W.; Song, Z.; Xu, Z.; Yang, L.; Wang, Y.; et al. Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy 2016, 12, 1575–1592. [Google Scholar] [CrossRef]
- Liu, C.; Wang, H.; Shang, Y.; Liu, W.; Song, Z.; Zhao, H.; Wang, L.; Jia, P.; Gao, F.; Xu, Z.; et al. Autophagy is required for ectoplasmic specialization assembly in sertoli cells. Autophagy 2016, 12, 814–832. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Li, G.; Liu, C.; Gao, H.; Wang, H.; Liu, W.; Chen, M.; Shang, Y.; Wang, L.; Shi, J.; et al. Autophagy regulates testosterone synthesis by facilitating cholesterol uptake in Leydig cells. J. Cell Biol. 2018, 217, 2103–2119. [Google Scholar] [CrossRef] [Green Version]
- Mancilla, H.; Maldonado, R.; Cereceda, K.; Villarroel-Espíndola, F.; de Oca, M.M.; Angulo, C.; Castro, M.A.; Slebe, J.C.; Vera, J.C.; Lavandero, S.; et al. Glutathione depletion induces spermatogonial cell autophagy. J. Cell. Biochem. 2015, 116, 2283–2292. [Google Scholar] [CrossRef]
- Zhang, M.; Jiang, M.; Bi, Y.; Zhu, H.; Zhou, Z.; Sha, J. Autophagy and apoptosis act as partners to induce germ cell death after heat stress in mice. PLoS ONE 2012, 7, e41412. [Google Scholar] [CrossRef] [Green Version]
- Duan, P.; Hu, C.; Quan, C.; Yu, T.; Huang, W.; Chen, W.; Tang, S.; Shi, Y.; Martin, F.L.; Yang, K. 4-Nonylphenol induces autophagy and attenuates mTOR-p70S6K/4EBP1 signaling by modulating AMPK activation in Sertoli cells. Toxicol. Lett. 2017, 267, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Luo, X.; Zhu, Y.; Zhao, L.; Li, L.; Peng, Q.; Ma, M.; Gao, Y. ATM signals to AMPK to promote autophagy and positively regulate DNA damage in response to cadmium-induced ROS in mouse spermatocytes. Environ. Pollut. 2017, 231, 1560–1568. [Google Scholar] [CrossRef]
- Bolaños, J.M.G.; Morán, Á.M.; da Silva, C.M.B.; Rodríguez, A.M.; Dávila, M.P.; Aparicio, I.M.; Tapia, J.A.; Ferrusola, C.O.; Peña, F.J. Autophagy and apoptosis have a role in the survival or death of stallion spermatozoa during conservation in refrigeration. PLoS ONE 2012, 7, e30688. [Google Scholar] [CrossRef] [Green Version]
- Kanninen, T.T.; de Andrade Ramos, B.R.; Witkin, S.S. The role of autophagy in reproduction from gametogenesis to parturition. Eur. J. Obstet. Gynecol. Reprod. Biol. 2013, 171, 3–8. [Google Scholar] [CrossRef]
- Aparicio, I.M.; Espino, J.; Bejarano, I.; Gallardo-Soler, A.; Campo, M.L.; Salido, G.M.; Pariente, J.A.; Peña, F.J.; Tapia, J.A. Autophagy-related proteins are functionally active in human spermatozoa and may be involved in the regulation of cell survival and motility. Sci. Rep. 2016, 6, 33647. [Google Scholar] [CrossRef]
- Aparicio, I.M.; Muñoz, P.M.; Salido, G.M.; Peña, F.J.; Tapia, J.A. The autophagy-related protein LC3 is processed in stallion spermatozoa during short-and long-term storage and the related stressful conditions. Animal 2016, 10, 1182–1191. [Google Scholar] [CrossRef]
- Sato, M.; Sato, K.; Tomura, K.; Kosako, H.; Sato, K. The autophagy receptor ALLO-1 and the IKKE-1 kinase control clearance of paternal mitochondria in Caenorhabditis elegans. Nat. Cell Biol. 2018, 20, 81–91. [Google Scholar] [CrossRef]
