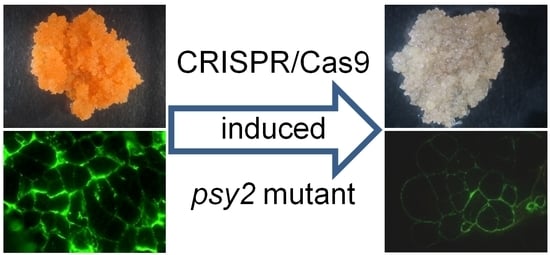
Inhibition of Carotenoid Biosynthesis by CRISPR/Cas9 Triggers Cell Wall Remodelling in Carrot
Abstract
:1. Introduction
2. Results
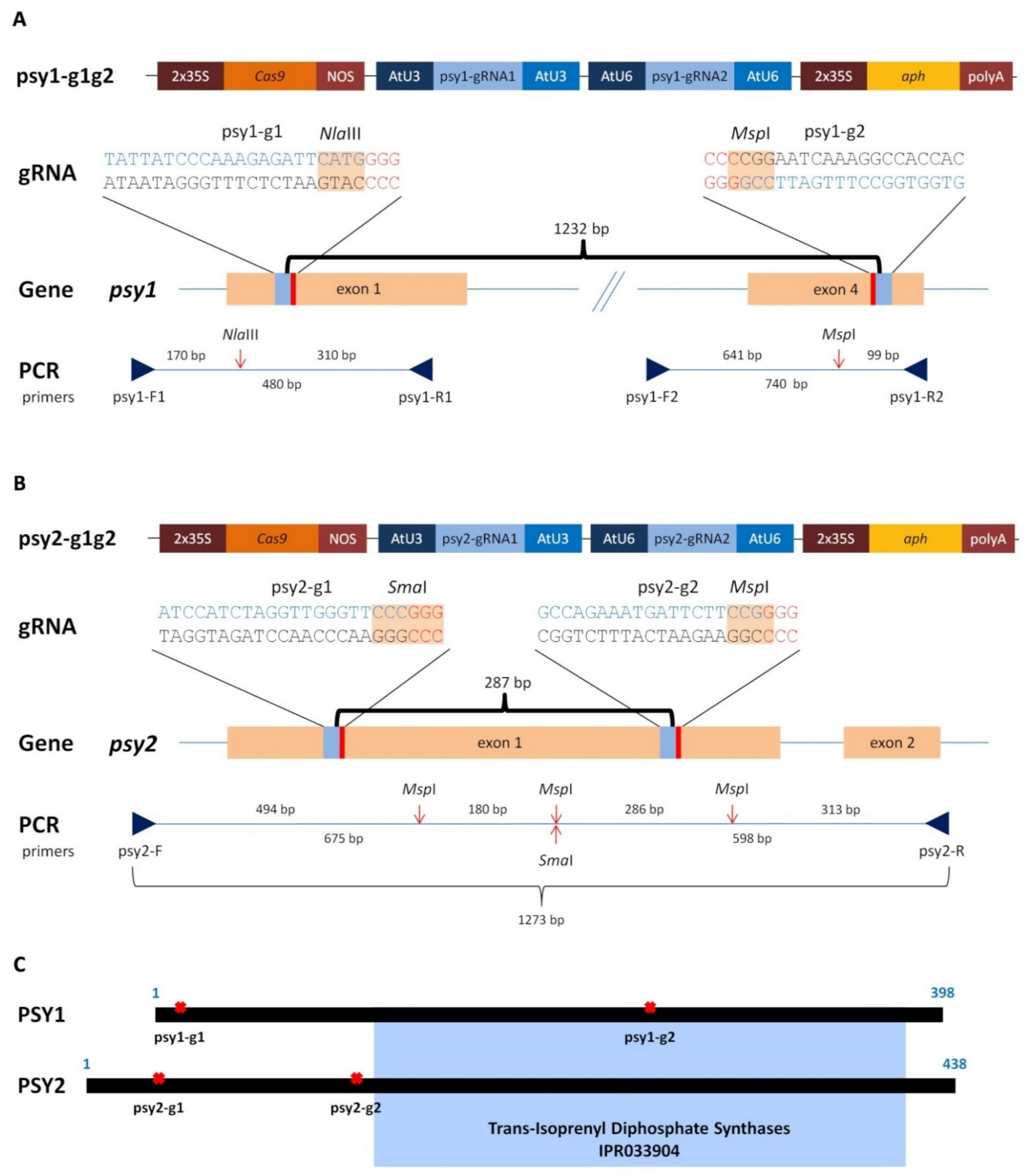
2.1. Generating of psy1 and psy2 Mutants by CRISPR/Cas9
2.2. Characterisation of the psy1 Mutants
2.3. Characterisation of the psy2 Mutants
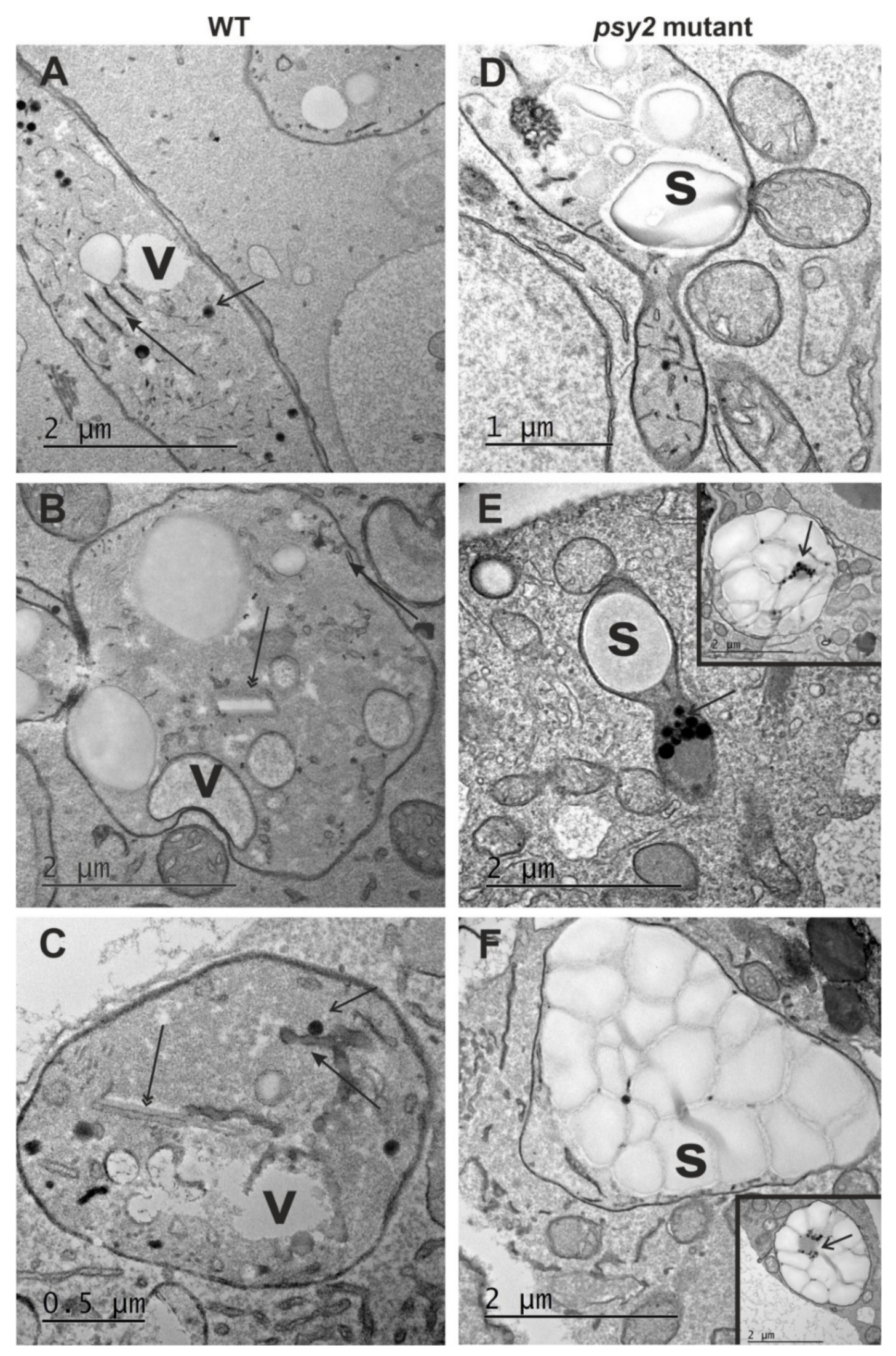
2.4. Altered Plastid Ultrastructure in the psy2 Mutants
2.5. Altered Cell Wall Composition in the psy2 Mutants
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Vector Construction
4.3. Agrobacterium-Mediated Callus Transformation
4.4. Molecular Identification of Mutants
4.5. Determination of Carotenoids Content
4.6. Gene Expression Analysis
4.7. Light Microscopy
4.8. Fluorescence Microscopy
4.9. Transmission Electron Microscopy (TEM)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosas-Saavedra, C.; Stange, C. Biosynthesis of carotenoids in plants: Enzymes and color. In Carotenoids in Nature: Biosynthesis, Regulation and Function; Stange, C., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 35–69. [Google Scholar] [CrossRef]
- Eggersdorfer, M.; Wyss, A. Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 2018, 652, 18–28. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Yuan, H.; Cao, H.; Yazdani, M.; Tadmor, Y.; Li, L. Carotenoid metabolism in plants: The role of plastids. Mol. Plant 2018, 11, 58–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egea, I.; Barsan, C.; Bian, W.; Purgatto, E.; Latché, A.; Chervin, C.; Bouzayen, M.; Pech, J.-C. Chromoplast differentiation: Current status and perspectives. Plant Cell Physiol. 2010, 51, 1601–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schweiggert, R.M.; Carle, R. Carotenoid deposition in plant and animal foods and its impact on bioavailability. Crit. Rev. Food Sci. 2017, 57, 1807–1830. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.-J.; Bressan, R.A.; Song, C.-P.; Zhu, J.-K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [Green Version]
- Faizan, M.; Faraz, A.; Sami, F.; Siddiqui, H.; Yusuf, M.; Gruszka, D.; Hayat, S. Role of strigolactones: Signalling and crosstalk with other phytohormones. Open Life Sci. 2020, 15, 217–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diretto, G.; Frusciante, S.; Fabbri, C.; Schauer, N.; Busta, L.; Wang, Z.; Matas, A.J.; Fiore, A.; Rose, J.K.C.; Fernie, A.R.; et al. Manipulation of β-carotene levels in tomato fruits results in increased ABA content and extended shelf life. Plant Biotechnol. J. 2020, 18, 1185–1199. [Google Scholar] [CrossRef]
- Qin, Y.; Woo, H.-J.; Shin, K.-S.; Lim, M.-H.; Lee, S.-K. Comparative transcriptome profiling of different tissues from beta-carotene-enhanced transgenic soybean and its non-transgenic counterpart. Plant Cell Tiss. Organ. Cult. 2020, 140, 341–356. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zhang, L.; Dong, C.; Guo, J.; Jin, L.; Wei, P.; Li, F.; Zhang, X.; Wang, R. Characterization and functional analysis of phytoene synthase gene family in tobacco. BMC Plant Biol. 2021, 21, 32. [Google Scholar] [CrossRef]
