Mechanisms by Which Kinesin-5 Motors Perform Their Multiple Intracellular Functions
Abstract
:1. Historical Outlook
2. Structural Features
2.1. The Homotetrameric Kinesin-5 Complex
2.2. The C-Terminal Tail Domain
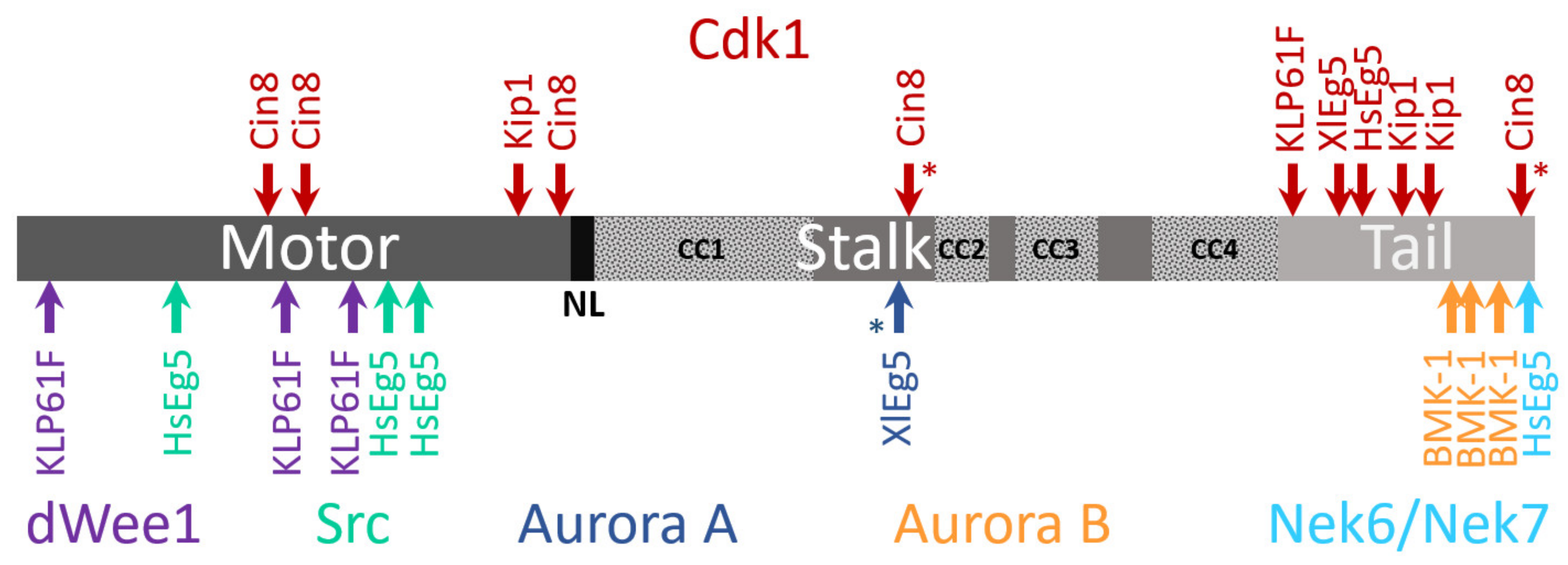
2.3. Neck Linker, Neck Cover Bundle and the N-Terminal Non-Motor Extension
2.4. Loop 5
2.5. Loop 8
3. Motor Activity
3.1. Velocity, Processivity and Anti-Parallel MT Sliding
3.2. Bidirectional Motility of Fungal Kinesin-5 Motors
3.3. Interaction with MT Ends
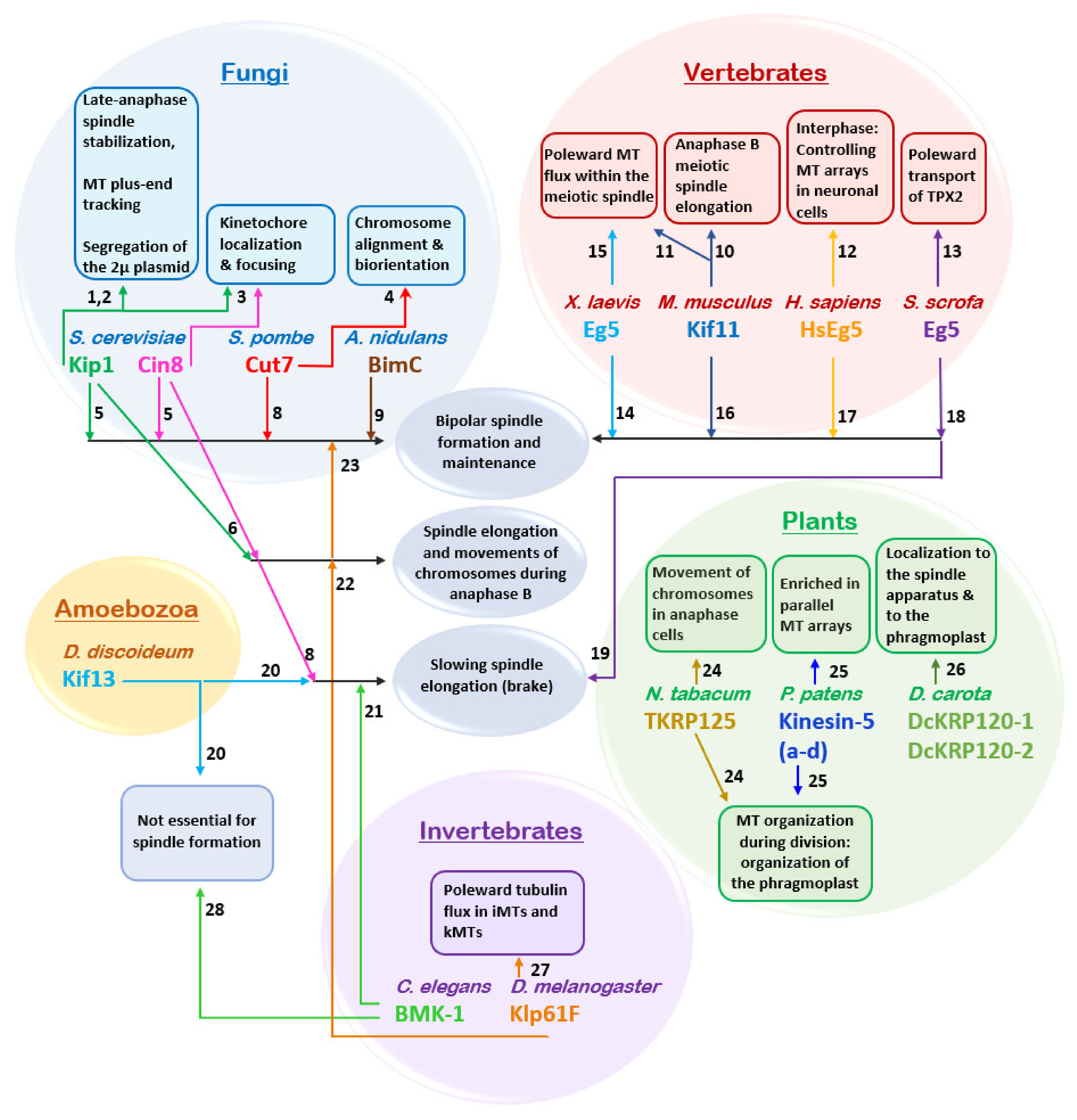
4. Intracellular Function
4.1. Roles in Dividing Cells
4.1.1. Structure and Dynamics of the Mitotic Spindle
4.1.2. Bipolar Spindle Assembly, Maintenance, and Elongation
4.1.3. Models for Maintaining Spindle Bipolarity
4.1.4. Effects on MT Turnover and Dynamics
4.1.5. Functions at the Spindle Poles

4.2. Roles in Non-Dividing Cells
4.3. Phosphoregulation
4.4. Other Post-Translation Modifications
5. Kinesin-5 Motors and Pathological Conditions
Author Contributions
Funding
Conflicts of Interest
References
- Vale, R.D.; Reese, T.S.; Sheetz, M.P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 1985, 42, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Enos, A.P.; Morris, N.R. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell 1990, 60, 1019–1027. [Google Scholar] [CrossRef]
- Hagan, I.; Yanagida, M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature 1990, 347, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Le Guellec, R.; Paris, J.; Couturier, A.; Roghi, C.; Philippe, M. Cloning by differential screening of a Xenopus cDNA that encodes a kinesin-related protein. Mol. Cell. Biol. 1991, 11, 3395–3398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawin, K.E.; LeGuellec, K.; Philippe, M.; Mitchison, T.J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 1992, 359, 540–543. [Google Scholar] [CrossRef] [PubMed]
- Bloom, G.S. Motor proteins for cytoplasmic microtubules. Curr. Opin. Cell Biol. 1992, 4, 66–73. [Google Scholar] [CrossRef]
- Hoyt, M.A.; He, L.; Loo, K.K.; Saunders, W.S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 1992, 118, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Roof, D.M.; Meluh, P.B.; Rose, M.D. Kinesin-related proteins required for assembly of the mitotic spindle. J. Cell Biol. 1992, 118, 95–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saunders, W.S.; Hoyt, M.A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 1992, 70, 451–458. [Google Scholar] [CrossRef]
- Singh, S.K.; Pandey, H.; Al-Bassam, J.; Gheber, L. Bidirectional motility of kinesin-5 motor proteins: Structural determinants, cumulative functions and physiological roles. Cell Mol. Life Sci. 2018, 1757–1771. [Google Scholar] [CrossRef]
- Adams, A.E.; Pringle, J.R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 1984, 98, 934–945. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allan, V.J. Cytoplasmic dynein. Biochem. Soc. Trans. 2011, 39, 1169–1178. [Google Scholar] [CrossRef]
- Brust-Mascher, I.; Sommi, P.; Cheerambathur, D.K.; Scholey, J.M. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol. Biol. Cell 2009, 20, 1749–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferenz, N.P.; Gable, A.; Wadsworth, P. Mitotic functions of kinesin-5. Semin. Cell Dev. Biol. 2010, 21, 255–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholey, J.M.; Civelekoglu-Scholey, G.; Brust-Mascher, I. Anaphase b. Biology 2016, 5, 51. [Google Scholar] [CrossRef] [Green Version]
- Heck, M.M.; Pereira, A.; Pesavento, P.; Yannoni, Y.; Spradling, A.C.; Goldstein, L.S. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J. Cell Biol. 1993, 123, 665–679. [Google Scholar] [CrossRef] [Green Version]
- Blangy, A.; Lane, H.A.; d’Herin, P.; Harper, M.; Kress, M.; Nigg, E.A. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 1995, 83, 1159–1169. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, C.J.; Dawe, R.K.; Christie, K.R.; Cleveland, D.W.; Dawson, S.C.; Endow, S.A.; Goldstein, L.S.; Goodson, H.V.; Hirokawa, N.; Howard, J.; et al. A standardized kinesin nomenclature. J. Cell Biol. 2004, 167, 19–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, D.G.; Saxton, W.M.; Sheehan, K.B.; Scholey, J.M. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J. Biol. Chem. 1994, 269, 22913–22916. [Google Scholar] [CrossRef]
- Kashina, A.S.; Baskin, R.J.; Cole, D.G.; Wedaman, K.P.; Saxton, W.M.; Scholey, J.M. A bipolar kinesin. Nature 1996, 379, 270–272. [Google Scholar] [CrossRef]
- Gordon, D.M.; Roof, D.M. The kinesin-related protein Kip1p of Saccharomyces cerevisiae is bipolar. J. Biol. Chem. 1999, 274, 28779–28786. [Google Scholar] [CrossRef] [Green Version]
- Kwok, B.H.; Kapitein, L.C.; Kim, J.H.; Peterman, E.J.; Schmidt, C.F.; Kapoor, T.M. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat. Chem. Biol. 2006, 2, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Kwok, B.H.; Yang, J.G.; Kapoor, T.M. The rate of bipolar spindle assembly depends on the microtubule-gliding velocity of the mitotic kinesin Eg5. Curr. Biol. 2004, 14, 1783–1788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, T.U.; Kapoor, T.M.; Haggarty, S.J.; King, R.W.; Schreiber, S.L.; Mitchison, T.J. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 1999, 286, 971–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, T.M.; Mayer, T.U.; Coughlin, M.L.; Mitchison, T.J. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 2000, 150, 975–988. [Google Scholar] [CrossRef]
- Beer, T.M.; Goldman, B.; Synold, T.W.; Ryan, C.W.; Vasist, L.S.; Van Veldhuizen, P.J., Jr.; Dakhil, S.R.; Lara, P.N., Jr.; Drelichman, A.; Hussain, M.H.; et al. Southwest oncology group phase II study of ispinesib in androgen-independent prostate cancer previously treated with taxanes. Clin. Genitourin Cancer 2008, 6, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.A.; Siu, L.L.; Chen, E.X.; Hotte, S.J.; Chia, S.; Schwarz, J.K.; Pond, G.R.; Johnson, C.; Colevas, A.D.; Synold, T.W.; et al. Phase II study of ispinesib in recurrent or metastatic squamous cell carcinoma of the head and neck. Investug. New Drugs 2008, 26, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Belanger, K.; Rao, S.C.; Petrella, T.M.; Tozer, R.G.; Wood, L.; Savage, K.J.; Eisenhauer, E.A.; Synold, T.W.; Wainman, N.; et al. A phase II study of ispinesib (SB-715992) in patients with metastatic or recurrent malignant melanoma: A National cancer institute of canada clinical trials group trial. Investig. New Drugs 2008, 26, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Lad, L.; Luo, L.; Carson, J.D.; Wood, K.W.; Hartman, J.J.; Copeland, R.A.; Sakowicz, R. Mechanism of inhibition of human KSP by ispinesib. Biochemistry 2008, 47, 3576–3585. [Google Scholar] [CrossRef]
- Sharp, D.J.; McDonald, K.L.; Brown, H.M.; Matthies, H.J.; Walczak, C.; Vale, R.D.; Mitchison, T.J.; Scholey, J.M. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 1999, 144, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Kapitein, L.C.; Peterman, E.J.; Kwok, B.H.; Kim, J.H.; Kapoor, T.M.; Schmidt, C.F. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 2005, 435, 114–118. [Google Scholar] [CrossRef]
- Gerson-Gurwitz, A.; Thiede, C.; Movshovich, N.; Fridman, V.; Podolskaya, M.; Danieli, T.; Lakamper, S.; Klopfenstein, D.R.; Schmidt, C.F.; Gheber, L. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 2011, 30, 4942–4954. [Google Scholar] [CrossRef] [Green Version]