- Sato, M.; Sato, K. Maternal inheritance of mitochondrial DNA: Degradation of paternal mitochondria by allogeneic organelle autophagy, allophagy. Autophagy 2012, 8, 424–425. [Google Scholar] [CrossRef] [Green Version]
- Song, W.H.; Yi, Y.J.; Sutovsky, M.; Meyers, S.; Sutovsky, P. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc. Natl. Acad. Sci. USA 2016, 113, E5261–E5270. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.M.; Ge, Z.J.; Wang, Z.W.; Jiang, Z.Z.; Wang, Z.B.; Ouyang, Y.C.; Hou, Y.; Schatten, H.; Sun, Q.Y. Unique insights into maternal mitochondrial inheritance in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 13038–13043. [Google Scholar] [CrossRef] [Green Version]
- Tsukamoto, S.; Tatsumi, T. Degradation of maternal factors during preimplantation embryonic development. J. Reprod. Dev. 2018, 64, 217–222. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.X.; Zheng, Z.; Yu, X.F.; Zhang, J.B.; Namgoong, S.; Cui, X.S.; Hyun, S.H.; Kim, N.H. Autophagy and ubiquitin-mediated proteolysis may not be involved in the degradation of spermatozoon mitochondria in mouse and porcine early embryos. Zygote 2016, 24, 31–41. [Google Scholar] [CrossRef]
- de Melo, K.P.; Camargo, M. Mechanisms for sperm mitochondrial removal in embryos. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118916. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Hwang, K.C.; Sun, S.C.; Xu, Y.N.; Kim, N.H. Modulation of autophagy influences development and apoptosis in mouse embryos developing in vitro. Mol. Reprod. Dev. 2011, 78, 498–509. [Google Scholar] [CrossRef] [PubMed]
- Song, B.S.; Yoon, S.B.; Kim, J.S.; Sim, B.W.; Kim, Y.H.; Cha, J.J.; Choi, S.A.; Min, H.K.; Lee, Y.; Huh, J.W.; et al. Induction of autophagy promotes preattachment development of bovine embryos by reducing endoplasmic reticulum stress. Biol. Reprod. 2012, 87, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.N.; Shen, X.H.; Lee, S.E.; Kwon, J.S.; Kim, D.J.; Heo, Y.T.; Cui, X.S.; Kim, N.H. Autophagy influences maternal mRNA degradation and apoptosis in porcine parthenotes developing in vitro. J. Reprod. Dev. 2012, 58, 576–584. [Google Scholar] [CrossRef] [Green Version]
- Adastra, K.L.; Chi, M.M.; Riley, J.K.; Moley, K.H. A differential autophagic response to hyperglycemia in the developing murine embryo. Reproduction 2011, 141, 607–615. [Google Scholar] [CrossRef] [Green Version]
- Wyman, A.; Pinto, A.B.; Sheridan, R.; Moley, K.H. One-cell zygote transfer from diabetic to nondiabetic mouse results in congenital malformations and growth retardation in offspring. Endocrinology 2008, 149, 466–469. [Google Scholar] [CrossRef]
- Lee, J.; Park, J.I.; Yun, J.I.; Lee, Y.; Yong, H.; Lee, S.T.; Park, C.K.; Hyun, S.H.; Lee, G.S.; Lee, E. Rapamycin treatment during in vitro maturation of oocytes improves embryonic development after parthenogenesis and somatic cell nuclear transfer in pigs. J. Vet. Sci. 2015, 16, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Zhang, N.; Wang, Z.; Bai, G.; Zheng, Z.; Gu, Y.; Wu, Y.; Liu, H.; Zhou, D.; Lei, L. Induction of autophagy improves embryo viability in cloned mouse embryos. Sci. Rep. 2015, 5, 17829. [Google Scholar] [CrossRef] [Green Version]