- Ramirez, V.; Xiong, G.; Mashiguchi, K.; Yamaguchi, S.; Pauly, M. Growth-and stress-related defects associated with wall hypoacetylation are strigolactone-dependent. Plant Direct 2018, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sola, M.A.; Rodriguez-Concepcion, M. Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arabidopsis Book 2012, 10, e0158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, M.-H.; Zhu, J.; Jiang, J.-G. Carotenoids biosynthesis and cleavage related genes from bacteria to plants. Crit. Rev. Food Sci. 2018, 58, 2314–2333. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, G. Plant carotenoids: Genomics meets multi-gene engineering. Curr. Opin. Plant Biol. 2014, 19, 111–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goo, Y.-M.; Kim, T.-W.; Ha, S.-H.; Back, K.-W.; Bae, J.-M.; Shin, Y.-W.; Lee, C.-H.; Ahn, M.-J.; Lee, S.-W. Expression profiles of genes involved in the carotenoid biosynthetic pathway in yellow-fleshed potato cultivars (Solanum tuberosum L.) from South Korea. J. Plant Biol. 2009, 52, 49–55. [Google Scholar] [CrossRef]
- Bartley, G.E.; Viitanen, P.V.; Bacot, K.O.; Scolnik, P.A. A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J. Biol. Chem. 1992, 267, 5036–5039. [Google Scholar] [CrossRef]
- Bartley, G.E.; Scolnik, P.A. cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J. Biol. Chem. 1993, 268, 25718–25721. [Google Scholar] [CrossRef]
- Stauder, R.; Welsch, R.; Camagna, M.; Kohlen, W.; Balcke, G.U.; Tissier, A.; Walter, M.H. Strigolactone levels in dicot roots are determined by an ancestral symbiosis-regulated clade of the PHYTOENE SYNTHASE gene family. Front. Plant Sci. 2018, 9, 255. [Google Scholar] [CrossRef]
- Qin, X.; Coku, A.; Inoue, K. Expression, subcellular localization, and cis-regulatory structure of duplicated phytoene synthase genes in melon (Cucumis melo L.). Planta 2011, 234, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Llorente, B.; Martinez-Garcia, J.F.; Stange, C.; Rodriguez-Concepcion, M. Illuminating colors: Regulation of carotenoid biosynthesis and accumulation by light. Curr. Opin. Plant Biol. 2017, 37, 49–55. [Google Scholar] [CrossRef]
- Maass, D.; Arango, J.; Wust, F.; Beyer, P.; Welsch, R. Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS ONE 2009, 4, e6373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.; Iovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, P.; Pizarro, L.; Moreno, J.C.; Handford, M.; Rodriguez-Concepcion, M.; Stange, C. Light-dependent changes in plastid differentiation influence carotenoid gene expression and accumulation in carrot roots. Plant Mol. Biol. 2012, 79, 47–59. [Google Scholar] [CrossRef]