- Roostalu, J.; Hentrich, C.; Bieling, P.; Telley, I.A.; Schiebel, E.; Surrey, T. Directional switching of the Kinesin cin8 through motor coupling. Science 2011, 332, 94–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fridman, V.; Gerson-Gurwitz, A.; Shapira, O.; Movshovich, N.; Lakamper, S.; Schmidt, C.F.; Gheber, L. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J. Cell Sci. 2013, 126, 4147–4159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edamatsu, M. Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochem. Biophys. Res. Commun. 2014, 446, 231–234. [Google Scholar] [CrossRef]
- Acar, S.; Carlson, D.B.; Budamagunta, M.S.; Yarov-Yarovoy, V.; Correia, J.J.; Ninonuevo, M.R.; Jia, W.; Tao, L.; Leary, J.A.; Voss, J.C.; et al. The bipolar assembly domain of the mitotic motor kinesin-5. Nat. Commun. 2013, 4, 1343. [Google Scholar] [CrossRef] [Green Version]
- Scholey, J.E.; Nithianantham, S.; Scholey, J.M.; Al-Bassam, J. Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. eLife 2014, 3, e02217. [Google Scholar] [CrossRef] [PubMed]
- Cochran, J.C. Kinesin Motor Enzymology: Chemistry, structure, and physics of nanoscale molecular machines. Biophys. Rev. 2015, 7, 269–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulet, A.; Moores, C. New insights into the mechanism of force generation by kinesin-5 molecular motors. Int. Rev. Cell Mol. Biol. 2013, 304, 419–466. [Google Scholar] [CrossRef] [PubMed]
- Morfini, G.; Schmidt, N.; Weissmann, C.; Pigino, G.; Kins, S. Conventional kinesin: Biochemical heterogeneity and functional implications in health and disease. Brain Res. Bull. 2016, 126, 347–353. [Google Scholar] [CrossRef]
- Waitzman, J.S.; Rice, S.E. Mechanism and regulation of kinesin-5, an essential motor for the mitotic spindle. Biol. Cell 2014, 106, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cross, R.A. Review: Mechanochemistry of the kinesin-1 ATPase. Biopolymers 2016, 105, 476–482. [Google Scholar] [CrossRef] [Green Version]
- Friel, C.T.; Howard, J. Coupling of kinesin ATP turnover to translocation and microtubule regulation: One engine, many machines. J. Muscle Res. Cell Motil. 2012, 33, 377–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Wildenberg, S.M.; Tao, L.; Kapitein, L.C.; Schmidt, C.F.; Scholey, J.M.; Peterman, E.J. The homotetrameric kinesin-5 KLP61F preferentially crosslinks m icrotubules into antiparallel orientations. Curr. Biol. 2008, 18, 1860–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildebrandt, E.R.; Gheber, L.; Kingsbury, T.; Hoyt, M.A. Homotetrameric form of Cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J. Biol. Chem. 2006, 281, 26004–26013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahu, J.; Surrey, T. Motile microtubule crosslinkers require distinct dynamic properties for correct functioning during spindle organization in Xenopus egg extract. J. Cell Sci. 2009, 122, 1295–1300. [Google Scholar] [CrossRef] [Green Version]
- Tao, L.; Mogilner, A.; Civelekoglu-Scholey, G.; Wollman, R.; Evans, J.; Stahlberg, H.; Scholey, J.M. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr. Biol. 2006, 16, 2293–2302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fakhri, N.; Schmidt, C.F. A surprising twist. eLife 2014, 3, e02715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hackney, D.D. Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature 1995, 377, 448–450. [Google Scholar] [CrossRef] [PubMed]
- Stock, M.F.; Guerrero, J.; Cobb, B.; Eggers, C.T.; Huang, T.G.; Li, X.; Hackney, D.D. Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J. Biol. Chem. 1999, 274, 14617–14623. [Google Scholar] [CrossRef] [Green Version]
- Kaan, H.Y.; Hackney, D.D.; Kozielski, F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science 2011, 333, 883–885. [Google Scholar] [CrossRef] [Green Version]
- Bodrug, T.; Wilson-Kubalek, E.M.; Nithianantham, S.; Thompson, A.F.; Alfieri, A.; Gaska, I.; Major, J.; Debs, G.; Inagaki, S.; Gutierrez, P.; et al. The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding. eLife 2020, 9, 51131. [Google Scholar] [CrossRef]
- Weinger, J.S.; Qiu, M.; Yang, G.; Kapoor, T.M. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr. Biol. 2011, 21, 154–160. [Google Scholar] [CrossRef] [Green Version]
- Duselder, A.; Fridman, V.; Thiede, C.; Wiesbaum, A.; Goldstein, A.; Klopfenstein, D.R.; Zaitseva, O.; Janson, M.E.; Gheber, L.; Schmidt, C.F. Deletion of the Tail Domain of the Kinesin-5 Cin8 affects its directionality. J. Biol. Chem. 2015, 19, 620799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawin, K.E.; Mitchison, T.J. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc. Natl. Acad. Sci. USA 1995, 92, 4289–4293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goshima, G.; Vale, R.D. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell 2005, 16, 3896–3907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avunie-Masala, R.; Movshovich, N.; Nissenkorn, Y.; Gerson-Gurwitz, A.; Fridman, V.; Koivomagi, M.; Loog, M.; Hoyt, M.A.; Zaritsky, A.; Gheber, L. Phospho-regulation of kinesin-5 during anaphase spindle elongation. J. Cell Sci. 2011, 124, 873–878. [Google Scholar] [CrossRef] [Green Version]
- Drummond, D.R.; Hagan, I.M. Mutations in the bimC box of Cut7 indicate divergence of regulation within the bimC family of kinesin related proteins. J. Cell Sci. 1998, 111 Pt 7, 853–865. [Google Scholar] [CrossRef]
- Goldstein, A.; Siegler, N.; Goldman, D.; Judah, H.; Valk, E.; Koivomagi, M.; Loog, M.; Gheber, L. Three Cdk1 sites in the kinesin-5 Cin8 catalytic domain coordinate motor localization and activity during anaphase. Cell Mol. Life Sci. 2017, 74, 3395–3412. [Google Scholar] [CrossRef]
- Chee, M.K.; Haase, S.B. B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast. PLoS Genet. 2010, 6, e1000935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapira, O.; Goldstein, A.; Al-Bassam, J.; Gheber, L. A potential physiological role for bi-directional motility and motor clustering of mitotic kinesin-5 Cin8 in yeast mitosis. J. Cell Sci. 2017, 130, 725–734. [Google Scholar] [CrossRef] [Green Version]
- Blangy, A.; Arnaud, L.; Nigg, E.A. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J. Biol. Chem. 1997, 272, 19418–19424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahu, J.; Olichon, A.; Hentrich, C.; Schek, H.; Drinjakovic, J.; Zhang, C.; Doherty-Kirby, A.; Lajoie, G.; Surrey, T. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS ONE 2008, 3, e3936. [Google Scholar] [CrossRef] [Green Version]
- Olsen, J.V.; Vermeulen, M.; Santamaria, A.; Kumar, C.; Miller, M.L.; Jensen, L.J.; Gnad, F.; Cox, J.; Jensen, T.S.; Nigg, E.A.; et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010, 3, ra3. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Hegarat, N.; Vesely, C.; Roseboom, I.; Larch, C.; Streicher, H.; Straatman, K.; Flynn, H.; Skehel, M.; Hirota, T.; et al. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 2011, 30, 2233–2245. [Google Scholar] [CrossRef] [Green Version]
- Garcia, K.; Stumpff, J.; Duncan, T.; Su, T.T. Tyrosines in the kinesin-5 head domain are necessary for phosphorylation by Wee1 and for mitotic spindle integrity. Curr. Biol. 2009, 19, 1670–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bickel, K.G.; Mann, B.J.; Waitzman, J.S.; Poor, T.A.; Rice, S.E.; Wadsworth, P. Src family kinase phosphorylation of the motor domain of the human kinesin-5, Eg5. Cytoskeleton 2017, 74, 317–330. [Google Scholar] [CrossRef]
