Preclinical Development of FA5, a Novel AMP-Activated Protein Kinase (AMPK) Activator as an Innovative Drug for the Management of Bowel Inflammation
Abstract
1. Introduction
2. Results
2.1. Western Blot Analysis
2.2. Protein Phosphorylation in Protein Extracts from C2C12 Cells
2.3. Gene Ontology and Pathways Analysis
2.4. In Vivo Experiments
2.4.1. Body and Spleen Weight
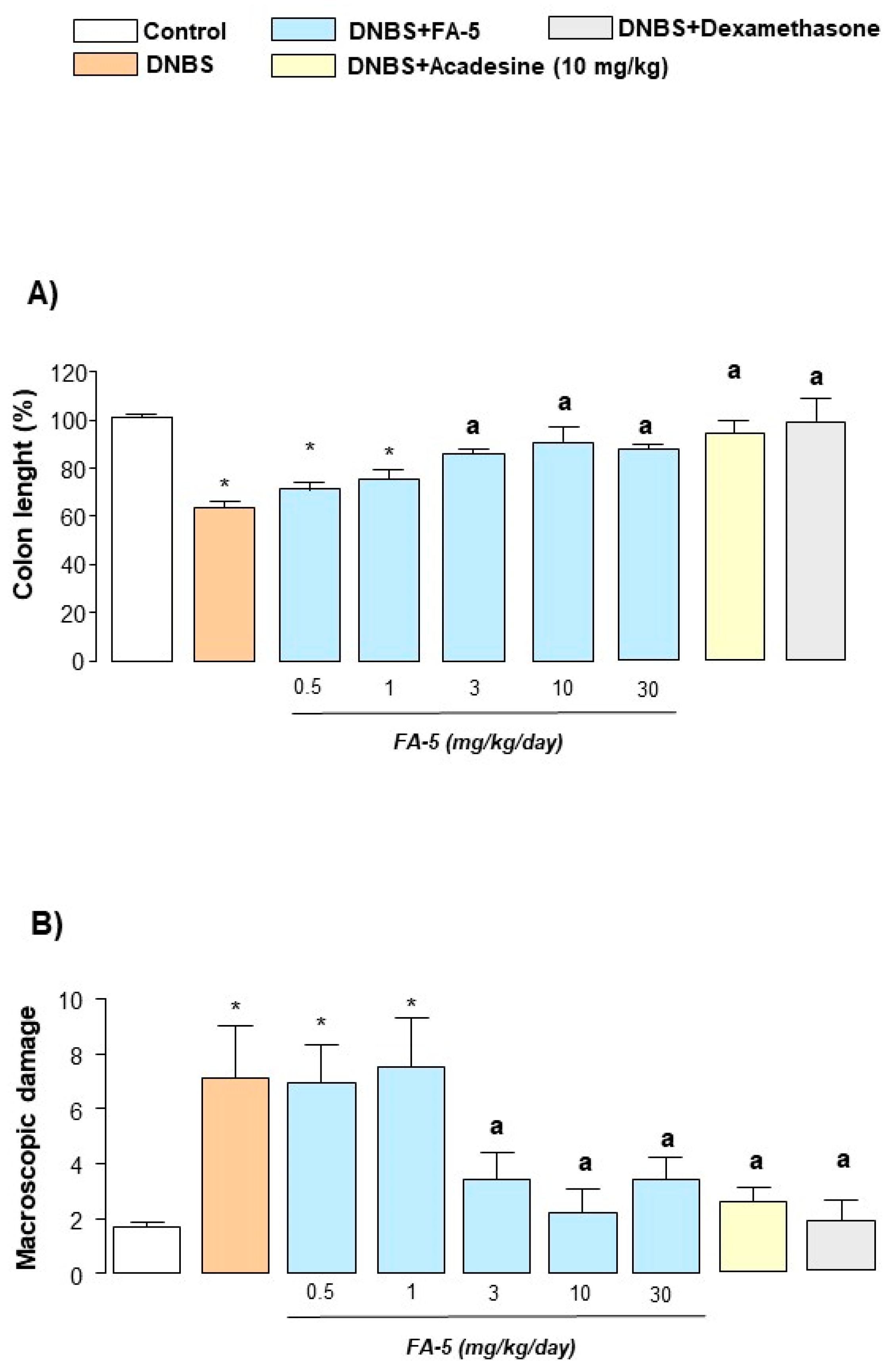
2.4.2. Colonic Length and Macroscopic Damage
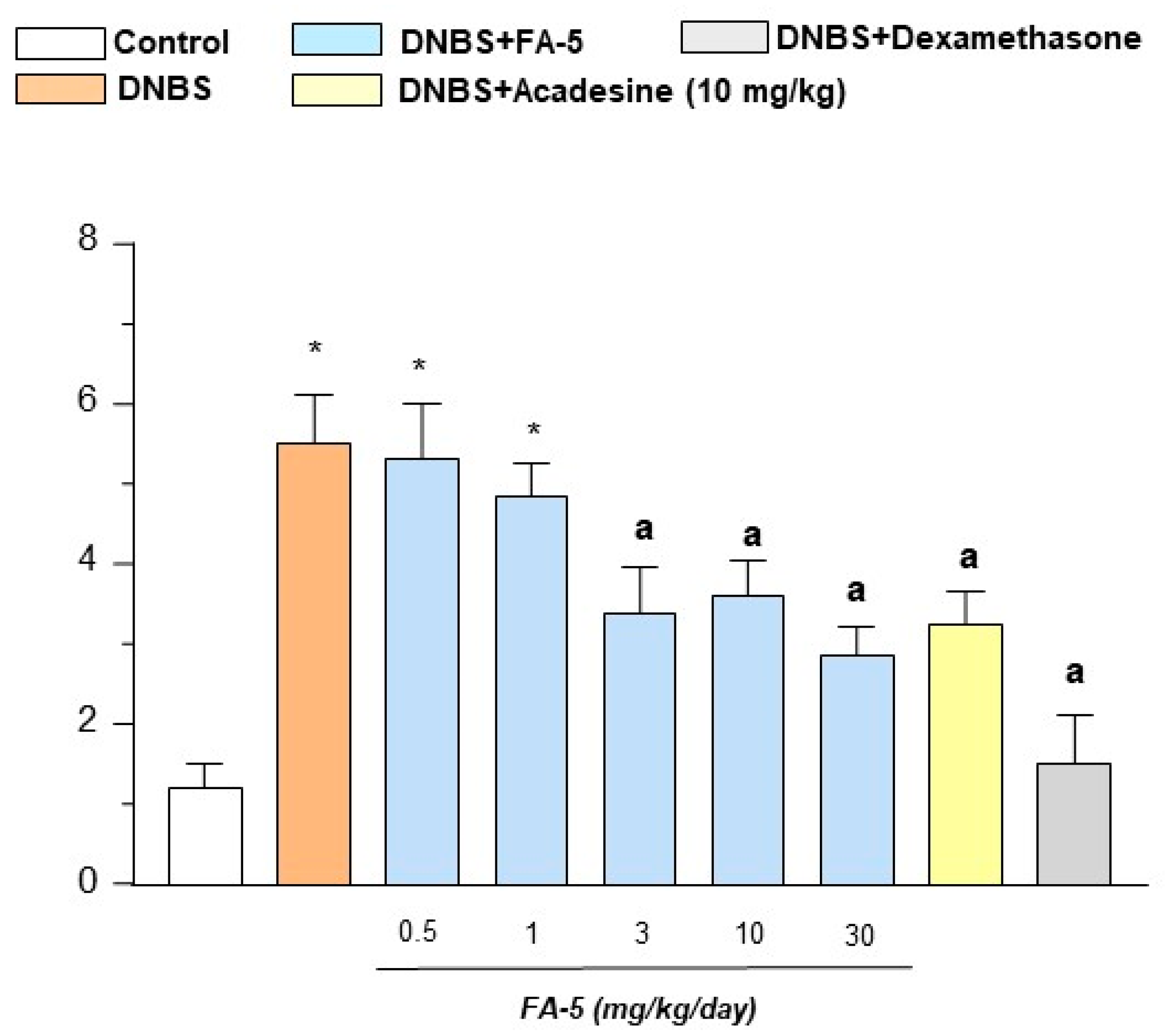
2.4.3. Histological Analysis
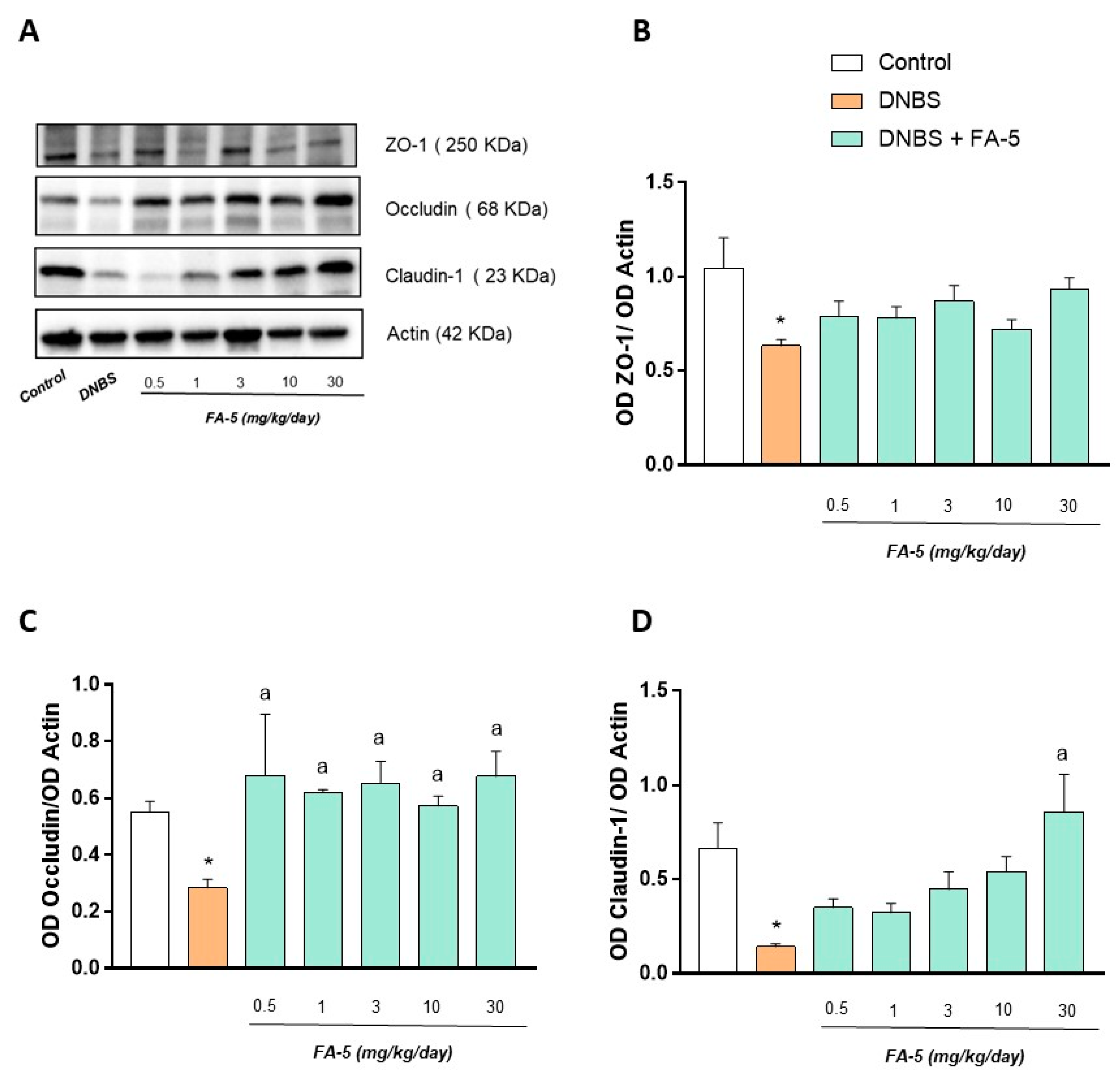
2.4.4. Western Blot Analysis
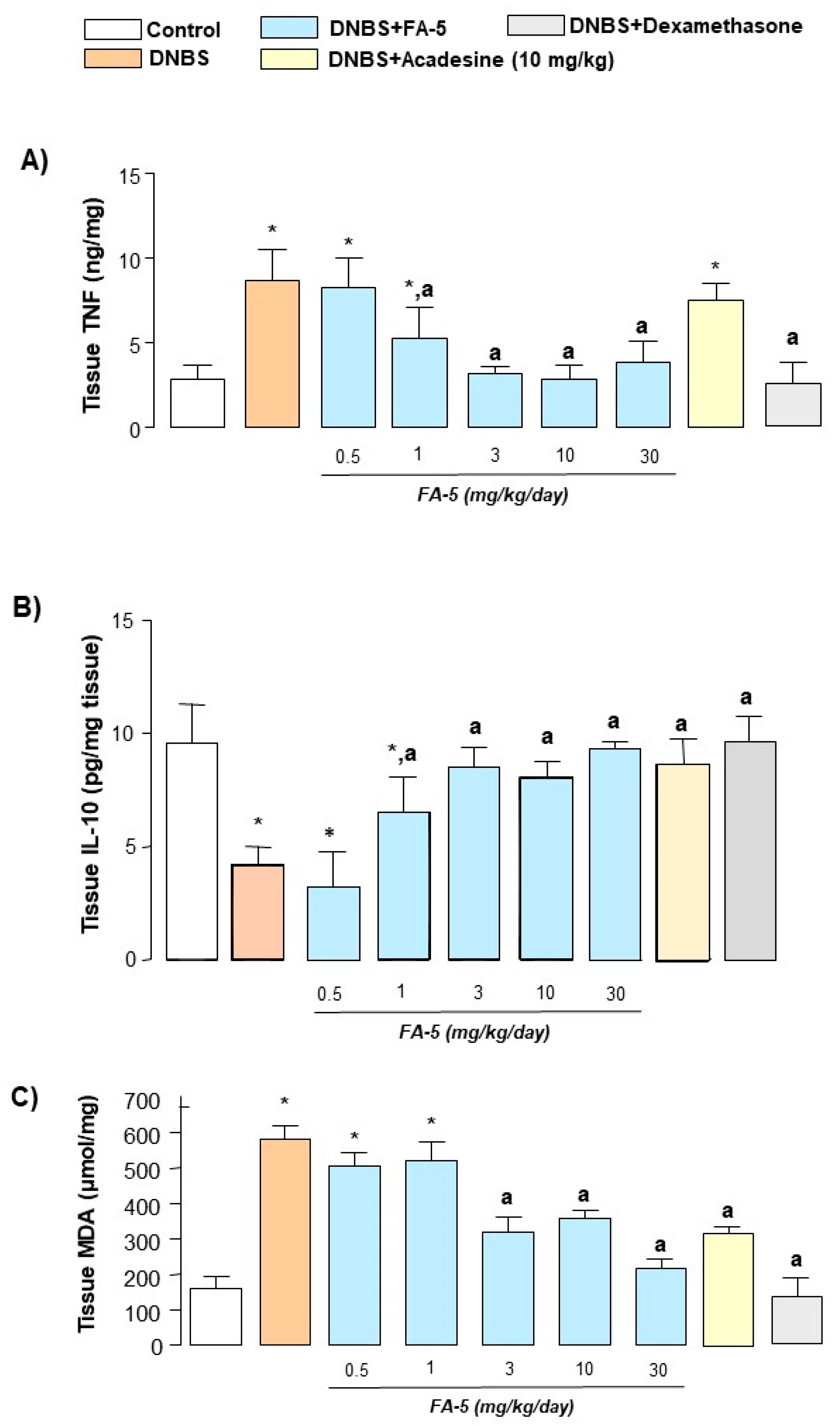
2.4.5. Tissue TNF and IL-10 Levels
2.4.6. MDA Assay
3. Discussion
4. Materials and Methods
4.1. Synthesis of 3-Amino-5-phenylbenzofuran-2-carboxamide, FA5
4.2. In Vitro Experiments
4.2.1. Cell Culture and Protein Extraction
4.2.2. Cytotoxicity Assay
4.2.3. Western Blot Analysis of AMPK Phosphorylation in Differentiated Mouse C2C12 Skeletal Myoblasts
4.2.4. Sirt1 Assay
4.2.5. Two-Dimensional Electrophoresis and Western Blot Analysis
4.2.6. Spot Digestion and Protein Identification
4.2.7. Ingenuity Pathway Analysis and Panther Analysis
4.3. In Vivo Experiments
4.3.1. Animals
4.3.2. Induction of Colitis and Drug Treatments
4.3.3. Assessment of Colitis
4.3.4. Histologic Damage Score
4.3.5. Western Blot Analysis
4.3.6. Cytokine Assays
4.3.7. Evaluation of Tissue Malondialdehyde
4.4. Drugs and Reagents
4.5. Proteomic Data Analysis
4.6. Statistical Analysis
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ahluwalia, B.; Moraes, L.; Magnusson, M.K.; Ohman, L. Immunopathogenesis of inflammatory bowel disease and mechanisms of biological therapies. Scand. J. Gastroenterol. 2018, 53, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Vancamelbeke, M.; Vanuytsel, T.; Farre, R.; Verstockt, S.; Ferrante, M.; Van Assche, G.; Rutgeerts, P.; Schuit, F.; Vermeire, S.; Arijs, I.; et al. Genetic and transcriptomic bases of intestinal epithelial barrier dysfunction in inflammatory bowel disease. Inflamm. Bowel Dis. 2017, 23, 1718–1729. [Google Scholar] [CrossRef]
- Kominsky, D.J.; Campbell, E.L.; Colgan, S.P. Metabolic shifts in immunity and inflammation. J. Immunol. 2010, 184, 4062–4068. [Google Scholar] [CrossRef] [PubMed]
- Vyas, S.P.; Goswami, R. A decade of th9 cells: Role of th9 cells in inflammatory bowel disease. Front. Immunol. 2018, 9, 1139. [Google Scholar] [CrossRef]
- Brand, S. Crohn’s disease: Th1, th17 or both? The change of a paradigm: New immunological and genetic insights implicate th17 cells in the pathogenesis of crohn’s disease. Gut 2009, 58, 1152–1167. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, Z. Inflammatory bowel disease related innate immunity and adaptive immunity. Am. J. Transl. Res. 2016, 8, 2490–2497. [Google Scholar] [PubMed]
- Pagnini, C.; Pizarro, T.T.; Cominelli, F. Novel pharmacological therapy in inflammatory bowel diseases: Beyond anti-tumor necrosis factor. Front. Pharmacol. 2019, 10, 671. [Google Scholar] [CrossRef]
