Free Radicals and ROS Induce Protein Denaturation by UV Photostability Assay
Abstract
:1. Introduction
2. Results and Discussion
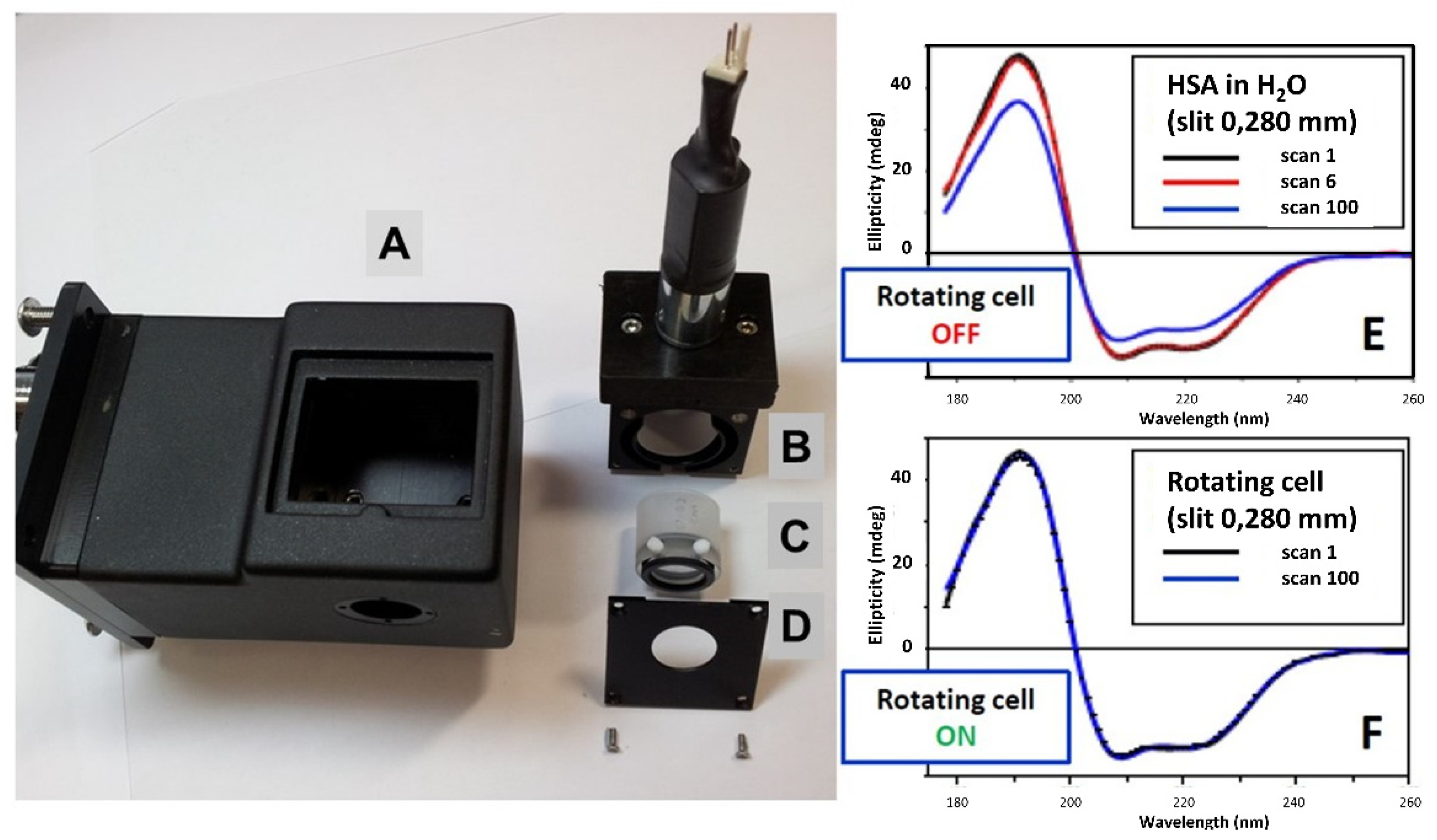
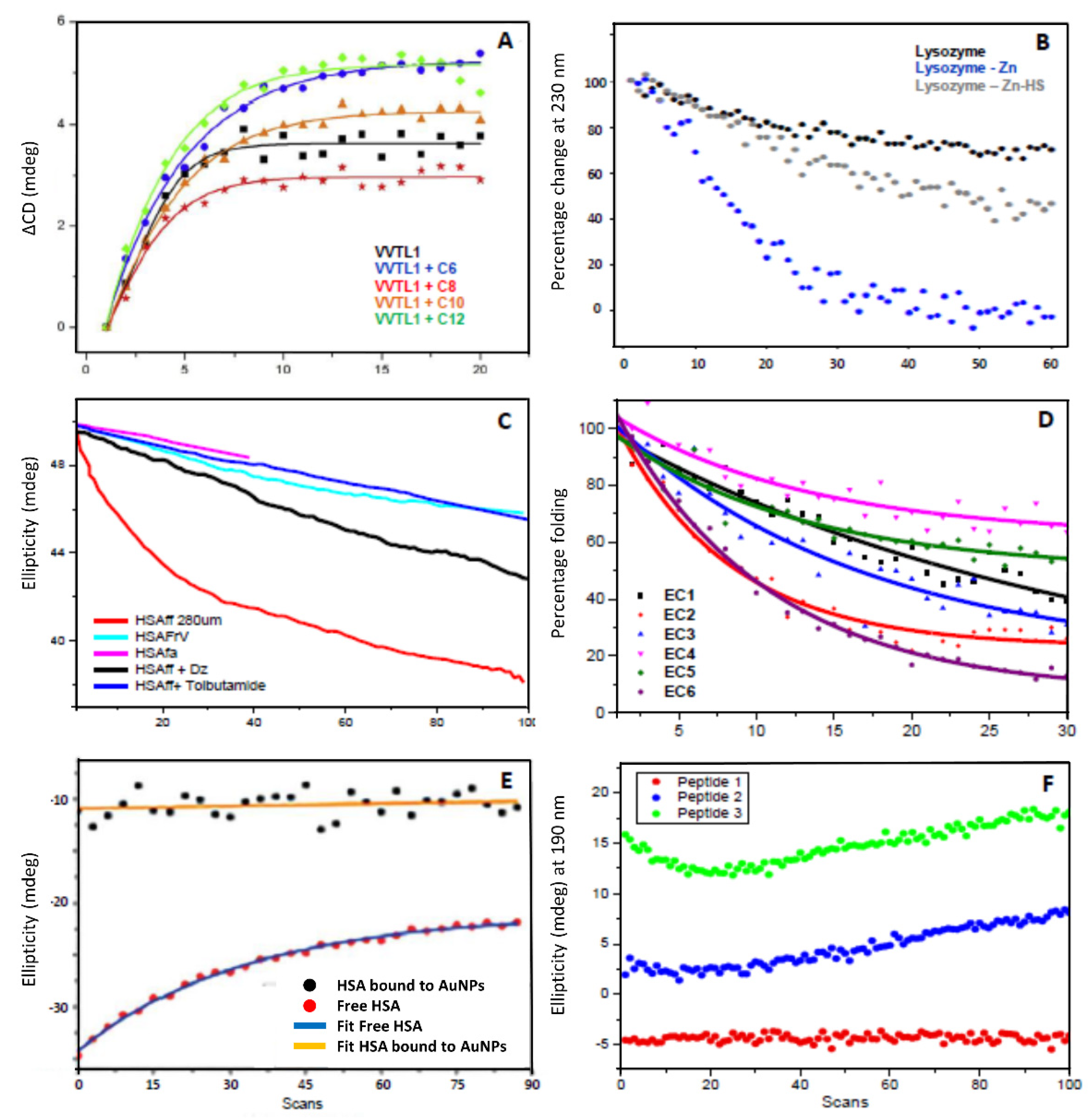
2.1. Assessment of the Hypothesis That Protein Denaturation Induced by UV Irradiation Is Originated by the Heating of Protein Bound Water Molecules
2.2. Assessment of the Hypothesis That the Protein Denaturation Induced by UV Irradiation Is Originated by the Formation of Free Radicals
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Tamizi, E.; Jouyban, A. Forced degradation studies of biopharmaceuticals: Selection of stress conditions. Eur. J. Pharm. Biopharm. 2016, 98, 26–46. [Google Scholar] [CrossRef]
- Johnson, W.C., Jr. Protein secondary structure and circular dichroism: A practical guide. Proteins 1990, 7, 205–214. [Google Scholar] [CrossRef]
- Johnson, W.C., Jr. Circular dichroism and its empirical application to biopolymers. Methods Biochem. Anal. 1985, 31, 61–163. [Google Scholar] [PubMed]
- Woody, R.W. Circular dichroism. Methods Enzym. 1995, 246, 34–71. [Google Scholar]
- Meierhenrich, U.J.; Filippi, J.J.; Meinert, C.; Bredehöft, J.H.; Takahashi, J.; Nahon, L.; Jones, N.C.; Hoffmann, S.V. Circular dichroism of amino acids in the vacuum-ultraviolet region. Angew. Chem. 2010, 49, 7799–7802. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.T.; Jones, G. CD12: A new high-flux beamline for ultraviolet and vacuum-ultraviolet circular dichroism on the SRS, Daresbury. J. Synchrot. Radiat. 2004, 11, 142–149. [Google Scholar] [CrossRef]
- Hussain, R.; Javorfi, T.S.; Siligardi, G. Circular dichroism beamline B23 at the Diamond Light Source. J. Synchrot. Radiat. 2012, 19, 132–135. [Google Scholar] [CrossRef]