- Bowman, M.J.; Willis, D.K.; Simon, P.W. Transcript abundance of phytoene synthase 1 and phytoene synthase 2 is associated with natural variation of storage root carotenoid pigmentation in carrot. J. Am. Soc. Hortic. Sci. 2014, 139, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Ou, C.-G.; Zhuang, F.-Y.; Ma, Z.-G. The dual role of phytoene synthase genes in carotenogenesis in carrot roots and leaves. Mol. Breed. 2014, 34, 2065–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrin, F.; Hartmann, L.; Dubois-Laurent, C.; Welsch, R.; Huet, S.; Hamama, L.; Briard, M.; Peltier, D.; Gagné, S.; Geoffriau, E. Carotenoid gene expression explains the difference of carotenoid accumulation in carrot root tissues. Planta 2017, 245, 737–747. [Google Scholar] [CrossRef] [Green Version]
- Simpson, K.; Fuentes, P.; Quiroz-Iturra, L.F.; Flores-Ortiz, C.; Contreras, R.; Handford, M.; Stange, C. Unraveling the induction of phytoene synthase 2 expression by salt stress and abscisic acid in Daucus carota. J. Exp. Bot. 2018, 69, 4113–4126. [Google Scholar] [CrossRef]
- Clotault, J.; Peltier, D.; Berruyer, R.; Thomas, M.; Briard, M.; Geoffriau, E. Expression of carotenoid biosynthesis genes during carrot root development. J. Exp. Bot. 2008, 59, 3563–3573. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Ahn, Y.O.; Ahn, M.-J.; Jeong, J.C.; Lee, H.-S.; Kwak, S.-S. Cloning and characterization of an Orange gene that increases carotenoid accumulation and salt stress tolerance in transgenic sweetpotato cultures. Plant Physiol. Biochem. 2013, 70, 445–454. [Google Scholar] [CrossRef]
- Schaub, P.; Rodriguez-Franco, M.; Cazzonelli, C.I.; Álvarez, D.; Wüst, F.; Welsch, R. Establishment of an Arabidopsis callus system to study the interrelations of biosynthesis, degradation and accumulation of carotenoids. PLoS ONE 2018, 13, e0192158. [Google Scholar] [CrossRef] [Green Version]
- Baranska, M.; Baranski, R.; Schulz, H.; Nothnagel, T. Tissue-specific accumulation of carotenoids in carrot roots. Plant a 2006, 224, 1028–1037. [Google Scholar] [CrossRef] [PubMed]
- Baranski, R.; Klocke, E.; Schumann, G. Green fluorescent protein as an efficient selection marker for Agrobacterium rhizogenes mediated carrot transformation. Plant Cell Rep. 2006, 25, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Oleszkiewicz, T.; Klimek-Chodacka, M.; Milewska-Hendel, A.; Zubko, M.; Stróż, D.; Kurczyńska, E.; Boba, A.; Szopa, J.; Baranski, R. Unique chromoplast organisation and carotenoid gene expression in carotenoid-rich carrot callus. Planta 2018, 248, 1455–1471. [Google Scholar] [CrossRef]
- Rygula, A.; Oleszkiewicz, T.; Grzebelus, E.; Pacia, M.Z.; Baranska, M.; Baranski, R. Raman, AFM and SNOM high resolution imaging of carotene crystals in a model carrot cell system. Spectrochim. Acta A 2018, 197, 47–55. [Google Scholar] [CrossRef]