- Chi, D.; Zeng, Y.; Xu, M.; Si, L.; Qu, X.; Liu, H.; Li, J. LC3-dependent autophagy in Pig 2-cell cloned embryos could influence the degradation of maternal mRNA and the regulation of epigenetic modification. Cell Reprogram. 2017, 19, 354–362. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Han, Z.; Fang, J.; Chen, H.; Guo, Z. Transcriptome analyses reveal effects of vitamin C-treated donor cells on cloned bovine embryo development. Int. J. Mol. Sci. 2019, 20, 2628. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Hiradate, Y.; Hoshino, Y.; Tanemura, K.; Sato, E. Quantitative analysis in LC3-II protein in vitro maturation of porcine oocyte. Zygote 2014, 22, 404–410. [Google Scholar] [CrossRef]
- Park, M.R.; Gupta, M.K.; Lee, H.R.; Das, Z.C.; Uhm, S.J.; Lee, H.T. Possible involvement of Class III phosphatidylinositol-3-kinase in meiotic progression of porcine oocytes beyond germinal vesicle stage. Theriogenology 2011, 75, 940–950. [Google Scholar] [CrossRef]
- Song, B.S.; Kim, J.S.; Kim, Y.H.; Sim, B.W.; Yoon, S.B.; Cha, J.J.; Choi, S.A.; Yang, H.J.; Mun, S.E.; Park, Y.H.; et al. Induction of autophagy during in vitro maturation improves the nuclear and cytoplasmic maturation of porcine oocytes. Reprod. Fertil. Dev. 2014, 26, 974–981. [Google Scholar] [CrossRef]
- Gaytán, M.; Morales, C.; Sánchez-Criado, J.E.; Gaytán, F. Immunolocalization of beclin 1, a bcl-2-binding, autophagy-related protein, in the human ovary: Possible relation to life span of corpus luteum. Cell Tissue Res. 2008, 331, 509–517. [Google Scholar] [CrossRef]
- Choi, J.Y.; Jo, M.W.; Lee, E.Y.; Yoon, B.K.; Choi, D.S. The role of autophagy in follicular development and atresia in rat granulosa cells. Fertil. Steril. 2010, 93, 2532–2537. [Google Scholar] [CrossRef]
- Gawriluk, T.R.; Hale, A.N.; Flaws, J.A.; Dillon, C.P.; Green, D.R.; Rucker, E.B., 3rd. Autophagy is a cell survival program for female germ cells in the murine ovary. Reproduction 2011, 141, 759–765. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Hwang, S.Y.; Min, K.S.; Yoon, J.T. Molecular cloning and expression analyses of porcine MAP1LC3A in the granulosa cells of normal and miniature pig. Reprod. Biol. Endocrinol. 2013, 11, 8. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.E.; Kim, E.Y.; Choi, H.Y.; Moon, J.J.; Park, M.J.; Lee, J.B.; Jeong, C.J.; Park, S.P. Rapamycin rescues the poor developmental capacity of aged porcine oocytes. Asian-Australas. J. Anim. Sci. 2014, 27, 635–647. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, M.; Kawahara-Miki, R.; Kawana, H.; Shirasuna, K.; Kuwayama, T.; Iwata, H. Resveratrol-induced mitochondrial synthesis and autophagy in oocytes derived from early antral follicles of aged cows. J. Reprod. Dev. 2015, 61, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.E.; Sun, S.C.; Choi, H.Y.; Uhm, S.J.; Kim, N.H. mTOR is required for asymmetric division through small GTPases in mouse oocytes. Mol. Reprod. Dev. 2012, 79, 356–366. [Google Scholar] [CrossRef]