- Hussain, R.; Jávorfi, T.; Siligardi, G. 8.23 Spectroscopic Analysis: Synchrotron Radiation Circular Dichroism. In Comprehensive Chirality; Carreira, E.M., Yamamoto, H., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 438–448. [Google Scholar]
- Di Gaspero, M.; Ruzza, P.; Hussain, R.; Vincenzi, S.; Biondi, B.; Gazzola, D.; Siligardi, G.; Curioni, A. Spectroscopy reveals that ethyl esters interact with proteins in wine. Food Chem. 2017, 217, 373–378. [Google Scholar] [CrossRef]
- Javorfi, T.; Hussain, R.; Myatt, D.; Siligardi, G. Measuring Circular Dichroism in a Capillary Cell Using the B23 Synchrotron Radiation CD Beamline at Diamond Light Source. Chirality 2010, 22, E149–E153. [Google Scholar] [CrossRef]
- Longo, E.; de Santis, E.; Hussain, R.; van der Walle, C.F.; Casas-Finet, J.; Uddin, S.; Dos Santos, A.; Siligardi, G. The effect of palmitoylation on the conformation and physical stability of a model peptide hormone. Int. J. Pharm. 2014, 472, 156–164. [Google Scholar] [CrossRef]
- Grosvenor, A.J.; Morton, J.D.; Dyer, J.M. Profiling of residue-level photo-oxidative damage in peptides. Amino Acids 2010, 39, 285–296. [Google Scholar] [CrossRef]
- Wien, F.; Miles, A.J.; Lees, J.G.; Hoffmann, S.V.; Wallace, B.A. VUV irradiation effects on proteins in high-flux synchrotron radiation circular dichroism spectroscopy. J. Synchrot. Radiat. 2005, 12 Pt 4, 517–523. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.J.; Hussain, R.; Cosentino, C.; Guerrini, M.; Siligardi, G.; Yates, E.A.; Rudd, T.R. A zinc complex of heparan sulfate destabilises lysozyme and alters its conformation. Biochem. Biophys. Res. Commun. 2012, 425, 794–799. [Google Scholar] [CrossRef]
- Siligardi, G.; Hussain, R. CD spectroscopy: An essential tool for quality control of protein folding. Methods Mol. Biol. 2015, 1261, 255–276. [Google Scholar]