- Bettencourt, I.A.; Powell, J.D. Targeting metabolism as a novel therapeutic approach to autoimmunity, inflammation, and transplantation. J. Immunol. 2017, 198, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Ma, E.H.; Poffenberger, M.C.; Wong, A.H.; Jones, R.G. The role of ampk in t cell metabolism and function. Curr. Opin. Immunol. 2017, 46, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Antonioli, L.; Colucci, R.; Pellegrini, C.; Giustarini, G.; Sacco, D.; Tirotta, E.; Caputi, V.; Marsilio, I.; Giron, M.C.; Nemeth, Z.H.; et al. The ampk enzyme-complex: From the regulation of cellular energy homeostasis to a possible new molecular target in the management of chronic inflammatory disorders. Expert Opin. Ther. Targets 2016, 20, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Di Fusco, D.; Dinallo, V.; Monteleone, I.; Laudisi, F.; Marafini, I.; Franze, E.; Di Grazia, A.; Dwairi, R.; Colantoni, A.; Ortenzi, A.; et al. Metformin inhibits inflammatory signals in the gut by controlling ampk and p38 map kinase activation. Clin. Sci. 2018, 132, 1155–1168. [Google Scholar] [CrossRef]
- Bai, A.; Yong, M.; Ma, A.G.; Ma, Y.; Weiss, C.R.; Guan, Q.; Bernstein, C.N.; Peng, Z. Novel anti-inflammatory action of 5-aminoimidazole-4-carboxamide ribonucleoside with protective effect in dextran sulfate sodium-induced acute and chronic colitis. J. Pharmacol. Exp. Ther. 2010, 333, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Bai, A.; Ma, A.G.; Yong, M.; Weiss, C.R.; Ma, Y.; Guan, Q.; Bernstein, C.N.; Peng, Z. Ampk agonist downregulates innate and adaptive immune responses in tnbs-induced murine acute and relapsing colitis. Biochem. Pharmacol. 2010, 80, 1708–1717. [Google Scholar] [CrossRef]
- Glazunova, V.A.; Lobanov, K.V.; Shakulov, R.S.; Mironov, A.S.; Shtil, A.A. Acadesine triggers non-apoptotic death in tumor cells. Acta Nat. 2013, 5, 74–78. [Google Scholar] [CrossRef]
- Bowser, J.L.; Phan, L.H.; Eltzschig, H.K. The hypoxia-adenosine link during intestinal inflammation. J. Immunol. 2018, 200, 897–907. [Google Scholar] [CrossRef]
- Yang, J.L.; Ha, T.K.; Dhodary, B.; Kim, K.H.; Park, J.; Lee, C.H.; Kim, Y.C.; Oh, W.K. Dammarane triterpenes as potential sirt1 activators from the leaves of panax ginseng. J. Nat. Prod. 2014, 77, 1615–1623. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Wu, J.; Otsuka, M.; Kishikawa, T.; Suzuki, N.; Takata, A.; Ohno, M.; Ishibashi, R.; Yamagami, M.; Nakagawa, R.; et al. Repression of microrna function mediates inflammation-associated colon tumorigenesis. Gastroenterology 2017, 152, 631–643. [Google Scholar] [CrossRef]
- Cseko, K.; Beckers, B.; Keszthelyi, D.; Helyes, Z. Role of trpv1 and trpa1 ion channels in inflammatory bowel diseases: Potential therapeutic targets? Pharmaceuticals 2019, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Maurino, J.J.; Furuzawa-Carballeda, J.; Villeda-Ramirez, M.A.; Fonseca-Camarillo, G.; Meza-Guillen, D.; Barreto-Zuniga, R.; Yamamoto-Furusho, J.K. The transient receptor potential vanilloid 1 is associated with active inflammation in ulcerative colitis. Mediat. Inflamm. 2018, 2018, 6570371. [Google Scholar] [CrossRef]
- Ciregia, F.; Bugliani, M.; Ronci, M.; Giusti, L.; Boldrini, C.; Mazzoni, M.R.; Mossuto, S.; Grano, F.; Cnop, M.; Marselli, L.; et al. Palmitate-induced lipotoxicity alters acetylation of multiple proteins in clonal beta cells and human pancreatic islets. Sci. Rep. 2017, 7, 13445. [Google Scholar] [CrossRef] [PubMed]
- Ciregia, F.; Giusti, L.; Ronci, M.; Bugliani, M.; Piga, I.; Pieroni, L.; Rossi, C.; Marchetti, P.; Urbani, A.; Lucacchini, A. Glucagon-like peptide 1 protects ins-1e mitochondria against palmitate-mediated beta-cell dysfunction: A proteomic study. Mol. Biosyst. 2015, 11, 1696–1707. [Google Scholar] [CrossRef]