- Dudek, M.; Machalska, E.; Oleszkiewicz, T.; Grzebelus, E.; Baranski, R.; Szcześniak, P.; Mlynarski, J.; Zajac, G.; Kaczor, A.; Baranska, M. Chiral amplification in nature: Studying cell-extracted chiral carotenoid microcrystals via the resonance Raman optical activity of model systems. Angew. Chem. Int. Ed. 2019, 58, 8383–8388. [Google Scholar] [CrossRef]
- Zhang, Y.; Malzahn, A.A.; Sretenovic, S.; Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 2019, 5, 778–794. [Google Scholar] [CrossRef]
- El-Mounadi, K.; Morales-Floriano, M.L.; Garcia-Ruiz, H. Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front. Plant Sci. 2020, 11, 56. [Google Scholar] [CrossRef]
- D’Ambrosio, C.; Stigliani, A.L.; Giorio, G. CRISPR/Cas9 editing of carotenoid genes in tomato. Transgenic Res. 2018, 27, 367–378. [Google Scholar] [CrossRef]
- Dahan-Meir, T.; Filler-Hayut, S.; Melamed-Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in planta gene targeting in tomato using gemiviral replicons and the CRISPR/Cas9 system. Plant J. 2018, 95, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Song, N.; Sun, S.; Yang, W.; Zhao, H.; Song, W.; Lai, J. Efficiency and Inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J. Genet. Genom. 2015, 43, 25–36. [Google Scholar] [CrossRef]
- Klimek-Chodacka, M.; Oleszkiewicz, T.; Qi, Y.; Baranski, R. Carrot genome editing using CRISPR-based systems. Acta Hortic. 2019, 1264, 53–66. [Google Scholar] [CrossRef]
- Klimek-Chodacka, M.; Oleszkiewicz, T.; Lowder, L.G.; Qi, Y.; Baranski, R. Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 2018, 37, 575–586. [Google Scholar] [CrossRef] [Green Version]
- Mikami, M.; Toki, S.; Endo, M. Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep. 2015, 34, 1807–1815. [Google Scholar] [CrossRef]
- Xu, Z.-S.; Feng, K.; Xiong, A.-S. CRISPR/Cas9-mediated multiply targeted mutagenesis in orange and purple carrot plants. Mol. Biotechnol. 2019, 61, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Zischewski, J.; Fischer, R.; Bortesi, L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol. Adv. 2017, 35, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Sentmanat, M.F.; Peters, S.T.; Floria, C.P.; Connelly, J.P.; Pruett-Miller, S.M. A survey of validating strategies for CRISPR-Cas9 editing. Sci. Rep. 2018, 8, 888. [Google Scholar] [CrossRef] [Green Version]
- Jang, G.; Lee, S.; Um, T.Y.; Chang, S.H.; Lee, H.Y.; Chung, P.J.; Kim, J.-K.; Choi, Y.D. Genetic chimerism of CRISPR/Cas9-mediated rice mutants. Plant Biotechnol. Rep. 2016, 10, 425–435. [Google Scholar] [CrossRef]