- Capomaccio, R.; Osório, I.; Ojea-Jiménez, I.; Ceccone, G.; Colpo, P.; Gilliland, D.; Hussain, R.; Siligardi, G.; Rossi, F.; Ricard-Blum, S.; et al. Gold nanoparticles increases UV and thermal stability of human serum albumin. Biointerphases 2016, 11, 04B310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Provencher, S.W.; Glockner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 1981, 20, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.D.; Hollaender, A.; Greenstein, J.P. Electrophoretic Patterns, Colloid Osmotic Pressure, And Viscosity Of Serum Denatured By Ultraviolet Radiation. J. Biol. Chem. 1942, 146, 663–671. [Google Scholar] [CrossRef]
- Neves-Petersen, M.; Petersen, S.; Prakash Gajula, G. UV Light Effects on Proteins: From Photochemistry to Nanomedicine. In Molecular Photochemistry-Various Aspects; Intech Open: Rijeka, Croatia, 2012; pp. 125–158. [Google Scholar]
- Xu, G.H.; Chance, M.R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 2007, 107, 3514–3543. [Google Scholar] [CrossRef]
- Rappole, C.A.; Mitra, K.; Wen, H. Dynamic fluorescence imaging of the free radical products of X-ray absorption in live cells. Opt. Nanoscopy 2012, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- He, Y.Y.; Hader, D.P. UV-B-induced formation of reactive oxygen species and oxidative damage of the cyanobacterium Anabaena sp.: Protective effects of ascorbic acid and N-acetyl-L-cysteine. J. Photochem. Photobiol. B Biol. 2002, 66, 115–124. [Google Scholar] [CrossRef]
- Kim, E.-H. Better Understanding of the Biological Effects of Radiation by Microscopic Approaches. Nucl. Eng. Technol. 2008, 40, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Roots, R.; Okada, S. Estimation of Life Times and Diffusion Distances of Radicals Involved in X-Ray-Induced DNA Strand Breaks or Killing of Mammalian Cells. Radiat. Res. 1975, 64, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J. Protein oxidation and peroxidation. Biochem. J. 2016, 473, 805–825. [Google Scholar] [CrossRef] [Green Version]