- Antonioli, L.; Fornai, M.; Colucci, R.; Ghisu, N.; Da Settimo, F.; Natale, G.; Kastsiuchenka, O.; Duranti, E.; Virdis, A.; Vassalle, C.; et al. Inhibition of adenosine deaminase attenuates inflammation in experimental colitis. J. Pharmacol. Exp. Ther. 2007, 322, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.A.; Vago, J.P.; Perretti, M.; Teixeira, M.M. Mediators of the resolution of the inflammatory response. Trends Immunol. 2019, 40, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Lyons, C.L.; Roche, H.M. Nutritional modulation of ampk-impact upon metabolic-inflammation. Int. J. Mol. Sci. 2018, 19, 3092. [Google Scholar] [CrossRef] [PubMed]
- Sag, D.; Carling, D.; Stout, R.D.; Suttles, J. Adenosine 5′-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J. Immunol. 2008, 181, 8633–8641. [Google Scholar] [CrossRef]
- Muller-Durovic, B.; Lanna, A.; Covre, L.P.; Mills, R.S.; Henson, S.M.; Akbar, A.N. Killer cell lectin-like receptor g1 inhibits nk cell function through activation of adenosine 5′-monophosphate-activated protein kinase. J. Immunol. 2016, 197, 2891–2899. [Google Scholar] [CrossRef] [PubMed]
- Gualdoni, G.A.; Mayer, K.A.; Goschl, L.; Boucheron, N.; Ellmeier, W.; Zlabinger, G.J. The amp analog aicar modulates the treg/th17 axis through enhancement of fatty acid oxidation. FASEB J. 2016, 30, 3800–3809. [Google Scholar] [CrossRef]
- Guigas, B.; Sakamoto, K.; Taleux, N.; Reyna, S.M.; Musi, N.; Viollet, B.; Hue, L. Beyond aica riboside: In search of new specific amp-activated protein kinase activators. IUBMB Life 2009, 61, 18–26. [Google Scholar] [CrossRef]
- Day, P.; Sharff, A.; Parra, L.; Cleasby, A.; Williams, M.; Horer, S.; Nar, H.; Redemann, N.; Tickle, I.; Yon, J. Structure of a cbs-domain pair from the regulatory gamma1 subunit of human ampk in complex with amp and zmp. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 587–596. [Google Scholar] [CrossRef]
- Niesler, C.U.; Myburgh, K.H.; Moore, F. The changing ampk expression profile in differentiating mouse skeletal muscle myoblast cells helps confer increasing resistance to apoptosis. Exp. Physiol. 2007, 92, 207–217. [Google Scholar] [CrossRef]
- Egawa, T.; Ohno, Y.; Goto, A.; Ikuta, A.; Suzuki, M.; Ohira, T.; Yokoyama, S.; Sugiura, T.; Ohira, Y.; Yoshioka, T.; et al. Aicar-induced activation of ampk negatively regulates myotube hypertrophy through the hsp72-mediated pathway in c2c12 skeletal muscle cells. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E344–E354. [Google Scholar] [CrossRef]
- Price, N.L.; Gomes, A.P.; Ling, A.J.; Duarte, F.V.; Martin-Montalvo, A.; North, B.J.; Agarwal, B.; Ye, L.; Ramadori, G.; Teodoro, J.S.; et al. Sirt1 is required for ampk activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012, 15, 675–690. [Google Scholar] [CrossRef] [PubMed]
- Ruderman, N.B.; Xu, X.J.; Nelson, L.; Cacicedo, J.M.; Saha, A.K.; Lan, F.; Ido, Y. Ampk and sirt1: A long-standing partnership? Am. J. Physiol. Endocrinol. Metab. 2010, 298, E751–E760. [Google Scholar] [CrossRef] [PubMed]
- Caruso, R.; Marafini, I.; Franze, E.; Stolfi, C.; Zorzi, F.; Monteleone, I.; Caprioli, F.; Colantoni, A.; Sarra, M.; Sedda, S.; et al. Defective expression of sirt1 contributes to sustain inflammatory pathways in the gut. Mucosal Immunol. 2014, 7, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhou, G.; Yu, T.; Chen, L.; Yu, L.; Guo, Y.; Cong, Y.; Liu, Z. Critical role of rock2 activity in facilitating mucosal cd4(+) t cell activation in inflammatory bowel disease. J. Autoimmun. 2018, 89, 125–138. [Google Scholar] [CrossRef]