- Huang, H.; Wu, Q. CRISPR double cutting through the labyrinthine architecture of 3D genomes. J. Genet. Genom. 2016, 43, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Welsch, R.; Arango, J.; Bär, C.; Salazar, B.; Al-Babili, S.; Beltrán, J.; Chavarriaga, P.; Ceballos, H.; Tohme, J.; Beyer, P. Provitamin A accumulation in Cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 2010, 22, 3348–3356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Moreno, J.C.; Pizarro, L.; Fuentes, P.; Handford, M.; Cifuentes, V.; Stange, C. Levels of lycopene β-cyclase 1 modulate carotenoid gene expression and accumulation in Daucus carota. PLoS ONE 2013, 8, e58144. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Villalón, A.; Gas, E.; Rodríguez-Concepción, M. Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 2009, 60, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Arango, J.; Jourdan, M.; Geoffriau, E.; Beyer, P.; Welsch, R. Carotene hydroxylase activity determines the levels of both α-carotene and total carotenoids in orange carrots. Plant Cell 2014, 26, 2223–2233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser, P.D.; Enfissi, E.M.A.; Halket, J.M.; Truesdale, M.R.; Yu, D.; Gerrish, C.; Bramley, P.M. Manipulation of phytoene levels in tomato fruit: Effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 2007, 19, 3194–3211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, K.; Quiroz, L.F.; Rodríguez-Concepción, M.; Stange, C.R. Differential contribution of the first two enzymes of the MEP pathway to the supply of metabolic precursors for carotenoid and chlorophyll biosynthesis in carrot (Daucus carota). Front. Plant Sci. 2016, 7, 1344. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.-J.; Jeong, H.-B.; Jung, A.; Kang, M.-Y.; Kim, S.; Ha, S.-H.; Kwon, J.-K.; Kang, B.-C. Phytoene synthase 2 can compensate for the absence of PSY1 in the control of color in Capsicum fruit. J. Exp. Bot. 2020, 71, 3417–3427. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Paolillo, D.J.; Parthasarathy, M.V.; DiMuzio, E.M.; Garvin, D.F. A novel gene mutation that confers abnormal patterns of β-carotene accumulation in cauliflower (Brassica oleracea var. botrytis). Plant J. 2001, 26, 59–67. [Google Scholar] [CrossRef]
- Bai, C.; Rivera, S.M.; Medina, V.; Alves, R.; Vilaprinyo, E.; Sorribas, A.; Canela, R.; Capell, T.; Sandmann, G.; Christou, P.; et al. An in vitro system for the rapid functional characterization of genes involved in carotenoid biosynthesis and accumulation. Plant J. 2014, 77, 464–475. [Google Scholar] [CrossRef]
- Zhou, X.; Welsch, R.; Yang, Y.; Álvarez, D.; Riediger, M.; Yuan, H.; Fish, T.; Liu, J.; Thannhauser, T.W.; Li, L. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 3558–3563. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.E.; Rensing, K.H.; Douglas, C.J.; Cheng, K.M. Chromoplasts ultrastructure and estimated carotene content in root secondary phloem of different carrot varieties. Plant a 2010, 231, 549–558. [Google Scholar] [CrossRef]
- Roman, M.; Marzec, K.M.; Grzebelus, E.; Simon, P.W.; Baranska, M.; Baranski, R. Composition and (in)homogeneity of carotenoid crystals in carrot cells revealed by high resolution Raman imaging. Spectrochim. Acta A 2015, 136, 1395–1400. [Google Scholar] [CrossRef] [PubMed]