- Giet, R.; Uzbekov, R.; Cubizolles, F.; Le Guellec, K.; Prigent, C. The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J. Biol. Chem. 1999, 274, 15005–15013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, J.D.; Han, Z.; Schumacher, J.M. The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol. Biol. Cell 2005, 16, 742–756. [Google Scholar] [CrossRef] [Green Version]
- Rapley, J.; Nicolas, M.; Groen, A.; Regue, L.; Bertran, M.T.; Caelles, C.; Avruch, J.; Roig, J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J. Cell Sci. 2008, 121, 3912–3921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertran, M.T.; Sdelci, S.; Regue, L.; Avruch, J.; Caelles, C.; Roig, J. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 2011, 30, 2634–2647. [Google Scholar] [CrossRef] [Green Version]
- Eibes, S.; Gallisa-Sune, N.; Rosas-Salvans, M.; Martinez-Delgado, P.; Vernos, I.; Roig, J. Nek9 phosphorylation defines a new role for TPX2 in Eg5-dependent centrosome separation before nuclear envelope breakdown. Curr. Biol. 2018, 28, 121–129.e124. [Google Scholar] [CrossRef] [Green Version]
- Case, R.B.; Rice, S.; Hart, C.L.; Ly, B.; Vale, R.D. Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr. Biol. 2000, 10, 157–160. [Google Scholar] [CrossRef] [Green Version]
- Endow, S.A.; Higuchi, H. A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature 2000, 406, 913–916. [Google Scholar] [CrossRef]
- Endow, S.A.; Waligora, K.W. Determinants of kinesin motor polarity. Science 1998, 281, 1200–1202. [Google Scholar] [CrossRef] [Green Version]
- Rice, S.; Lin, A.W.; Safer, D.; Hart, C.L.; Naber, N.; Carragher, B.O.; Cain, S.M.; Pechatnikova, E.; Wilson-Kubalek, E.M.; Whittaker, M.; et al. A structural change in the kinesin motor protein that drives motility. Nature 1999, 402, 778–784. [Google Scholar] [CrossRef]
- Vinogradova, M.V.; Reddy, V.S.; Reddy, A.S.; Sablin, E.P.; Fletterick, R.J. Crystal structure of kinesin regulated by Ca(2+)-calmodulin. J. Biol. Chem. 2004, 279, 23504–23509. [Google Scholar] [CrossRef] [Green Version]
- Goulet, A.; Behnke-Parks, W.M.; Sindelar, C.V.; Major, J.; Rosenfeld, S.S.; Moores, C.A. The structural basis of force generation by the mitotic motor kinesin-5. J. Biol. Chem. 2012, 287, 44654–44666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sablin, E.P.; Case, R.B.; Dai, S.C.; Hart, C.L.; Ruby, A.; Vale, R.D.; Fletterick, R.J. Direction determination in the minus-end-directed kinesin motor ncd. Nature 1998, 395, 813–816. [Google Scholar] [CrossRef] [PubMed]
- Duselder, A.; Thiede, C.; Schmidt, C.F.; Lakamper, S. Neck-linker length dependence of processive kinesin-5 motility. J. Mol. Biol. 2012, 423, 159–168. [Google Scholar] [CrossRef]
- Endow, S.A. Determinants of molecular motor directionality. Nat. Cell Biol. 1999, 1, E163–E167. [Google Scholar] [CrossRef]
- Thiede, C.; Lakämper, S.; Wessel, A.D.; Kramer, S.; Schmidt, C.F. A chimeric kinesin-1 head/kinesin-5 tail motor switches between diffusive and processive motility. Biophys. J. 2013, 104, 432–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henningsen, U.; Schliwa, M. Reversal in the direction of movement of a molecular motor. Nature 1997, 389, 93–96. [Google Scholar] [CrossRef]
- Wade, R.H.; Kozielski, F. Structural links to kinesin directionality and movement. Nat. Struct. Biol. 2000, 7, 456–460. [Google Scholar] [CrossRef]
- Case, R.B.; Pierce, D.W.; Hom-Booher, N.; Hart, C.L.; Vale, R.D. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell 1997, 90, 959–966. [Google Scholar] [CrossRef] [Green Version]
- Shastry, S.; Hancock, W.O. Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. USA 2011, 108, 16253–16258. [Google Scholar] [CrossRef] [Green Version]
- Sindelar, C.V.; Downing, K.H. An atomic-level mechanism for activation of the kinesin molecular motors. Proc. Natl. Acad. Sci. USA 2010, 107, 4111–4116. [Google Scholar] [CrossRef] [Green Version]
- Rosenfeld, S.S.; Jefferson, G.M.; King, P.H. ATP reorients the neck linker of kinesin in two sequential steps. J. Biol. Chem. 2001, 276, 40167–40174. [Google Scholar] [CrossRef] [Green Version]
- Rosenfeld, S.S.; Xing, J.; Jefferson, G.M.; King, P.H. Docking and rolling, a model of how the mitotic motor Eg5 works. J. Biol. Chem. 2005, 280, 35684–35695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, J.; Anderson, R.; Guo, J.; Beraud, C.; Fletterick, R.; Sakowicz, R. Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J. Biol. Chem. 2001, 276, 25496–25502. [Google Scholar] [CrossRef] [Green Version]
- Larson, A.G.; Naber, N.; Cooke, R.; Pate, E.; Rice, S.E. The conserved L5 loop establishes the pre-powerstroke conformation of the Kinesin-5 motor, eg5. Biophys. J. 2010, 98, 2619–2627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulet, A.; Major, J.; Jun, Y.; Gross, S.P.; Rosenfeld, S.S.; Moores, C.A. Comprehensive structural model of the mechanochemical cycle of a mitotic motor highlights molecular adaptations in the kinesin family. Proc. Natl. Acad. Sci. USA 2014, 111, 1837–1842. [Google Scholar] [CrossRef] [Green Version]
- Khalil, A.S.; Appleyard, D.C.; Labno, A.K.; Georges, A.; Karplus, M.; Belcher, A.M.; Hwang, W.; Lang, M.J. Kinesin’s cover-neck bundle folds forward to generate force. Proc. Natl. Acad. Sci. USA 2008, 105, 19247–19252. [Google Scholar] [CrossRef] [Green Version]
- Hwang, W.; Lang, M.J.; Karplus, M. Force generation in kinesin hinges on cover-neck bundle formation. Structure 2008, 16, 62–71. [Google Scholar] [CrossRef] [Green Version]
- Geng, Y.-Z.; Li, T.; Ji, Q.; Yan, S. Simulation study of interactions between kinesin’s neck linker and motor domain. Cell. Mol. Bioeng. 2014, 7, 99–105. [Google Scholar] [CrossRef]
- Yi-Zhao, G.; Qing, J.; Shu-Xia, L.; Shi-Wei, Y. Initial conformation of kinesin’s neck linker. Chin. Phys. B 2014, 23, 108701. [Google Scholar]
- Muretta, J.M.; Jun, Y.; Gross, S.P.; Major, J.; Thomas, D.D.; Rosenfeld, S.S. The structural kinetics of switch-1 and the neck linker explain the functions of kinesin-1 and Eg5. Proc. Natl. Acad. Sci. USA 2015, 112, E6606–E6613. [Google Scholar] [CrossRef] [Green Version]
- Von Loeffelholz, O.; Pena, A.; Drummond, D.R.; Cross, R.; Moores, C.A. Cryo-EM structure (4.5-A) of yeast kinesin-5-microtubule complex reveals a distinct binding footprint and mechanism of drug resistance. J. Mol. Biol. 2019, 431, 864–872. [Google Scholar] [CrossRef]
- Behnke-Parks, W.M.; Vendome, J.; Honig, B.; Maliga, Z.; Moores, C.; Rosenfeld, S.S. Loop L5 acts as a conformational latch in the mitotic kinesin Eg5. J. Biol. Chem. 2011, 286, 5242–5253. [Google Scholar] [CrossRef] [Green Version]
- Waitzman, J.S.; Larson, A.G.; Cochran, J.C.; Naber, N.; Cooke, R.; Jon Kull, F.; Pate, E.; Rice, S.E. The loop 5 element structurally and kinetically coordinates dimers of the human kinesin-5, Eg5. Biophys. J. 2011, 101, 2760–2769. [Google Scholar] [CrossRef] [Green Version]
- Cochran, J.C.; Gilbert, S.P. ATPase mechanism of Eg5 in the absence of microtubules: Insight into microtubule activation and allosteric inhibition by monastrol. Biochemistry 2005, 44, 16633–16648. [Google Scholar] [CrossRef] [Green Version]
- Maliga, Z.; Xing, J.; Cheung, H.; Juszczak, L.J.; Friedman, J.M.; Rosenfeld, S.S. A pathway of structural changes produced by monastrol binding to Eg5. J. Biol. Chem. 2006, 281, 7977–7982. [Google Scholar] [CrossRef] [Green Version]
- Krzysiak, T.C.; Gilbert, S.P. Dimeric Eg5 maintains processivity through alternating-site catalysis with rate-limiting ATP hydrolysis. J. Biol. Chem. 2006, 281, 39444–39454. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Sardana, V.; Xu, B.; Homnick, C.; Halczenko, W.; Buser, C.A.; Schaber, M.; Hartman, G.D.; Huber, H.E.; Kuo, L.C. Inhibition of a mitotic motor protein: Where, how, and conformational consequences. J. Mol. Biol. 2004, 335, 547–554. [Google Scholar] [CrossRef] [Green Version]