- Horowitz, S.; Binion, D.G.; Nelson, V.M.; Kanaa, Y.; Javadi, P.; Lazarova, Z.; Andrekopoulos, C.; Kalyanaraman, B.; Otterson, M.F.; Rafiee, P. Increased arginase activity and endothelial dysfunction in human inflammatory bowel disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G1323–G1336. [Google Scholar] [CrossRef]
- Jin, Y.; Blikslager, A.T. The regulation of intestinal mucosal barrier by myosin light chain kinase/rho kinases. Int. J. Mol. Sci. 2020, 21, 3550. [Google Scholar] [CrossRef]
- Novak, E.A.; Mollen, K.P. Mitochondrial dysfunction in inflammatory bowel disease. Front. Cell Dev. Biol. 2015, 3, 62. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. Ampk: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, S.E.; Sena, L.A.; Chandel, N.S. Mitochondria in the regulation of innate and adaptive immunity. Immunity 2015, 42, 406–417. [Google Scholar] [CrossRef]
- Chang, C.H.; Curtis, J.D.; Maggi, L.B., Jr.; Faubert, B.; Villarino, A.V.; O’Sullivan, D.; Huang, S.C.; van der Windt, G.J.; Blagih, J.; Qiu, J.; et al. Posttranscriptional control of t cell effector function by aerobic glycolysis. Cell 2013, 153, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Palsson-McDermott, E.M.; O’Neill, L.A.J. Targeting immunometabolism as an anti-inflammatory strategy. Cell Res. 2020, 30, 300–314. [Google Scholar] [CrossRef]
- Viola, A.; Munari, F.; Sanchez-Rodriguez, R.; Scolaro, T.; Castegna, A. The metabolic signature of macrophage responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [PubMed]
- Ursini, F.; Russo, E.; Pellino, G.; D’Angelo, S.; Chiaravalloti, A.; De Sarro, G.; Manfredini, R.; De Giorgio, R. Metformin and autoimmunity: A “new deal” of an old drug. Front. Immunol. 2018, 9, 1236. [Google Scholar] [CrossRef]
- Schuiveling, M.; Vazirpanah, N.; Radstake, T.; Zimmermann, M.; Broen, J.C.A. Metformin, a new era for an old drug in the treatment of immune mediated disease? Curr. Drug Targets 2018, 19, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Morampudi, V.; Bhinder, G.; Wu, X.; Dai, C.; Sham, H.P.; Vallance, B.A.; Jacobson, K. Dnbs/tnbs colitis models: Providing insights into inflammatory bowel disease and effects of dietary fat. J. Vis. Exp. JoVE 2014, 58, e51297. [Google Scholar] [CrossRef]
- Barone, M.; Chain, F.; Sokol, H.; Brigidi, P.; Bermudez-Humaran, L.G.; Langella, P.; Martin, R. A versatile new model of chemically induced chronic colitis using an outbred murine strain. Front. Microbiol. 2018, 9, 565. [Google Scholar] [CrossRef]
- Antonioli, L.; Fornai, M.; Colucci, R.; Awwad, O.; Ghisu, N.; Tuccori, M.; Da Settimo, F.; La Motta, C.; Natale, G.; Duranti, E.; et al. The blockade of adenosine deaminase ameliorates chronic experimental colitis through the recruitment of adenosine a2a and a3 receptors. J. Pharmacol. Exp. Ther. 2010, 335, 434–442. [Google Scholar] [CrossRef]
- Arab, H.H.; Al-Shorbagy, M.Y.; Saad, M.A. Activation of autophagy and suppression of apoptosis by dapagliflozin attenuates experimental inflammatory bowel disease in rats: Targeting ampk/mtor, hmgb1/rage and nrf2/ho-1 pathways. Chemico-Biol. Interact. 2021, 335, 109368. [Google Scholar] [CrossRef]
- Deng, Z.; Ni, J.; Wu, X.; Wei, H.; Peng, J. Gpa peptide inhibits nlrp3 inflammasome activation to ameliorate colitis through ampk pathway. Aging 2020, 12, 18522–18544. [Google Scholar] [CrossRef] [PubMed]