- Hussain, R.; Benning, K.; Javorfi, T.; Longo, E.; Rudd, T.R.; Pulford, B.; Siligardi, G. CDApps: Integrated software for experimental planning and data processing at beamline B23, Diamond Light Source. J. Synchrot. Radiat. 2015, 22, 465–468. [Google Scholar] [CrossRef] [Green Version]




| % α-Helix (n° Helices *; aa Length) | % β-Strand (n° Strands *; aa Length) | % Turns | |
|---|---|---|---|
| HSAff 5 µM and 10 µM, 1st at 20 °C | 53.0 (4.7; 11.2) | 8.4 (1.8; 4.7) | 14.8 |
| HSAff 5 µM, 50th at 20 °C | 31.4 (3.7; 8.5) | 18.1 (3.6; 5.1) | 22.0 |
| HSAff 10 µM, 50th at 20 °C | 37.4 (4.2; 8.9) | 12.3 (2.9; 4.2) | 22.2 |
| HSAff 5 µM and 10 µM at 90 °C | 26.6 (3.3; 7.9) | 23.7 (4.4; 5.4) | 21.5 |
| HSAff 5 µM and 10 µM, back to 20 °C | 45.8 (4.4; 10.5) | 8.8 (2.1; 4.3) | 15.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruzza, P.; Honisch, C.; Hussain, R.; Siligardi, G. Free Radicals and ROS Induce Protein Denaturation by UV Photostability Assay. Int. J. Mol. Sci. 2021, 22, 6512. https://doi.org/10.3390/ijms22126512
Ruzza P, Honisch C, Hussain R, Siligardi G. Free Radicals and ROS Induce Protein Denaturation by UV Photostability Assay. International Journal of Molecular Sciences. 2021; 22(12):6512. https://doi.org/10.3390/ijms22126512
Chicago/Turabian StyleRuzza, Paolo, Claudia Honisch, Rohanah Hussain, and Giuliano Siligardi. 2021. "Free Radicals and ROS Induce Protein Denaturation by UV Photostability Assay" International Journal of Molecular Sciences 22, no. 12: 6512. https://doi.org/10.3390/ijms22126512