- Cochran, J.C.; Krzysiak, T.C.; Gilbert, S.P. Pathway of ATP hydrolysis by monomeric kinesin Eg5. Biochemistry 2006, 45, 12334–12344. [Google Scholar] [CrossRef] [Green Version]
- Lakamper, S.; Thiede, C.; Duselder, A.; Reiter, S.; Korneev, M.J.; Kapitein, L.C.; Peterman, E.J.; Schmidt, C.F. The effect of monastrol on the processive motility of a dimeric kinesin-5 head/kinesin-1 stalk chimera. J. Mol. Biol. 2010, 399, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Asraf, H.; Avunie-Masala, R.; Hershfinkel, M.; Gheber, L. Mitotic slippage and expression of survivin are linked to differential sensitivity of human cancer cell-lines to the Kinesin-5 inhibitor monastrol. PLoS ONE 2015, 10, e0129255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leizerman, I.; Avunie-Masala, R.; Elkabets, M.; Fich, A.; Gheber, L. Differential effects of monastrol in two human cell lines. Cell Mol. Life Sci. 2004, 61, 2060–2070. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.G.; Torrent, M.; Williams, O.; Hamilton, K.A.; Buser, C.A. Characterization of inhibitor binding to human kinesin spindle protein by site-directed mutagenesis. Arch. Biochem. Biophys. 2009, 484, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Brier, S.; Lemaire, D.; DeBonis, S.; Forest, E.; Kozielski, F. Molecular dissection of the inhibitor binding pocket of mitotic kinesin Eg5 reveals mutants that confer resistance to antimitotic agents. J. Mol. Biol. 2006, 360, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Maliga, Z.; Mitchison, T.J. Small-molecule and mutational analysis of allosteric Eg5 inhibition by monastrol. BMC Chem. Biol. 2006, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.D.; Buckley, R.; Learman, S.; Richard, J.; Parke, C.; Worthylake, D.K.; Wojcik, E.J.; Walker, R.A.; Kim, S. Allosteric drug discrimination is coupled to mechanochemical changes in the kinesin-5 motor core. J. Biol. Chem. 2010, 285, 18650–18661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodey, A.J.; Kikkawa, M.; Moores, C.A. 9-Angstrom structure of a microtubule-bound mitotic motor. J. Mol. Biol. 2009, 388, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Sablin, E.P.; Kull, F.J.; Cooke, R.; Vale, R.D.; Fletterick, R.J. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature 1996, 380, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Kozielski, F.; Sack, S.; Marx, A.; Thormahlen, M.; Schonbrunn, E.; Biou, V.; Thompson, A.; Mandelkow, E.M.; Mandelkow, E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell 1997, 91, 985–994. [Google Scholar] [CrossRef]
- Gigant, B.; Wang, W.; Dreier, B.; Jiang, Q.; Pecqueur, L.; Pluckthun, A.; Wang, C.; Knossow, M. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat. Struct. Mol. Biol. 2013, 20, 1001–1007. [Google Scholar] [CrossRef]
- Bell, K.M.; Cha, H.K.; Sindelar, C.V.; Cochran, J.C. The yeast kinesin-5 Cin8 interacts with the microtubule in a noncanonical manner. J. Biol. Chem. 2017, 12, 797662. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, M.; Maruyama, Y.; Sugawa, M.; Yajima, J. Characterization of the motility of monomeric kinesin-5/Cin8. Biochem. Biophys. Res. Commun. 2021, 555, 115–120. [Google Scholar] [CrossRef]
- Cochran, J.C.; Sontag, C.A.; Maliga, Z.; Kapoor, T.M.; Correia, J.J.; Gilbert, S.P. Mechanistic analysis of the mitotic kinesin Eg5. J. Biol. Chem. 2004, 279, 38861–38870. [Google Scholar] [CrossRef] [Green Version]
- Cross, R.A. The kinetic mechanism of kinesin. Trends Biochem. Sci. 2004, 29, 301–309. [Google Scholar] [CrossRef]
- Mann, B.J.; Wadsworth, P. Kinesin-5 regulation and function in mitosis. Trends Cell Biol. 2019, 29, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, E.J.; Buckley, R.S.; Richard, J.; Liu, L.; Huckaba, T.M.; Kim, S. Kinesin-5: Cross-bridging mechanism to targeted clinical therapy. Gene 2013, 531, 133–149. [Google Scholar] [CrossRef] [Green Version]
- Kaseda, K.; Crevel, I.; Hirose, K.; Cross, R.A. Single-headed mode of kinesin-5. EMBO Rep. 2008, 9, 761–765. [Google Scholar] [CrossRef] [Green Version]
- Sadakane, K.; Takaichi, M.; Maruta, S. Photo-control of the mitotic kinesin Eg5 using a novel photochromic inhibitor composed of a spiropyran derivative. J. Biochem. 2018, 164, 239–246. [Google Scholar] [CrossRef]
- Yajima, J.; Mizutani, K.; Nishizaka, T. A torque component present in mitotic kinesin Eg5 revealed by three-dimensional tracking. Nat. Struct. Mol. Biol. 2008, 15, 1119–1121. [Google Scholar] [CrossRef]
- Gheber, L.; Kuo, S.C.; Hoyt, M.A. Motile properties of the kinesin-related Cin8p spindle motor extracted from Saccharomyces cerevisiae cells. J. Biol. Chem. 1999, 274, 9564–9572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapitein, L.C.; Kwok, B.H.; Weinger, J.S.; Schmidt, C.F.; Kapoor, T.M.; Peterman, E.J. Microtubule cross-linking triggers the directional motility of kinesin-5. J. Cell Biol. 2008, 182, 421–428. [Google Scholar] [CrossRef] [Green Version]
- Jaud, J.; Bathe, F.; Schliwa, M.; Rief, M.; Woehlke, G. Flexibility of the neck domain enhances Kinesin-1 motility under load. Biophys. J. 2006, 91, 1407–1412. [Google Scholar] [CrossRef] [Green Version]
- Cohn, S.A.; Ingold, A.L.; Scholey, J.M. Quantitative analysis of sea urchin egg kinesin-driven microtubule motility. J. Biol. Chem. 1989, 264, 4290–4297. [Google Scholar] [CrossRef]
- Howard, J.; Hudspeth, A.J.; Vale, R.D. Movement of microtubules by single kinesin molecules. Nature 1989, 342, 154–158. [Google Scholar] [CrossRef]
- Chen, Y.; Hancock, W.O. Kinesin-5 is a microtubule polymerase. Nat. Commun. 2015, 6, 8160. [Google Scholar] [CrossRef] [Green Version]
- Valentine, M.T.; Fordyce, P.M.; Krzysiak, T.C.; Gilbert, S.P.; Block, S.M. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 2006, 8, 470–476. [Google Scholar] [CrossRef]
- Vale, R.D.; Funatsu, T.; Pierce, D.W.; Romberg, L.; Harada, Y.; Yanagida, T. Direct observation of single kinesin molecules moving along microtubules. Nature 1996, 380, 451–453. [Google Scholar] [CrossRef] [Green Version]
- Valentine, M.T.; Block, S.M. Force and premature binding of ADP can regulate the processivity of individual Eg5 dimers. Biophys. J. 2009, 97, 1671–1677. [Google Scholar] [CrossRef] [Green Version]
- Shimamoto, Y.; Forth, S.; Kapoor, T.M. Measuring pushing and braking forces generated by ensembles of kinesin-5 crosslinking two microtubules. Dev. Cell 2015, 34, 669–681. [Google Scholar] [CrossRef] [Green Version]
- Crevel, I.M.; Alonso, M.C.; Cross, R.A. Monastrol stabilises an attached low-friction mode of Eg5. Curr. Biol. 2004, 14, R411–R412. [Google Scholar] [CrossRef] [Green Version]
- Civelekoglu-Scholey, G.; Scholey, J.M. Mitotic motors: Kinesin-5 takes a brake. Curr. Biol. 2007, 17, R544–R547. [Google Scholar] [CrossRef] [Green Version]
- Rozelle, D.K.; Hansen, S.D.; Kaplan, K.B. Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. J. Cell Biol. 2011, 193, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Saunders, A.M.; Powers, J.; Strome, S.; Saxton, W.M. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr. Biol. 2007, 17, R453–R454. [Google Scholar] [CrossRef] [Green Version]
- Edamatsu, M. Molecular properties of the N-terminal extension of the fission yeast kinesin-5, Cut7. Genet. Mol. Res. 2016, 15, 15017799. [Google Scholar] [CrossRef] [PubMed]