- Takahara, M.; Takaki, A.; Hiraoka, S.; Adachi, T.; Shimomura, Y.; Matsushita, H.; Nguyen, T.T.T.; Koike, K.; Ikeda, A.; Takashima, S.; et al. Berberine improved experimental chronic colitis by regulating interferon-gamma- and il-17a-producing lamina propria cd4(+) t cells through ampk activation. Sci. Rep. 2019, 9, 11934. [Google Scholar] [CrossRef] [PubMed]
- Lechuga, S.; Ivanov, A.I. Disruption of the epithelial barrier during intestinal inflammation: Quest for new molecules and mechanisms. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef]
- Guragain, D.; Gurung, P.; Chang, J.H.; Katila, N.; Chang, H.W.; Jeong, B.S.; Choi, D.Y.; Kim, J.A. Ampk is essential for il-10 expression and for maintaining balance between inflammatory and cytoprotective signaling. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129631. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.P.; Brown, J.R.; Sag, D.; Zhang, L.; Suttles, J. Adenosine 5′-monophosphate-activated protein kinase regulates il-10-mediated anti-inflammatory signaling pathways in macrophages. J. Immunol. 2015, 194, 584–594. [Google Scholar] [CrossRef]
- Wang, S.; Song, P.; Zou, M.H. Amp-activated protein kinase, stress responses and cardiovascular diseases. Clin. Sci. 2012, 122, 555–573. [Google Scholar] [CrossRef]
- Rabinovitch, R.C.; Samborska, B.; Faubert, B.; Ma, E.H.; Gravel, S.P.; Andrzejewski, S.; Raissi, T.C.; Pause, A.; St-Pierre, J.; Jones, R.G. Ampk maintains cellular metabolic homeostasis through regulation of mitochondrial reactive oxygen species. Cell Rep. 2017, 21, 1–9. [Google Scholar] [CrossRef]
- Kilkenny, C.; Browne, W.; Cuthill, I.C.; Emerson, M.; Altman, D.G. NC3Rs Reporting Guidelines Working Group. Animal research: Reporting in vivo experiments: The arrive guidelines. J. Gene Med. 2010, 12, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Fabian, T.C.; Fabian, M.J.; Yockey, J.M.; Proctor, K.G. Acadesine and lipopolysaccharide-evoked pulmonary dysfunction after resuscitation from traumatic shock. Surgery 1996, 119, 302–315. [Google Scholar] [CrossRef]
- Antonioli, L.; Fornai, M.; Awwad, O.; Giustarini, G.; Pellegrini, C.; Tuccori, M.; Caputi, V.; Qesari, M.; Castagliuolo, I.; Brun, P.; et al. Role of the a(2b) receptor-adenosine deaminase complex in colonic dysmotility associated with bowel inflammation in rats. Br. J. Pharmacol. 2014, 171, 1314–1329. [Google Scholar] [CrossRef] [PubMed]
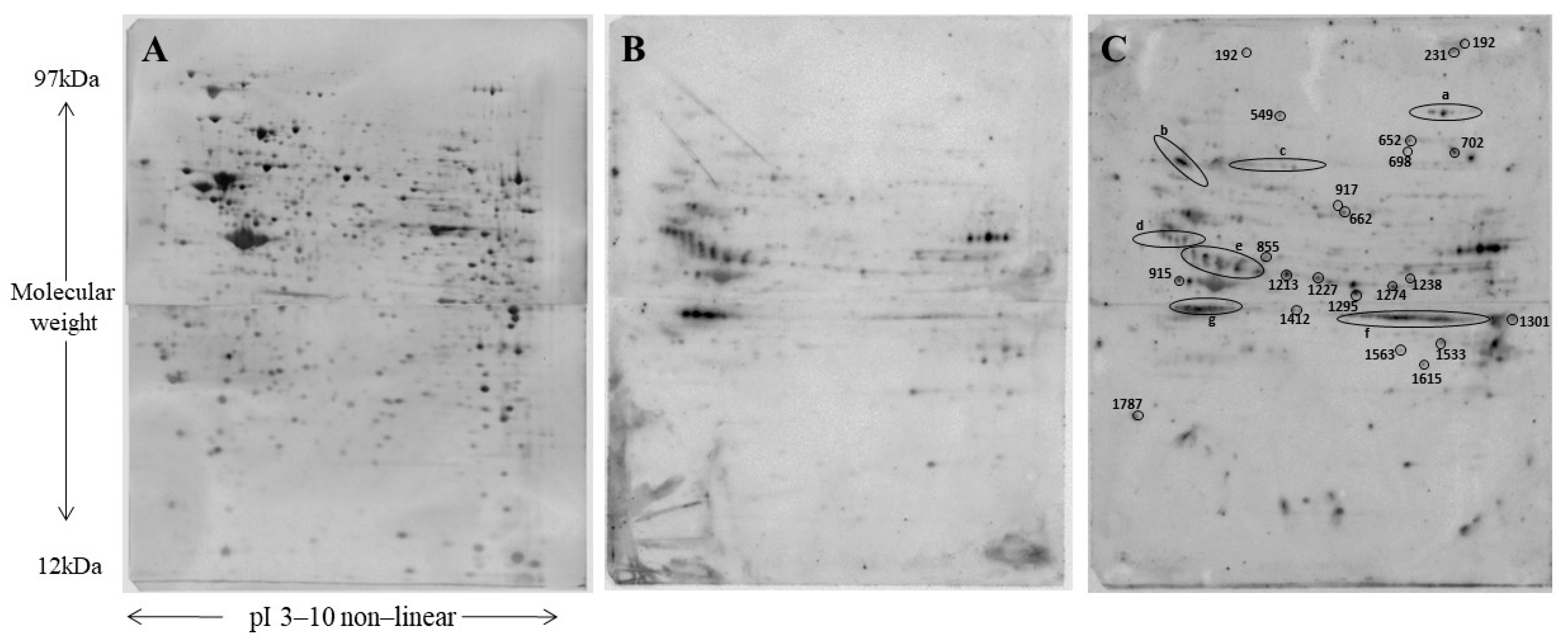


| Spot n°/Position | ID | Protein Name | Gene Name | Coverage (%) | Unique Peptides | MW (th) | pI (th) | Ratio (FA5/CTRL) | ANOVA (p-Value) | Fold Phosphorylation |
|---|---|---|---|---|---|---|---|---|---|---|
| 192 | Q8CGC7 | Bifunctional glutamate/proline—tRNA ligase | Eprs | 3 | 4 | 170,078 | 7.75 | 6.3 | 0.03 | 6.3 |
| 197 | Q6PDG5 | SWI/SNF complex subunit SMARCC2 | Smarcc2 | 2 | 2 | 132,604 | 5.41 | 4.7 | 0.013 | 4.7 |
| 201 | Q02053 | Ubiquitin-like modifier-activating enzyme 1 | Uba1 | 21 | 18 | 117,809 | 5.43 | 2.2 | 0.021 | 2.2 |
| 231 | Q8VDJ3 | Vigilin | Hdlbp | 3 | 4 | 141,742 | 6.43 | 2.5 | 0.023 | 2.5 |
| 247/a | P58252 | Elongation factor 2 | Eef2 | 25 | 21 | 95,314 | 6.41 | 5.2 | 0.002 | 5.2 |
| 270/a | P49717 | DNA replication licensing factor MCM4 | Mcm4 | 9 | 9 | 96,736 | 6.77 | 5.2 | 0.002 | 5.2 |