- Schell, C.; Kretz, O.; Liang, W.; Kiefer, B.; Schneider, S.; Sellung, D.; Bork, T.; Leiber, C.; Rüegg, M.A.; Mallidis, C.; et al. The rapamycin-sensitive complex of mammalian target of rapamycin is essential to maintain male fertility. Am. J. Pathol. 2016, 186, 324–336. [Google Scholar] [CrossRef] [Green Version]
- Horibe, A.; Eid, N.; Ito, Y.; Otsuki, Y.; Kondo, Y. Ethanol-induced autophagy in sertoli cells is specifically marked at androgen-dependent stages of the spermatogenic cycle: Potential mechanisms and implications. Int. J. Mol. Sci. 2019, 20, 184. [Google Scholar] [CrossRef] [Green Version]
- Rojansky, R.; Cha, M.Y.; Chan, D.C. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife 2016, 5, e17896. [Google Scholar] [CrossRef]
- Liu, S.; Sun, Y.; Li, Z. Resveratrol protects Leydig cells from nicotine-induced oxidative damage through enhanced autophagy. Clin. Exp. Pharmacol. Physiol. 2018, 45, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, K.; Ling, X.; Wang, Z.; Zou, P.; Wang, X.; Gao, J.; Yin, L.; Zhang, X.; Liu, J.; et al. DBP-induced endoplasmic reticulum stress in male germ cells causes autophagy, which has a cytoprotective role against apoptosis in vitro and in vivo. Toxicol. Lett. 2016, 245, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Bustamante-Marín, X.; Quiroga, C.; Lavandero, S.; Reyes, J.G.; Moreno, R.D. Apoptosis, necrosis and autophagy are influenced by metabolic energy sources in cultured rat spermatocytes. Apoptosis 2012, 17, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Yamaguchi, K. Epigenetic modifications and reprogramming in paternal pronucleus: Sperm, preimplantation embryo, and beyond. Cell. Mol. Life Sci. 2017, 74, 1957–1967. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Kuma, A.; Mizushima, N. The role of autophagy during the oocyte-to-embryo transition. Autophagy 2008, 4, 1076–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukamoto, S.; Hara, T.; Yamamoto, A.; Ohta, Y.; Wada, A.; Ishida, Y.; Kito, S.; Nishikawa, T.; Minami, N.; Sato, K.; et al. Functional analysis of lysosomes during mouse preimplantation embryo development. J. Reprod. Dev. 2013, 59, 33–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, A.; Mizushima, N.; Tsukamoto, S. Fertilization-induced autophagy in mouse embryos is independent of mTORC1. Biol. Reprod. 2014, 91, 7. [Google Scholar] [CrossRef]
- Xu, Y.N.; Cui, X.S.; Sun, S.C.; Lee, S.E.; Li, Y.H.; Kwon, J.S.; Lee, S.H.; Hwang, K.C.; Kim, N.H. Mitochondrial dysfunction influences apoptosis and autophagy in porcine parthenotes developing in vitro. J. Reprod. Dev. 2011, 57, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.J.; Nie, Z.W.; Shin, K.T.; Zhou, W.; Cui, X.S. PINK1 regulates mitochondrial morphology via promoting mitochondrial fission in porcine preimplantation embryos. FASEB J. 2019, 33, 7882–7895. [Google Scholar] [CrossRef]
- Lee, H.R.; Gupta, M.K.; Kim, D.H.; Hwang, J.H.; Kwon, B.; Lee, H.T. Poly(ADP-ribosyl)ation is involved in pro-survival autophagy in porcine blastocysts. Mol. Reprod. Dev. 2016, 83, 37–49. [Google Scholar] [CrossRef]