- Camara, B.; Hugueney, P.; Bouvier, F.; Kuntz, M.; Monéger, R. Biochemistry and molecular biology of chromoplast development. Int. Rev. Cytol. 1995, 163, 175–247. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Bender, L.; Neumann, K.H. Growth regulation, plastid differentiation and the development of a photosynthetic system in cultured carrot root explants as influenced by exogenous sucrose and various phytohormones. Plant Cell Tissue Org. Cult. 1984, 3, 11–28. [Google Scholar] [CrossRef]
- Hempel, J.; Amrehn, E.; Quesada, S.; Esquivel, P.; Jiménez, V.M.; Heller, A.; Carle, R.; Schweiggert, R.M. Lipid-dissolved γ-carotene, β-carotene, and lycopene in globular chromoplasts of peach palm (Bactris gasipaes Kunth) fruits. Planta 2014, 240, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Concepcion, M.; Stange, C. Biosynthesis of carotenoids in carrot: An underground story comes to light. Arch. Biochem. Biophys. 2013, 539, 110–116. [Google Scholar] [CrossRef]
- Sun, L.; Sun, Y.; Zhang, M.; Wang, L.; Ren, J.; Cui, M.; Wang, Y.; Ji, K.; Li, P.; Li, Q.; et al. Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol. 2012, 158, 283–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, N.; Feng, H.; Meng, X.; Li, D.; Yang, D.; Wu, C.; Meng, Q. Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 2014, 14, 351. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Zhao, W.; Qu, H.; Wang, Q.; Zhao, L. The yellow-fruited tomato 1 (yft1) mutant has altered fruit carotenoid accumulation and reduced ethylene production as a result of a genetic lesion in ETHYLENE INSENSITIVE2. Theor. Appl. Genet. 2016, 129, 717–728. [Google Scholar] [CrossRef]
- Li, L.; Zhao, W.; Feng, X.; Chen, L.; Zhang, L.; Zhao, L. Changes in fruit firmness, cell wall composition, and transcriptional profile in the yellow fruit tomato 1 (yft1) mutant. J. Agric. Food Chem. 2019, 67, 463–472. [Google Scholar] [CrossRef]
- Zhao, W.; Gao, L.; Li, Y.; Wang, M.; Zhang, L.; Zhao, L. Yellow-fruited phenotype is caused by 573 bp insertion at 5’ UTR of YFT1 allele in yft1 mutant tomato. Plant Sci. 2020, 300, 110637. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, F.; Zheng, X.; Xie, Z.; Ye, J.; Cheng, Y.; Deng, X.; Zeng, Y. Investigation of chromoplast ultrastructure and tissue-specific accumulation of carotenoids in citrus flesh. Sci. Hortic. 2019, 256, 108547. [Google Scholar] [CrossRef]
- Otaka, J.; Seo, S.; Nishimura, M. Lutein, a natural carotenoid, induces α-1,3-glucan accumulation on the cell wall surface of fungal plant pathogens. Molecules 2016, 21, 980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosgrove, D.J.; Anderson, C.T. Plant cell growth: Do pectins drive lobe formation in Arabidopsis pavement cells? Curr. Biol. 2020, 30, R660–R662. [Google Scholar] [CrossRef]