- Britto, M.; Goulet, A.; Rizvi, S.; von Loeffelholz, O.; Moores, C.A.; Cross, R.A. Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism. Proc. Natl. Acad. Sci. USA 2016, 113, E7483–E7489. [Google Scholar] [CrossRef] [Green Version]
- Roccuzzo, M.; Visintin, C.; Tili, F.; Visintin, R. FEAR-mediated activation of Cdc14 is the limiting step for spindle elongation and anaphase progression. Nat. Cell Biol. 2015, 17, 251–261. [Google Scholar] [CrossRef]
- Shapira, O.; Gheber, L. Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain. Sci. Rep. 2016, 6, 25597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fallesen, T.; Roostalu, J.; Duellberg, C.; Pruessner, G.; Surrey, T. Ensembles of bidirectional kinesin cin8 produce additive forces in both directions of movement. Biophys. J. 2017, 113, 2055–2067. [Google Scholar] [CrossRef]
- Saito, N.; Kaneko, K. Embedding dual function into molecular motors through collective motion. Sci. Rep. 2017, 7, 44288. [Google Scholar] [CrossRef] [Green Version]
- Thiede, C.; Fridman, V.; Gerson-Gurwitz, A.; Gheber, L.; Schmidt, C.F. Regulation of bi-directional movement of single kinesin-5 Cin8 molecules. Bioarchitecture 2012, 2, 70–74. [Google Scholar] [CrossRef] [Green Version]
- Pandey, H.; Reithmann, E.; Goldstein-Levitin, A.; Al-Bassam, J.; Frey, E.; Gheber, L. Drag-induced directionality switching of kinesin-5 Cin8 revealed by cluster-motility analysis. Sci. Adv. 2021, 7, eabc1687. [Google Scholar] [CrossRef]
- Chen, G.Y.; Kang, Y.J.; Gayek, A.S.; Youyen, W.; Tuzel, E.; Ohi, R.; Hancock, W.O. Eg5 inhibitors have contrasting effects on microtubule stability and metaphase spindle integrity. ACS Chem. Biol. 2017, 12, 1038–1046. [Google Scholar] [CrossRef]
- Fridman, V.; Gerson-Gurwitz, A.; Movshovich, N.; Kupiec, M.; Gheber, L. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 2009, 10, 387–393. [Google Scholar] [CrossRef] [Green Version]
- Gardner, M.K.; Bouck, D.C.; Paliulis, L.V.; Meehl, J.B.; O’Toole, E.T.; Haase, J.; Soubry, A.; Joglekar, A.P.; Winey, M.; Salmon, E.D.; et al. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell 2008, 135, 894–906. [Google Scholar] [CrossRef] [Green Version]
- Tubman, E.; He, Y.; Hays, T.S.; Odde, D.J. Kinesin-5 mediated chromosome congression in insect spindles. Cell. Mol. Bioeng. 2018, 11, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Walczak, C.E.; Heald, R. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 2008, 265, 111–158. [Google Scholar] [CrossRef]
- Dumont, S.; Mitchison, T.J. Force and length in the mitotic spindle. Curr. Biol. 2009, 19, R749–R761. [Google Scholar] [CrossRef] [Green Version]
- Prosser, S.L.; Pelletier, L. Mitotic spindle assembly in animal cells: A fine balancing act. Nat. Rev. Mol. Cell Biol. 2017, 18, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Maddox, P.; Straight, A.; Coughlin, P.; Mitchison, T.J.; Salmon, E.D. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: Implications for spindle mechanics. J. Cell Biol. 2003, 162, 377–382. [Google Scholar] [CrossRef] [Green Version]
- McDonald, K.L.; O’Toole, E.T.; Mastronarde, D.N.; McIntosh, J.R. Kinetochore microtubules in PTK cells. J. Cell Biol. 1992, 118, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Grill, S.W.; Howard, J.; Schaffer, E.; Stelzer, E.H.; Hyman, A.A. The distribution of active force generators controls mitotic spindle position. Science 2003, 301, 518–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connell, C.B.; Wang, Y.L. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol. Biol. Cell 2000, 11, 1765–1774. [Google Scholar] [CrossRef] [Green Version]
- Sheeman, B.; Carvalho, P.; Sagot, I.; Geiser, J.; Kho, D.; Hoyt, M.A.; Pellman, D. Determinants of S. cerevisiae dynein localization and activation: Implications for the mechanism of spindle positioning. Curr. Biol. 2003, 13, 364–372. [Google Scholar] [CrossRef] [Green Version]
- Mastronarde, D.N.; McDonald, K.L.; Ding, R.; McIntosh, J.R. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 1993, 123, 1475–1489. [Google Scholar] [CrossRef]
- Winey, M.; Mamay, C.L.; O’Toole, E.T.; Mastronarde, D.N.; Giddings, T.H., Jr.; McDonald, K.L.; McIntosh, J.R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995, 129, 1601–1615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazarova, E.; O’Toole, E.; Kaitna, S.; Francois, P.; Winey, M.; Vogel, J. Distinct roles for antiparallel microtubule pairing and overlap during early spindle assembly. Mol. Biol. Cell 2013, 24, 3238–3250. [Google Scholar] [CrossRef]
- Kapoor, T.M. Metaphase spindle assembly. Biology 2017, 6, 8. [Google Scholar] [CrossRef]
- Tanenbaum, M.E.; Medema, R.H. Mechanisms of centrosome separation and bipolar spindle assembly. Dev. Cell 2010, 19, 797–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, H.H.; Zhang, T.; Surana, U. Regulation of centrosome separation in yeast and vertebrates: Common threads. Trends Cell Biol. 2009, 19, 325–333. [Google Scholar] [CrossRef]
- Rincon, S.A.; Lamson, A.; Blackwell, R.; Syrovatkina, V.; Fraisier, V.; Paoletti, A.; Betterton, M.D.; Tran, P.T. Kinesin-5-independent mitotic spindle assembly requires the antiparallel microtubule crosslinker Ase1 in fission yeast. Nat. Commun. 2017, 8, 15286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sazer, S.; Lynch, M.; Needleman, D. Deciphering the evolutionary history of open and closed mitosis. Curr. Biol. 2014, 24, R1099–R1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boettcher, B.; Barral, Y. The cell biology of open and closed mitosis. Nucleus 2013, 4, 160–165. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y. A membranous spindle matrix orchestrates cell division. Nat. Rev. Mol. Cell Biol. 2010, 11, 529–535. [Google Scholar] [CrossRef] [Green Version]
- Vaisberg, E.A.; Koonce, M.P.; McIntosh, J.R. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J. Cell Biol. 1993, 123, 849–858. [Google Scholar] [CrossRef] [Green Version]
- Gönczy, P.; Pichler, S.; Kirkham, M.; Hyman, A.A. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage caenorhabditis elegans embryo. J. Cell Biol. 1999, 147, 135–150. [Google Scholar] [CrossRef]
- Robinson, J.T.; Wojcik, E.J.; Sanders, M.A.; McGrail, M.; Hays, T.S. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 1999, 146, 597–608. [Google Scholar] [CrossRef] [Green Version]
- Sharp, D.J.; Rogers, G.C.; Scholey, J.M. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2000, 2, 922–930. [Google Scholar] [CrossRef]
- Salina, D.; Bodoor, K.; Eckley, D.M.; Schroer, T.A.; Rattner, J.B.; Burke, B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 2002, 108, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Cytrynbaum, E.N.; Scholey, J.M.; Mogilner, A. A force balance model of early spindle pole separation in Drosophila embryos. Biophys. J. 2003, 84, 757–769. [Google Scholar] [CrossRef] [Green Version]
- Surana, U.; Amon, A.; Dowzer, C.; McGrew, J.; Byers, B.; Nasmyth, K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993, 12, 1969–1978. [Google Scholar] [CrossRef]
- Alexandru, G.; Uhlmann, F.; Mechtler, K.; Poupart, M.A.; Nasmyth, K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 2001, 105, 459–472. [Google Scholar] [CrossRef] [Green Version]