- Kim, D.H.; Lee, H.R.; Kim, M.G.; Lee, J.S.; Jin, S.J.; Lee, H.T. The effect of poly(ADP-ribosyl)ation inhibition on the porcine cumulus-oocyte complex during in vitro maturation. Biochem. Biophys. Res. Commun. 2017, 483, 752–758. [Google Scholar] [CrossRef]
- Ng, F.; Tang, B.L. Sirtuins’ modulation of autophagy. J. Cell. Physiol. 2013, 228, 2262–2270. [Google Scholar] [CrossRef]
- Tatsumi, T.; Takayama, K.; Ishii, S.; Yamamoto, A.; Hara, T.; Minami, N.; Miyasaka, N.; Kubota, T.; Matsuura, A.; Itakura, E.; et al. Forced lipophagy reveals that lipid droplets are required for early embryonic development in mouse. Development 2018, 145, dev161893. [Google Scholar] [CrossRef] [Green Version]
- Cooper, A.S.R.; Boehle, K.; Riley, J.; Moley, K. Autophagy is a physiologic process regulated by glucose availability in the murine preimplantation blastocyst. Reprod. Sci. 2008, 15, 45. [Google Scholar]
- Ptak, G.; Zacchini, F.; Czernik, M.; Fidanza, A.; Palmieri, C.; Della Salda, L.; Scapolo, P.A.; Loi, P. A short exposure to polychlorinated biphenyls deregulates cellular autophagy in mammalian blastocyst in vitro. Hum. Reprod. 2012, 27, 1034–1042. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, A.; Aoki, A.; Kusabiraki, T.; Shima, T.; Yoshino, O.; Cheng, S.B.; Sharma, S.; Saito, S. Role of autophagy in oocytogenesis, embryogenesis, implantation, and pathophysiology of pre-eclampsia. J. Obstet. Gynaecol. Res. 2017, 43, 633–643. [Google Scholar] [CrossRef] [Green Version]
- Kawai, K.; Harada, T.; Ishikawa, T.; Sugiyama, R.; Kawamura, T.; Yoshida, A.; Tsutsumi, O.; Ishino, F.; Kubota, T.; Kohda, T. Parental age and gene expression profiles in individual human blastocysts. Sci. Rep. 2018, 8, 2380. [Google Scholar] [CrossRef] [Green Version]
- Xue, Z.; Huang, K.; Cai, C.; Cai, L.; Jiang, C.Y.; Feng, Y.; Liu, Z.; Zeng, Q.; Cheng, L.; Sun, Y.E.; et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013, 500, 593–597. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.G.; Heo, Y.T.; Lee, S.E.; Jang, W.I.; Min, S.G.; Uhm, S.J.; Kim, N.H. A new modified cut standard straw vitrification technique reduces the apoptosis of mouse blastocysts and generates more live mouse offspring. Cryo-Letters 2013, 34, 598–607. [Google Scholar]
- Gurdon, J.B.; Melton, D.A. Nuclear reprogramming in cells. Science 2008, 322, 1811–1815. [Google Scholar] [CrossRef] [Green Version]
- Moura, M.T.; Badaraco, J.; Sousa, R.V.; Lucci, C.M.; Rumpf, R. Improved functional oocyte enucleation by actinomycin D for bovine somatic cell nuclear transfer. Reprod. Fertil. Dev. 2019, 31, 1321–1329. [Google Scholar] [CrossRef]
- Keefer, C.L. Artificial cloning of domestic animals. Proc. Natl. Acad. Sci. USA 2015, 112, 8874–8878. [Google Scholar] [CrossRef] [Green Version]
- Jeong, P.S.; Sim, B.W.; Park, S.H.; Kim, M.J.; Kang, H.G.; Nanjidsuren, T.; Lee, S.; Song, B.S.; Koo, D.B.; Kim, S.U. Chaetocin improves pig cloning efficiency by enhancing epigenetic reprogramming and autophagic activity. Int. J. Mol. Sci. 2020, 21, 4836. [Google Scholar] [CrossRef]