| 272/a | P49717 | DNA replication licensing factor MCM4 | Mcm4 | 5 | 5 | 96,736 | 6.77 | 5.8 | 0.002 | 5.8 |
| 279/a | P58252 | Elongation factor 2 | Eef2 | 25 | 20 | 95,314 | 6.41 | 3.9 | 0.00008 | 3.9 |
| 283/a | P58252 | Elongation factor 2 | Eef2 | 28 | 24 | 95,314 | 6.41 | 2.2 | 0.00009 | 2.2 |
| 294/a | P58252 | Elongation factor 2 | Eef2 | 37 | 35 | 95,314 | 6.41 | 5.8 | 0.002 | 5.8 |
| 296/a | P49717 | DNA replication licensing factor MCM4 | Mcm4 | 5 | 5 | 96,736 | 6.77 | 5.4 | 0.001 | 5.4 |
| 296/a | P58252 | Elongation factor 2 | Eef2 | 41 | 40 | 95,314 | 6.41 | 5.4 | 0.001 | 5.4 |
| 302 | P13020 | Gelsolin | Gsn | 24 | 16 | 85,942 | 5.83 | 2.7 | 0.013 | 2.7 |
| 302 | Q9ERG0 | LIM domain and actin-binding protein 1 | Lima 1 | 7 | 6 | 84,060 | 6.18 | 2.7 | 0.013 | 2.7 |
| 315 | P13020 | Gelsolin | Gsn | 33 | 21 | 85,942 | 5.83 | 3.9 | 0.017 | 3.9 |
| 315 | Q9ERG0 | LIM domain and actin-binding protein 1 | Lima 1 | 20 | 11 | 84,060 | 6.18 | 3.9 | 0.017 | 3.9 |
| 390 | P20029 | 78 kDa glucose-regulated protein | Hspa5 | 58 | 50 | 72,422 | 5.07 | 4.5 | 0.00038 | 4.5 |
| 400 | P14824 | Annexin A6 | Anxa6 | 34 | 20 | 75,885 | 5.34 | 8.2 | 0.003 | 8.2 |
| 453 | P63017 | Heat shock cognate 71 kDa protein | Hspa8 | 60 | 31 | 70,871 | 5.37 | 2.8 | 0.00006 | 2.8 |
| 464 | Q9CZD3 | Glycine—tRNA ligase | Gars | 18 | 13 | 81,878 | 6.24 | 4.5 | 0.001 | 4.5 |
| 478 | Q9CZD3 | Glycine—tRNA ligase | Gars | 26 | 20 | 81,878 | 6.24 | 6.2 | 0.001 | 6.2 |
| 492 | Q9CZD3 | Glycine—tRNA ligase | Gars | 26 | 23 | 81,878 | 6.24 | 4.8 | 0.00005 | 4.8 |
| 494 | P17156 | Heat shock-related 70 kDa protein 2 | Hspa2 | 21 | 2 | 69,642 | 5.5 | 1.63 | 0.00066 | 2.0 |
| 494 | P63017 | Heat shock cognate 71 kDa protein | Hspa8 | 59 | 32 | 70,871 | 5.37 | 1.63 | 0.00066 | 2.0 |
| 504 | P48678 | Prelamin-A/C | Lmna | 15 | 9 | 74,238 | 6.54 | 0.34 | 0.0003 | 2.9 |
| 520/a | P49717 | DNA replication licensing factor MCM4 | Mcm4 | 16 | 15 | 96,736 | 6.77 | 6.5 | 0.00049 | 6.5 |
| 520/a | P58252 | Elongation factor 2 | Eef2 | 41 | 38 | 95,314 | 6.41 | 6.5 | 0.00049 | 6.5 |
| 521/a | P58252 | Elongation factor 2 | Eef2 | 26 | 22 | 95,314 | 6.41 | 4.4 | 0.003 | 4.4 |
| 526/a | P49717 | DNA replication licensing factor MCM4 | Mcm4 | 5 | 4 | 96,736 | 6.77 | 3.7 | 0.002 | 3.7 |
| 526/a | P58252 | Elongation factor 2 | Eef2 | 22 | 17 | 95,314 | 6.41 | 3.7 | 0.002 | 3.7 |
| 538/a | P58252 | Elongation factor 2 | Eef2 | 33 | 27 | 95,314 | 6.41 | 6.8 | 0.001 | 6.8 |
| 549 | Q8K009 | Mitochondrial 10-formyltetrahydrofolate dehydrogen | Aldh1l2 | 32 | 24 | 101,590 | 5.93 | 7.2 | 0.03 | 7.2 |
| 594 | P61979 | Heterogeneous nuclear ribonucleoprotein K | Hnrnpk | 5 | 2 | 50,976 | 5.39 | 0.15 | 0.000086 | 6.5 |
| 652 | Q61881 | DNA replication licensing factor MCM7 | Mcm7 | 33 | 21 | 81,211 | 5.98 | 2.1 | 0.028 | 2.1 |
| 662 | Q91YW3 | DnaJ homolog subfamily C member 3 | Dnajc3 | 4 | 2 | 57,464 | 5.61 | 2.9 | 0.001 | 2.9 |
| 698 | Q8R550 | SH3 domain-containing kinase-binding protein 1 | Sh3kbp1 | 9 | 5 | 78,170 | 7.15 | 3.5 | 0.018 | 3.5 |
| 702 | P26041 | Moesin | Msn | 3 | 2 | 67,767 | 6.22 | 11 | 0.00057 | 10.9 |
| 770/d | P31324 | cAMP-dependent protein kinase type II-beta regulatory | Prkar2b | 16 | 5 | 46,167 | 4.9 | 0.41 | 0.00065 | 2.4 |
| 772/d | P99024 | Tubulin beta-5 chain | Tubb5 | 30 | 2 | 49,671 | 4.78 | 0.37 | 0.000055 | 2.7 |
| 777/c | Q8C1A5 | Thimet oligopeptidase | Thop1 | 36 | 23 | 78,026 | 5.72 | 2.3 | 0.00023 | 2.3 |