- Bidhendi, A.J.; Geitmann, A. Relating the mechanics of the primary plant cell wall to morphogenesis. J. Exp. Bot. 2016, 67, 449–461. [Google Scholar] [CrossRef] [Green Version]
- Brulé, V.; Rafsanjani, A.; Pasini, D.; Western, T.L. Hierarchies of plant stiffness. Plant Sci. 2016, 250, 79–96. [Google Scholar] [CrossRef] [Green Version]
- Braybrook, S.A.; Peaucelle, A. Mechano-Chemical Aspects of Organ Formation in Arabidopsis thaliana: The relationship between auxin and pectin. PLoS ONE 2013, 8, e57813. [Google Scholar] [CrossRef] [Green Version]
- Sala, K.; Malarz, K.; Barlow, P.W.; Kurczyńska, E.U. Distribution of some pectic and arabinogalactan protein epitopes during Solanum lycopersicum (L.) adventitious root development. BMC Plant Biol. 2017, 17, 25. [Google Scholar] [CrossRef] [Green Version]
- Milewska-Hendel, A.; Zubko, M.; Karcz, J.; Stróż, D.; Kurczyńska, E. Fate of neutral-charged gold nanoparticles in the roots of the Hordeum vulgare L. cultivar Karat. Sci. Rep. 2017, 7, 3014. [Google Scholar] [CrossRef] [Green Version]
- Showalter, A.M. Structure and function of plant-cell wall proteins. Plant Cell 1993, 5, 9–23. [Google Scholar] [CrossRef]
- Nothnagel, E.A. Proteoglycans and related components in plant cells. Int. Rev. Cytol. 1997, 174, 195–291. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M. Arabinogalactan-proteins: Structure, expression and function. Cell. Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.; Egelund, J.; Schultz, C.J.; Bacic, A. Arabinogalactan-proteins: Key regulators at the cell surface? Plant Physiol. 2010, 153, 403–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, M.; Showalter, A.M. Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. Plant J. 1999, 19, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Mareri, L.; Romi, M.; Cai, G. Arabinogalactan proteins: Actors or spectators during abiotic and biotic stress in plants? Plant Biosyst. 2019, 153, 173–185. [Google Scholar] [CrossRef]
- Park, M.H.; Suzuki, Y.; Chono, M.; Knox, J.P.; Yamaguchi, I. CsAGP1, a gibberellin-responsive gene from cucumber hypocotyls, encodes a classical arabinogalactan protein and is involved in stem elongation. Plant Physiol. 2003, 131, 1450–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.L.; Li, Y.Y.; Zhang, Y.J.; Zhang, S.S.; Wu, Y.R.; Wu, P.; Zheng, S.J. Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol. 2008, 146, 602–611. [Google Scholar] [CrossRef]
- Mareri, L.; Faleri, C.; Romi, M.; Mariani, C.; Cresti, M.; Cai, G. Heat stress affects the distribution of JIM8-labelled arabinogalactan proteins in pistils of Solanum lycopersicum cv Micro-Tom. Acta Physiol. Plant 2016, 38, 184. [Google Scholar] [CrossRef]
- Smallwood, M.; Yates, E.A.; Willats, W.G.T.; Martin, H.; Knox, J.P. Immunochemical comparison of membrane-associated and secreted arabinogalactan-proteins in rice and carrot. Plant a 1996, 198, 452–459. [Google Scholar] [CrossRef]