- D’Amours, D.; Amon, A. At the interface between signaling and executing anaphase--Cdc14 and the FEAR network. Genes Dev. 2004, 18, 2581–2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asbury, C.L. Anaphase A: Disassembling microtubules move chromosomes toward spindle poles. Biology 2017, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Meadows, J.C.; Millar, J.B. Sharpening the anaphase switch. Biochem. Soc. Trans 2015, 43, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Winey, M.; Bloom, K. Mitotic spindle form and function. Genetics 2012, 190, 1197–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannigan, A.; Scheible, W.R.; Lukowitz, W.; Fagerstrom, C.; Wadsworth, P.; Somerville, C.; Baskin, T.I. A conserved role for kinesin-5 in plant mitosis. J. Cell Sci. 2007, 120, 2819–2827. [Google Scholar] [CrossRef] [Green Version]
- Castillo, A.; Justice, M.J. The kinesin related motor protein, Eg5, is essential for maintenance of pre-implantation embryogenesis. Biochem. Biophys. Res. Commun. 2007, 357, 694–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusan, N.M.; Tulu, U.S.; Fagerstrom, C.; Wadsworth, P. Reorganization of the microtubule array in prophase/prometaphase requires cytoplasmic dynein-dependent microtubule transport. J. Cell Biol. 2002, 158, 997–1003. [Google Scholar] [CrossRef] [Green Version]
- Crasta, K.; Huang, P.; Morgan, G.; Winey, M.; Surana, U. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 2006, 25, 2551–2563. [Google Scholar] [CrossRef] [Green Version]
- Tikhonenko, I.; Nag, D.K.; Martin, N.; Koonce, M.P. Kinesin-5 is not essential for mitotic spindle elongation in Dictyostelium. Cell Motil. Cytoskelet. 2008, 65, 853–862. [Google Scholar] [CrossRef]
- Miki, T.; Naito, H.; Nishina, M.; Goshima, G. Endogenous localizome identifies 43 mitotic kinesins in a plant cell. Proc. Natl. Acad. Sci. USA 2014, 111, E1053–E1061. [Google Scholar] [CrossRef] [Green Version]
- Saunders, W.S.; Koshland, D.; Eshel, D.; Gibbons, I.R.; Hoyt, M.A. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 1995, 128, 617–624. [Google Scholar] [CrossRef] [Green Version]
- Movshovich, N.; Fridman, V.; Gerson-Gurwitz, A.; Shumacher, I.; Gertsberg, I.; Fich, A.; Hoyt, M.A.; Katz, B.; Gheber, L. Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. J. Cell Sci. 2008, 121, 2529–2539. [Google Scholar] [CrossRef] [Green Version]
- Gerson-Gurwitz, A.; Movshovich, N.; Avunie, R.; Fridman, V.; Moyal, K.; Katz, B.; Hoyt, M.A.; Gheber, L. Mid-anaphase arrest in S. cerevisiae cells eliminated for the function of Cin8 and dynein. Cell Mol. Life Sci. 2009, 66, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Straight, A.F.; Sedat, J.W.; Murray, A.W. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 1998, 143, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Sharp, D.J.; Yu, K.R.; Sisson, J.C.; Sullivan, W.; Scholey, J.M. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1999, 1, 51–54. [Google Scholar] [CrossRef]
- Zhang, T.; Nirantar, S.; Lim, H.H.; Sinha, I.; Surana, U. DNA damage checkpoint maintains CDH1 in an active state to inhibit anaphase progression. Dev. Cell 2009, 17, 541–551. [Google Scholar] [CrossRef] [Green Version]
- Collins, E.; Mann, B.J.; Wadsworth, P. Eg5 restricts anaphase B spindle elongation in mammalian cells. Cytoskeleton 2014, 71, 136–144. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Ghosh, S.K.; Jayaram, M. The selfish yeast plasmid uses the nuclear motor Kip1p but not Cin8p for its localization and equal segregation. J. Cell Biol. 2009, 185, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, H.K.; Rizvi, S.M.; Rathore, I.; Ghosh, S.K. Microtubule-associated proteins, Bik1 and Bim1, are required for faithful partitioning of the endogenous 2 micron plasmids in budding yeast. Mol. Microbiol. 2017, 103, 1046–1064. [Google Scholar] [CrossRef] [Green Version]
- Rizvi, S.M.A.; Prajapati, H.K.; Ghosh, S.K. The 2 micron plasmid: A selfish genetic element with an optimized survival strategy within Saccharomyces cerevisiae. Curr. Genet. 2017, 8, 17–719. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Gupta, A.; Long, S.K.; Evans, R.; Badger, B.L.; Salmon, E.D.; Biggins, S.; Bloom, K. A Kinesin-5, Cin8, recruits protein phosphatase 1 to kinetochores and regulates chromosome segregation. Curr. Biol. 2018, 28, 2697–2704.e2693. [Google Scholar] [CrossRef] [PubMed]
- Wargacki, M.M.; Tay, J.C.; Muller, E.G.; Asbury, C.L.; Davis, T.N. Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle 2010, 9, 2581–2588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tytell, J.D.; Sorger, P.K. Analysis of kinesin motor function at budding yeast kinetochores. JCB 2006, 172, 861–874. [Google Scholar] [CrossRef] [Green Version]
- De Wulf, P.; McAinsh, A.D.; Sorger, P.K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003, 17, 2902–2921. [Google Scholar] [CrossRef] [Green Version]
- Akera, T.; Goto, Y.; Sato, M.; Yamamoto, M.; Watanabe, Y. Mad1 promotes chromosome congression by anchoring a kinesin motor to the kinetochore. Nat. Cell Biol. 2015, 17, 1124–1133. [Google Scholar] [CrossRef]
- Hoyt, M.A.; He, L.; Totis, L.; Saunders, W.S. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics 1993, 135, 35–44. [Google Scholar] [CrossRef]
- Hagan, I.; Yanagida, M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature 1992, 356, 74–76. [Google Scholar] [CrossRef] [PubMed]
- FitzHarris, G. Anaphase B precedes anaphase A in the mouse egg. Curr. Biol. 2012, 22, 437–444. [Google Scholar] [CrossRef] [Green Version]
- FitzHarris, G. A shift from kinesin 5-dependent metaphase spindle function during preimplantation development in mouse. Development 2009, 136, 2111–2119. [Google Scholar] [CrossRef] [Green Version]
- Haque, S.A.; Hasaka, T.P.; Brooks, A.D.; Lobanov, P.V.; Baas, P.W. Monastrol, a prototype anti-cancer drug that inhibits a mitotic kinesin, induces rapid bursts of axonal outgrowth from cultured postmitotic neurons. Cell Motil. Cytoskelet. 2004, 58, 10–16. [Google Scholar] [CrossRef]
- Myers, K.A.; Baas, P.W. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J. Cell Biol. 2007, 178, 1081–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferenz, N.P.; Paul, R.; Fagerstrom, C.; Mogilner, A.; Wadsworth, P. Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr. Biol. 2009, 19, 1833–1838. [Google Scholar] [CrossRef] [Green Version]
- Ma, N.; Tulu, U.S.; Ferenz, N.P.; Fagerstrom, C.; Wilde, A.; Wadsworth, P. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol. Biol. Cell 2010, 21, 979–988. [Google Scholar] [CrossRef]
- Mitchison, T.J. Mechanism and function of poleward flux in Xenopus extract meiotic spindles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 623–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, D.T.; Perlman, Z.E.; Burbank, K.S.; Groen, A.C.; Mitchison, T.J. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J. Cell Biol. 2004, 167, 813–818. [Google Scholar] [CrossRef]
- Blangy, A.; Chaussepied, P.; Nigg, E.A. Rigor-type mutation in the kinesin-related protein HsEg5 changes its subcellular localization and induces microtubule bundling. Cell Motil. Cytoskelet. 1998, 40, 174–182. [Google Scholar] [CrossRef]
- Nahaboo, W.; Zouak, M.; Askjaer, P.; Delattre, M. Chromatids segregate without centrosomes during Caenorhabditis elegans mitosis in a Ran- and CLASP-dependent manner. Mol. Biol. Cell 2015, 26, 2020–2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brust-Mascher, I.; Civelekoglu-Scholey, G.; Kwon, M.; Mogilner, A.; Scholey, J.M. Model for anaphase B: Role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc. Natl. Acad. Sci. USA 2004, 101, 15938–15943. [Google Scholar] [CrossRef] [Green Version]
- Asada, T.; Kuriyama, R.; Shibaoka, H. TKRP125, a kinesin-related protein involved in the centrosome- independent organization of the cytokinetic apparatus in tobacco BY-2 cells. J. Cell Sci. 1997, 110, 179–189. [Google Scholar] [CrossRef]