- Elahi, F.; Lee, H.; Lee, J.; Lee, S.T.; Park, C.K.; Hyun, S.H.; Lee, E. Effect of rapamycin treatment during post-activation and/or in vitro culture on embryonic development after parthenogenesis and in vitro fertilization in pigs. Reprod. Domest. Anim. 2017, 52, 741–748. [Google Scholar] [CrossRef]
- Li, H.; Song, M.; Yang, W.; Cao, P.; Zheng, L.; Zuo, Y. A comparative analysis of single-cell transcriptome identifies reprogramming driver factors for efficiency improvement. Mol. Ther. Nucleic Acids 2020, 19, 1053–1064. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T. How to interpret LC3 immunoblotting. Autophagy 2007, 3, 542–545. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Hara, T.; Yamamoto, A.; Kito, S.; Minami, N.; Kubota, T.; Sato, K.; Kokubo, T. Fluorescence-based visualization of autophagic activity predicts mouse embryo viability. Sci. Rep. 2014, 4, 4533. [Google Scholar] [CrossRef] [Green Version]

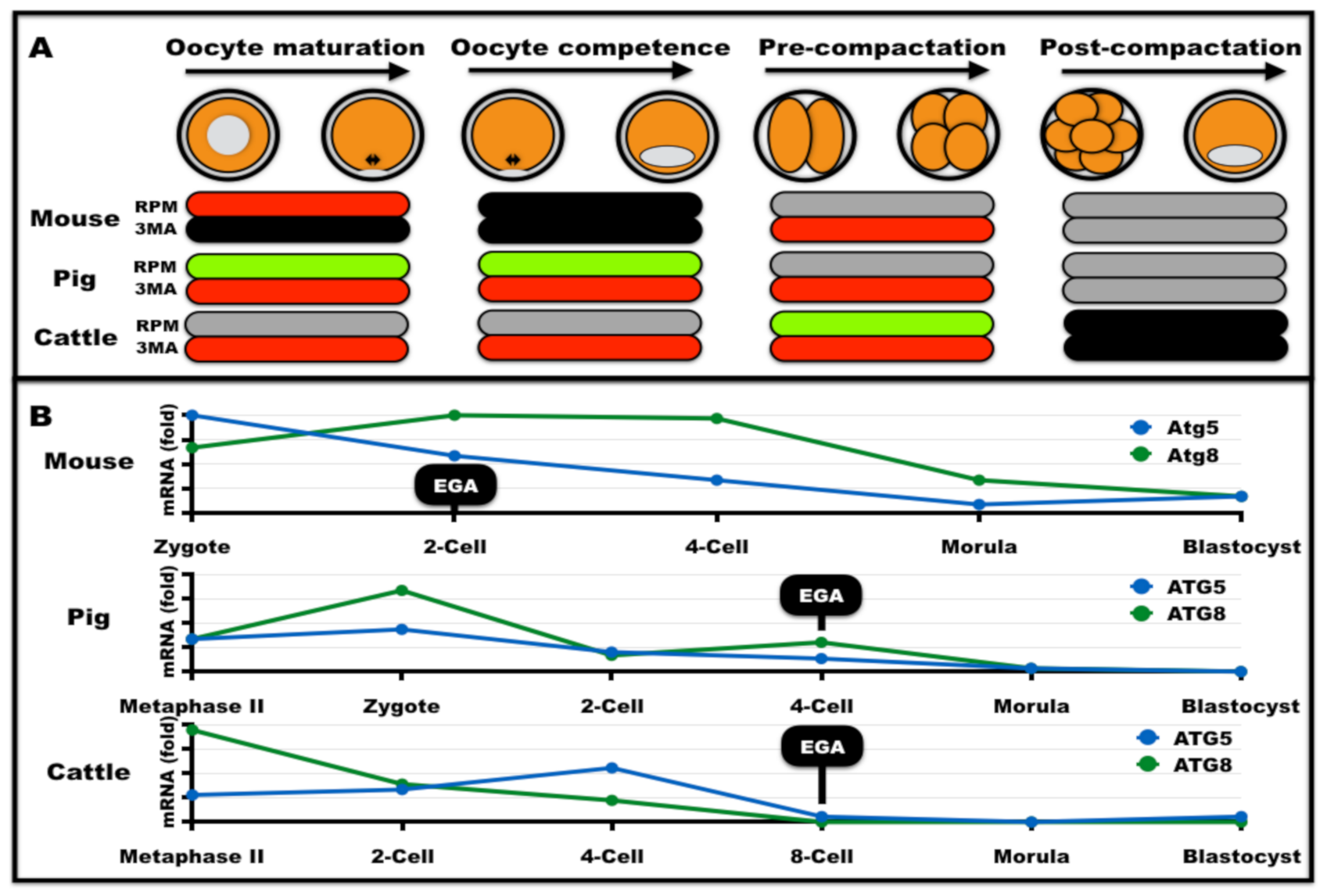

| Event | Physiological Role | Stress | ARTs |
|---|---|---|---|
| Oogenesis | Required for mouse oogenesis (except for oocyte maturation) [46,60] and for bovine and pig oocyte maturation [61,62] | Activated during oocyte aging [63] and environmental stress (e.g., heat stress) [62,64] | Contributes to oocyte survival after vitrification [65,66,67] but rapamycin was detrimental to developmental competence after warming [68] |
| Spermatogenesis | Required for testosterone production, sperm and acrosome formation, motility, and fertility [69,70,71,72,73] | Activated during environmental stress [74,75,76,77] | Activated by sperm cooling and freezing [78,79]. Autophagy inhibition improved sperm viability [80,81] |
| Fertilization | Ongoing debate if it recycles sperm-borne mitochondria [82,83,84,85,86,87,88] | - | - |
| Preimplantation embryonic development | Required for post-compactation embryogenesis in the mouse [45,89], cattle [90], and pigs [91] | Activated under environmental stress [92,93] | Decreased or delayed activation in SCNT embryos [94,95,96]. Autophagy inducers increased SCNT reprogramming to blastocysts in mice [95], pigs [94,96], and cattle [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moura, M.T.; Latorraca, L.B.; Paula-Lopes, F.F. Contextualizing Autophagy during Gametogenesis and Preimplantation Embryonic Development. Int. J. Mol. Sci. 2021, 22, 6313. https://doi.org/10.3390/ijms22126313
Moura MT, Latorraca LB, Paula-Lopes FF. Contextualizing Autophagy during Gametogenesis and Preimplantation Embryonic Development. International Journal of Molecular Sciences. 2021; 22(12):6313. https://doi.org/10.3390/ijms22126313
Chicago/Turabian StyleMoura, Marcelo T., Laís B. Latorraca, and Fabíola F. Paula-Lopes. 2021. "Contextualizing Autophagy during Gametogenesis and Preimplantation Embryonic Development" International Journal of Molecular Sciences 22, no. 12: 6313. https://doi.org/10.3390/ijms22126313
APA StyleMoura, M. T., Latorraca, L. B., & Paula-Lopes, F. F. (2021). Contextualizing Autophagy during Gametogenesis and Preimplantation Embryonic Development. International Journal of Molecular Sciences, 22(12), 6313. https://doi.org/10.3390/ijms22126313