| 784 | P56480 | ATP synthase subunit beta, mitochondrial | Atp5b | 20 | 8 | 56,301 | 5.19 | 2.1 | 0.00015 | 2.1 |
| 847 | Q99L45 | Eukaryotic translation initiation factor 2 subunit 2 | Eif2s2 | 50 | 17 | 38,092 | 5.61 | 0.13 | 0.001 | 7.3 |
| 855 | P60843 | Eukaryotic initiation factor 4A-I | Eif4a1 | 34 | 7 | 46,154 | 5.32 | 2.1 | 0.00021 | 2.1 |
| 909 | Q9Z1D1 | Eukaryotic translation initiation factor 3 subunit G | Eif3g | 7 | 2 | 35,638 | 5.7 | 0.47 | 0.00078 | 2.1 |
| 915 | Q9JKV1 | Proteasomal ubiquitin receptor ADRM1 | Adrm1 | 7 | 3 | 42,060 | 4.94 | 0.45 | 0.001 | 2.4 |
| 917 | P11983 | T-complex protein 1 subunit alpha | Tcp1 | 51 | 17 | 60,449 | 5.82 | 2.9 | 0.008 | 2.9 |
| 917 | P63038 | 60 kDa heat shock protein, mitochondrial | Hspd1 | 27 | 14 | 60,956 | 5.91 | 2.9 | 0.008 | 2.9 |
| 931 | Q04447 | Creatine kinase B-type | Ckb | 34 | 11 | 42,713 | 5.4 | 0.41 | 0.001 | 2.4 |
| 1060/d | P54775 | 26S protease regulatory subunit 6B | Psmc4 | 45 | 20 | 47,408 | 5.0 | 0.36 | 0.003 | 2.7 |
| 1066 | Q99KV1 | DnaJ homolog subfamily B member 11 | Dnajb11 | 28 | 6 | 40.555 | 5.92 | 0.23 | 0.0000009 | 4.3 |
| 1070/d | P20152 | Vimentin | Vim | 33 | 14 | 53,688 | 5.05 | 0.19 | 0.00004 | 5.1 |
| 1096/e | Q3UM45 | Protein phosphatase 1 regulatory subunit 7 | Ppp1r7 | 5 | 2 | 41,292 | 4.85 | 0.12 | 0.00005 | 8 |
| 1072/e | P20152 | Vimentin | Vim | 35 | 14 | 53,688 | 5.05 | 0.18 | 0.000003 | 5.5 |
| 1078/e | P60843 | Eukaryotic initiation factor 4A-I | Eif4a1 | 23 | 9 | 46,154 | 5.32 | 0.36 | 0.004 | 2.8 |
| 1113/e | Q9Z2X1 | Heterogeneous nuclear ribonucleoprotein F | Hnrnpf | 17 | 6 | 45,730 | 5.3 | 0.36 | 0.003 | 2.7 |
| 1119/e | Q3UM45 | Protein phosphatase 1 regulatory subunit 7 | Ppp1r7 | 11 | 4 | 41,292 | 4.85 | 0.12 | 0.00005 | 8 |
| 1213 | Q62433 | Protein NDRG1 | Ndrg1 | 32 | 8 | 43,009 | 5.69 | 4.9 | 0.00017 | 4.9 |
| 1227 | Q62433 | Protein NDRG1 | Ndrg1 | 32 | 8 | 43,009 | 5.69 | 2.2 | 0.002 | 2.2 |
| 1238 | P50580 | Proliferation-associated protein 2G4 | Pa2g4 | 33 | 14 | 43,699 | 6.41 | 2.3 | 0.005 | 2.3 |
| 1274 | Q9DCL9 | Multifunctional protein ADE2 | Paics | 13 | 6 | 47,006 | 6.94 | 5.0 | 0.043 | 5 |
| 1295 | P35486 | Pyruvate dehydrogenase E1 subunit alpha, mitoch | Pdha1 | 30 | 12 | 43,232 | 8.49 | 10 | 0.00005 | 10.4 |
| 1301 | P51174 | Long-chain specific acyl-CoA dehydrogenase, mitoch | Acadl | 34 | 13 | 47,908 | 8.53 | 2.6 | 0.00009 | 2.6 |
| 1313/f | P60335 | Poly(rC)-binding protein 1 | Pcbp1 | 7 | 2 | 37,498 | 6.66 | 3.4 | 0.002 | 3.4 |
| 1313/f | Q91WK2 | Eukaryotic translation initiation factor 3 subunit H | Eif3h | 39 | 14 | 39,832 | 6.19 | 3.4 | 0.002 | 3.4 |
| 1326/f | P60335 | Poly(rC)-binding protein 1 | Pcbp1 | 10 | 3 | 37,498 | 6.66 | 2.7 | 0.001 | 2.7 |
| 1329/f | P60335 | Poly(rC)-binding protein 1 | Pcbp1 | 40 | 7 | 37,498 | 6.66 | 5.5 | 0.0005 | 5.5 |
| 1330/f | Q91WK2 | Eukaryotic translation initiation factor 3 subunit H | Eif3h | 9 | 2 | 39,832 | 6.19 | 5.7 | 0.00011 | 5.7 |
| 1335/f | Q91YR1 | Twinfilin-1 | Twf1 | 37 | 11 | 40,079 | 6.2 | 5.7 | 0.00011 | 5.7 |
| 1408/g | Q9CX34 | Suppressor of G2 allele of SKP1 homolog | Sugt1 | 26 | 7 | 38,159 | 5.32 | 0.29 | 0.0001 | 3.4 |
| 1408/g | Q9R1T2 | SUMO-activating enzyme subunit 1 | Sae1 | 15 | 6 | 38,620 | 5.24 | 0.29 | 0.0001 | 3.4 |
| 1409/g | Q9CX34 | Suppressor of G2 allele of SKP1 homolog | Sugt1 | 56 | 18 | 38,159 | 5.32 | 0.31 | 0.0005 | 3.2 |