- Barroso, C.; Chan, J.; Allan, V.; Doonan, J.; Hussey, P.; Lloyd, C. Two kinesin-related proteins associated with the cold-stable cytoskeleton of carrot cells: Characterization of a novel kinesin, DcKRP120-2. Plant J. 2000, 24, 859–868. [Google Scholar] [CrossRef]
- Saunders, W.; Lengyel, V.; Hoyt, M.A. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell 1997, 8, 1025–1033. [Google Scholar] [CrossRef] [Green Version]
- Civelekoglu-Scholey, G.; Tao, L.; Brust-Mascher, I.; Wollman, R.; Scholey, J.M. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J. Cell Biol. 2010, 188, 49–68. [Google Scholar] [CrossRef] [Green Version]
- Loughlin, R.; Heald, R.; Nedelec, F. A computational model predicts Xenopus meiotic spindle organization. J. Cell Biol. 2010, 191, 1239–1249. [Google Scholar] [CrossRef] [Green Version]
- Schaffner, S.C.; Jose, J.V. Biophysical model of self-organized spindle formation patterns without centrosomes and kinetochores. Proc. Natl. Acad. Sci. USA 2006, 103, 11166–11171. [Google Scholar] [CrossRef] [Green Version]
- Rogers, G.C.; Rogers, S.L.; Sharp, D.J. Spindle microtubules in flux. J. Cell Sci. 2005, 118, 1105–1116. [Google Scholar] [CrossRef] [Green Version]
- Maddox, P.S.; Bloom, K.S.; Salmon, E.D. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2000, 2, 36–41. [Google Scholar] [CrossRef]
- Mallavarapu, A.; Sawin, K.; Mitchison, T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr. Biol. 1999, 9, 1423–1426. [Google Scholar] [CrossRef] [Green Version]
- Gable, A.; Qiu, M.; Titus, J.; Balchand, S.; Ferenz, N.P.; Ma, N.; Collins, E.S.; Fagerstrom, C.; Ross, J.L.; Yang, G.; et al. Dynamic reorganization of Eg5 in the mammalian spindle throughout mitosis requires dynein and TPX2. Mol. Biol. Cell 2012, 23, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Uteng, M.; Hentrich, C.; Miura, K.; Bieling, P.; Surrey, T. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J. Cell Biol. 2008, 182, 715–726. [Google Scholar] [CrossRef] [Green Version]
- Cheerambathur, D.K.; Brust-Mascher, I.; Civelekoglu-Scholey, G.; Scholey, J.M. Dynamic partitioning of mitotic kinesin-5 cross-linkers between microtubule-bound and freely diffusing states. J. Cell Biol. 2008, 182, 429–436. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, T.M.; Mitchison, T.J. Eg5 is static in bipolar spindles relative to tubulin: Evidence for a static spindle matrix. J. Cell Biol. 2001, 154, 1125–1133. [Google Scholar] [CrossRef] [Green Version]
- Wittmann, T.; Wilm, M.; Karsenti, E.; Vernos, I. TPX2, A novel xenopus MAP involved in spindle pole organization. J. Cell Biol. 2000, 149, 1405–1418. [Google Scholar] [CrossRef]
- Helmke, K.J.; Heald, R. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J. Cell Biol. 2014, 206, 385–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, N.; Titus, J.; Gable, A.; Ross, J.L.; Wadsworth, P. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J. Cell Biol. 2011, 195, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Blackwell, R.; Edelmaier, C.; Sweezy-Schindler, O.; Lamson, A.; Gergely, Z.R.; O’Toole, E.; Crapo, A.; Hough, L.E.; McIntosh, J.R.; Glaser, M.A.; et al. Physical determinants of bipolar mitotic spindle assembly and stability in fission yeast. Sci. Adv. 2017, 3, e1601603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winters, L.; Ban, I.; Prelogović, M.; Kalinina, I.; Pavin, N.; Tolić, I.M. Pivoting of microtubules driven by minus-end-directed motors leads to spindle assembly. BMC Biol. 2019, 17, 42. [Google Scholar] [CrossRef] [PubMed]
- Fong, K.K.; Davis, T.N.; Asbury, C.L. Microtubule pivoting enables mitotic spindle assembly in S. cerevisiae. J. Cell Biol. 2021, 220, 202007193. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, E.T.; Winey, M.; McIntosh, J.R. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 1999, 10, 2017–2031. [Google Scholar] [CrossRef]
- Stoica, C.; Park, J.; Pare, J.M.; Willows, S.; Hobman, T.C. The Kinesin motor protein Cut7 regulates biogenesis and function of Ago1-complexes. Traffic 2010, 11, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, C.M.; Rattner, J.B. Expanding the role of HsEg5 within the mitotic and post-mitotic phases of the cell cycle. J. Cell Sci. 1998, 111, 2551–2561. [Google Scholar] [CrossRef]
- Bartoli, K.M.; Jakovljevic, J.; Woolford, J.L., Jr.; Saunders, W.S. Kinesin molecular motor Eg5 functions during polypeptide synthesis. Mol. Biol. Cell 2011, 22, 3420–3430. [Google Scholar] [CrossRef]
- Lin, S.; Liu, M.; Mozgova, O.I.; Yu, W.; Baas, P.W. Mitotic motors coregulate microtubule patterns in axons and dendrites. J. Neurosci. 2012, 32, 14033–14049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadar, V.C.; Ketschek, A.; Myers, K.A.; Gallo, G.; Baas, P.W. Kinesin-5 is essential for growth-cone turning. Curr. Biol. 2008, 18, 1972–1977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falnikar, A.; Tole, S.; Baas, P.W. Kinesin-5, a mitotic microtubule-associated motor protein, modulates neuronal migration. Mol. Biol. Cell 2011, 22, 1561–1574. [Google Scholar] [CrossRef]
- Kahn, O.I.; Sharma, V.; Gonzalez-Billault, C.; Baas, P.W. Effects of kinesin-5 inhibition on dendritic architecture and microtubule organization. Mol. Biol. Cell 2015, 26, 66–77. [Google Scholar] [CrossRef]
- Liu, M.; Ran, J.; Zhou, J. Non-canonical functions of the mitotic kinesin Eg5. Thorac. Cancer 2018, 9, 904–910. [Google Scholar] [CrossRef]
- Nasa, I.; Kettenbach, A.N. Coordination of protein kinase and phosphoprotein phosphatase activities in mitosis. Front. Cell Dev. Biol. 2018, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.T.; Poon, R.Y. How protein kinases co-ordinate mitosis in animal cells. Biochem. J. 2011, 435, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Fisher, D.; Krasinska, L.; Coudreuse, D.; Novak, B. Phosphorylation network dynamics in the control of cell cycle transitions. J. Cell Sci. 2012, 125, 4703–4711. [Google Scholar] [CrossRef] [Green Version]
- Murray, A.W. Recycling the cell cycle: Cyclins revisited. Cell 2004, 116, 221–234. [Google Scholar] [CrossRef] [Green Version]
- Stegmeier, F.; Amon, A. Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004, 38, 203–232. [Google Scholar] [CrossRef]
- Sullivan, M.; Morgan, D.O. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007, 8, 894–903. [Google Scholar] [CrossRef]
- Hemsley, R.; McCutcheon, S.; Doonan, J.; Lloyd, C. P34(cdc2) kinase is associated with cortical microtubules from higher plant protoplasts. FEBS Lett. 2001, 508, 157–161. [Google Scholar] [CrossRef] [Green Version]
- Hemerly, A.S.; Ferreira, P.; de Almeida Engler, J.; Van Montagu, M.; Engler, G.; Inze, D. cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 1993, 5, 1711–1723. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.R.; Giang, H.M.; Liu, B. A novel plant kinesin-related protein specifically associates with the phragmoplast organelles. Plant Cell 2001, 13, 2427–2439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, N.R.; Pereira, A.J.; Goldstein, L.S. Motor activity and mitotic spindle localization of the Drosophila kinesin-like protein KLP61F. Mol. Biol. Cell 1995, 6, 1563–1574. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, A.; Goldman, D.; Valk, E.; Loog, M.; Holt, L.J.; Gheber, L. Synthetic-evolution reveals narrow paths to regulation of the saccharomyces cerevisiae mitotic kinesin-5 Cin8. Int. J. Biol. Sci. 2019, 15, 1125–1138. [Google Scholar] [CrossRef]