| 1409/g | Q9R1T2 | SUMO-activating enzyme subunit 1 | Sae1 | 37 | 12 | 38,620 | 5.24 | 0.31 | 0.0005 | 3.2 |
| 1412 | Q9Z0S1 | 3′(2′),5′-bisphosphate nucleotidase 1 | Bpnt1 | 25 | 7 | 33,196 | 5.54 | 6.5 | 0.011 | 6.5 |
| 1419/g | O35295 | Transcriptional activator protein Pur-beta | Purb | 18 | 4 | 33,901 | 5.33 | 0.35 | 0.001 | 2.9 |
| 1435/f | P60335 | Poly(rC)-binding protein 1 | Pcbp1 | 37 | 9 | 37,498 | 6.66 | 3.0 | 0.001 | 3 |
| 1441/f | P60335 | Poly(rC)-binding protein 1 | Pcbp1 | 36 | 6 | 37,498 | 6.66 | 8.3 | 0.001 | 8.3 |
| 1448/f | P60335 | Poly(rC)-binding protein 1 | Pcbp1 | 18 | 5 | 37,498 | 6.66 | 4.7 | 0.013 | 4.7 |
| 1449/f | Q91YR1 | Twinfilin-1 | Twf1 | 34 | 11 | 40,079 | 6.2 | 8.6 | 0.003 | 8.6 |
| 1450/f | Q91WK2 | Eukaryotic translation initiation factor 3 subunit H | Eif3h | 37 | 12 | 39,832 | 6.19 | 5.7 | 0.0005 | 5.7 |
| 1462 | P61982 | 14-3-3 protein gamma | Ywhag | 43 | 7 | 28,303 | 4.8 | 0.23 | 0.04 | 4.3 |
| 1471 | P63101 | 14-3-3 protein zeta/delta | Ywhaz | 73 | 15 | 27,771 | 4.73 | 4.5 | 0.00039 | 4.5 |
| 1533 | Q9Z130 | Heterogeneous nuclear ribonucleoprotein D-like | Hnrnpdl | 20 | 4 | 33,559 | 6.85 | 4.4 | 0.003 | 4.4 |
| 1563 | Q9D883 | Splicing factor U2AF 35 kDa subunit | U2af1 | 14 | 3 | 27,815 | 9.09 | 2.9 | 0.000535 | 2.9 |
| 1615 | P47962 | 60S ribosomal protein L5 | Rpl5 | 9 | 2 | 34,401 | 9.78 | 9.7 | 0.00034 | 9.7 |
| 1787 | P62259 | 14-3-3 protein epsilon | Ywhae | 75 | 23 | 29,174 | 4.63 | 0.32 | 0.00009 | 3.1 |
| 2489/b | P20029 | 78 kDa glucose-regulated protein | Hspa5 | 42 | 27 | 72,422 | 5.07 | 9.3 | 0.001 | 9.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonioli, L.; Pellegrini, C.; Fornai, M.; Benvenuti, L.; D’Antongiovanni, V.; Colucci, R.; Bertani, L.; Di Salvo, C.; Semeghini, G.; La Motta, C.; et al. Preclinical Development of FA5, a Novel AMP-Activated Protein Kinase (AMPK) Activator as an Innovative Drug for the Management of Bowel Inflammation. Int. J. Mol. Sci. 2021, 22, 6325. https://doi.org/10.3390/ijms22126325
Antonioli L, Pellegrini C, Fornai M, Benvenuti L, D’Antongiovanni V, Colucci R, Bertani L, Di Salvo C, Semeghini G, La Motta C, et al. Preclinical Development of FA5, a Novel AMP-Activated Protein Kinase (AMPK) Activator as an Innovative Drug for the Management of Bowel Inflammation. International Journal of Molecular Sciences. 2021; 22(12):6325. https://doi.org/10.3390/ijms22126325
Chicago/Turabian StyleAntonioli, Luca, Carolina Pellegrini, Matteo Fornai, Laura Benvenuti, Vanessa D’Antongiovanni, Rocchina Colucci, Lorenzo Bertani, Clelia Di Salvo, Giorgia Semeghini, Concettina La Motta, and et al. 2021. "Preclinical Development of FA5, a Novel AMP-Activated Protein Kinase (AMPK) Activator as an Innovative Drug for the Management of Bowel Inflammation" International Journal of Molecular Sciences 22, no. 12: 6325. https://doi.org/10.3390/ijms22126325
APA StyleAntonioli, L., Pellegrini, C., Fornai, M., Benvenuti, L., D’Antongiovanni, V., Colucci, R., Bertani, L., Di Salvo, C., Semeghini, G., La Motta, C., Giusti, L., Zallocco, L., Ronci, M., Quattrini, L., Angelucci, F., Coviello, V., Oh, W.-K., Ha, Q. T. K., Németh, Z. H., ... Blandizzi, C. (2021). Preclinical Development of FA5, a Novel AMP-Activated Protein Kinase (AMPK) Activator as an Innovative Drug for the Management of Bowel Inflammation. International Journal of Molecular Sciences, 22(12), 6325. https://doi.org/10.3390/ijms22126325