- Muretta, J.M.; Reddy, B.J.N.; Scarabelli, G.; Thompson, A.F.; Jariwala, S.; Major, J.; Venere, M.; Rich, J.N.; Willard, B.; Thomas, D.D.; et al. A posttranslational modification of the mitotic kinesin Eg5 that enhances its mechanochemical coupling and alters its mitotic function. Proc. Natl. Acad. Sci. USA 2018, 115, E1779–E1788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olziersky, A.-M.; Labidi-Galy, S.I. Clinical development of anti-mitotic drugs in cancer. In Cell Division Machinery and Disease; Gotta, M., Meraldi, P., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 125–152. [Google Scholar]
- Mills, C.C.; Kolb, E.A.; Sampson, V.B. Recent Advances of Cell-Cycle Inhibitor Therapies for Pediatric Cancer. Cancer Res. 2017, 77, 6489–6498. [Google Scholar] [CrossRef] [Green Version]
- Myers, S.M.; Collins, I. Recent findings and future directions for interpolar mitotic kinesin inhibitors in cancer therapy. Future Med. Chem. 2016, 8, 463–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, M.A.; Wilson, L. Microtubules and actin filaments: Dynamic targets for cancer chemotherapy. Curr. Opin. Cell Biol. 1998, 10, 123–130. [Google Scholar] [CrossRef]
- Kaestner, P.; Bastians, H. Mitotic drug targets. J. Cell Biochem. 2010, 111, 258–265. [Google Scholar] [CrossRef]
- Liu, M.; Wang, X.; Yang, Y.; Li, D.; Ren, H.; Zhu, Q.; Chen, Q.; Han, S.; Hao, J.; Zhou, J. Ectopic expression of the microtubule-dependent motor protein Eg5 promotes pancreatic tumourigenesis. J. Pathol. 2010, 221, 221–228. [Google Scholar] [CrossRef]
- Exertier, P.; Javerzat, S.; Wang, B.; Franco, M.; Herbert, J.; Platonova, N.; Winandy, M.; Pujol, N.; Nivelles, O.; Ormenese, S.; et al. Impaired angiogenesis and tumor development by inhibition of the mitotic kinesin Eg5. Oncotarget 2013, 4, 2302–2316. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Lu, J.; Niu, Z.; Ding, K.; Bi, D.; Liu, S.; Li, J.; Wu, F.; Zhang, H.; Zhao, Z.; et al. A potent chemotherapeutic strategy with eg5 inhibitor against gemcitabine resistant bladder cancer. PLoS ONE 2015, 10, e0144484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wu, X.; Du, M.; Chen, X.; Ning, X.; Chen, H.; Wang, S.; Liu, J.; Liu, Z.; Li, R.; et al. Eg5 inhibitor YL001 induces mitotic arrest and inhibits tumor proliferation. Oncotarget 2017, 8, 42510–42524. [Google Scholar] [CrossRef] [Green Version]
- Pei, Y.Y.; Li, G.C.; Ran, J.; Wei, F.X. Kinesin family member 11 contributes to the progression and prognosis of human breast cancer. Oncol. Lett. 2017, 14, 6618–6626. [Google Scholar] [CrossRef] [Green Version]
- Daigo, K.; Takano, A.; Thang, P.M.; Yoshitake, Y.; Shinohara, M.; Tohnai, I.; Murakami, Y.; Maegawa, J.; Daigo, Y. Characterization of KIF11 as a novel prognostic biomarker and therapeutic target for oral cancer. Int. J. Oncol. 2018, 52, 155–165. [Google Scholar] [CrossRef] [Green Version]
- Muretta, J.M.; Behnke-Parks, W.M.; Major, J.; Petersen, K.J.; Goulet, A.; Moores, C.A.; Thomas, D.D.; Rosenfeld, S.S. Loop L5 assumes three distinct orientations during the ATPase cycle of the mitotic kinesin Eg5: A transient and time-resolved fluorescence study. J. Biol. Chem. 2013, 288, 34839–34849. [Google Scholar] [CrossRef] [Green Version]
- Gartner, M.; Sunder-Plassmann, N.; Seiler, J.; Utz, M.; Vernos, I.; Surrey, T.; Giannis, A. Development and biological evaluation of potent and specific inhibitors of mitotic Kinesin Eg5. ChemBioChem 2005, 6, 1173–1177. [Google Scholar] [CrossRef]
- Huszar, D.; Theoclitou, M.E.; Skolnik, J.; Herbst, R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 2009, 28, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gong, H.; Huang, K. Oncogenic role of kinesin proteins and targeting kinesin therapy. Cancer Sci. 2013, 104, 651–656. [Google Scholar] [CrossRef] [Green Version]
- Brier, S.; Lemaire, D.; Debonis, S.; Forest, E.; Kozielski, F. Identification of the protein binding region of S-trityl-L-cysteine, a new potent inhibitor of the mitotic kinesin Eg5. Biochemistry 2004, 43, 13072–13082. [Google Scholar] [CrossRef]
- Sherin, L.; Farwa, S.; Sohail, A.; Li, Z.; Beg, O.A. Cancer drug therapy and stochastic modeling of “nano-motors”. Int. J. Nanomed. 2018, 13, 6429–6440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Zhai, L.; Lu, W.; Boohaker, R.J.; Padmalayam, I.; Li, Y. Discovery of novel allosteric eg5 inhibitors through structure-based virtual screening. Chem. Biol. Drug Des. 2016, 88, 178–187. [Google Scholar] [CrossRef] [Green Version]
- Sarli, V.; Giannis, A. Targeting the kinesin spindle protein: Basic principles and clinical implications. Clin. Cancer Res. 2008, 14, 7583–7587. [Google Scholar] [CrossRef] [Green Version]
- Shah, J.J.; Kaufman, J.L.; Zonder, J.A.; Cohen, A.D.; Bensinger, W.I.; Hilder, B.W.; Rush, S.A.; Walker, D.H.; Tunquist, B.J.; Litwiler, K.S.; et al. A Phase 1 and 2 study of Filanesib alone and in combination with low-dose dexamethasone in relapsed/refractory multiple myeloma. Cancer 2017, 123, 4617–4630. [Google Scholar] [CrossRef]
- Chan, K.S.; Koh, C.G.; Li, H.Y. Mitosis-targeted anti-cancer therapies: Where they stand. Cell Death Dis. 2012, 3, e411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rath, O.; Kozielski, F. Kinesins and cancer. Nat. Rev. Cancer 2012, 12, 527–539. [Google Scholar] [CrossRef]
- El-Nassan, H.B. Advances in the discovery of kinesin spindle protein (Eg5) inhibitors as antitumor agents. Eur. J. Med. Chem. 2013, 62, 614–631. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, G.; Tatrai, P.; Gergely, F. Hitting the brakes: Targeting microtubule motors in cancer. Br. J. Cancer 2015, 113, 693–698. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Garcia, S.; San-Segundo, L.; Gonzalez-Mendez, L.; Corchete, L.A.; Misiewicz-Krzeminska, I.; Martin-Sanchez, M.; Lopez-Iglesias, A.A.; Algarin, E.M.; Mogollon, P.; Diaz-Tejedor, A.; et al. The kinesin spindle protein inhibitor filanesib enhances the activity of pomalidomide and dexamethasone in multiple myeloma. Haematologica 2017, 102, 2113–2124. [Google Scholar] [CrossRef]
- Sturgill, E.G.; Norris, S.R.; Guo, Y.; Ohi, R. Kinesin-5 inhibitor resistance is driven by kinesin-12. J. Cell Biol. 2016, 213, 213–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milic, B.; Chakraborty, A.; Han, K.; Bassik, M.C.; Block, S.M. KIF15 nanomechanics and kinesin inhibitors, with implications for cancer chemotherapeutics. Proc. Natl. Acad. Sci. USA 2018, 115, E4613–E4622. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, H.; Popov, M.; Goldstein-Levitin, A.; Gheber, L. Mechanisms by Which Kinesin-5 Motors Perform Their Multiple Intracellular Functions. Int. J. Mol. Sci. 2021, 22, 6420. https://doi.org/10.3390/ijms22126420
Pandey H, Popov M, Goldstein-Levitin A, Gheber L. Mechanisms by Which Kinesin-5 Motors Perform Their Multiple Intracellular Functions. International Journal of Molecular Sciences. 2021; 22(12):6420. https://doi.org/10.3390/ijms22126420
Chicago/Turabian StylePandey, Himanshu, Mary Popov, Alina Goldstein-Levitin, and Larisa Gheber. 2021. "Mechanisms by Which Kinesin-5 Motors Perform Their Multiple Intracellular Functions" International Journal of Molecular Sciences 22, no. 12: 6420. https://doi.org/10.3390/ijms22126420
APA StylePandey, H., Popov, M., Goldstein-Levitin, A., & Gheber, L. (2021). Mechanisms by Which Kinesin-5 Motors Perform Their Multiple Intracellular Functions. International Journal of Molecular Sciences, 22(12), 6420. https://doi.org/10.3390/ijms22126420