- Herger, A.; Dünser, K.; Kleine-Vehn, J.; Ringli, C. Leucine-rich repeat extensin proteins and their role in cell wall sensing. Curr. Biol. 2019, 29, R851–R858. [Google Scholar] [CrossRef] [PubMed]
- Lamport, D.T.A.; Kieliszewski, M.J.; Chen, Y.; Cannon, M.C. Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 2011, 156, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Varner, J.E. Isolation and characterization of cDNA clones for carrot extensin and a proline-rich 33-kDa protein. Proc. Natl. Acad. Sci. USA 1985, 82, 4399–4403. [Google Scholar] [CrossRef] [Green Version]
- Stafstrom, J.P.; Staehelin, L.A. A second extensin-Like hydroxyproline-rich glycoprotein from carrot cell walls. Plant Physiol. 1987, 84, 820–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sala, K.; Karcz, J.; Rypień, A.; Kurczyńska, E.U. Unmethyl-esterified homogalacturonan and extensins seal Arabidopsis graft union. BMC Plant Biol. 2019, 19, 151. [Google Scholar] [CrossRef] [PubMed]
- Popielarska-Konieczna, M.; Sala, K.; Abdullah, M.; Tuleja, M.; Kurczyńska, E. Extracellular matrix and wall composition are diverse in the organogenic and non-organogenic calli of Actinidia arguta. Plant Cell Rep. 2020, 39, 779–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godel-Jędrychowska, K.; Maćkowska, K.; Kurczyńska, E.; Grzebelus, E. Composition of the reconstituted cell wall in protoplast-derived cells of Daucus is affected by phytosulfokine (PSK). Int. J. Mol. Sci. 2019, 20, 5490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, S.; Park, J.; Kim, J.-S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef] [Green Version]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef] [Green Version]
- Main, G.D.; Reynolds, S.; Gartland, J.S. Electroporation protocols for Agrobacterium. In Agrobacterium Protocols: Methods in Molecular Biology; Gartland, K.M.A., Davey, M.R., Eds.; Springer: Totowa, NJ, USA, 1995; Volume 44, pp. 405–412. [Google Scholar]
- Potocka, I.; Godel, K.; Dobrowolska, I.; Kurczyńska, E.U. Spatio-temporal localization of selected pectic and arabinogalactan protein epitopes and the ultrastructural characteristics of explant cells that accompany the changes in the cell fate during somatic embryogenesis in Arabidopsis thaliana. Plant Physiol. Biochem. 2018, 127, 573–589. [Google Scholar] [CrossRef]
- Clausen, M.H.; Madsen, R. Synthesis of hexasaccharide fragments of pectin. Chem. Eur. J. 2003, 9, 3821–3832. [Google Scholar] [CrossRef] [PubMed]
- Yates, E.A.; Valdor, J.F.; Haslam, S.M.; Morris, H.R.; Dell, A.; Mackie, W.; Knox, J.P. Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 1996, 6, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knox, J.P.; Linstead, P.J.; Cooper, J.P.C.; Roberts, K. Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. Plant J. 1991, 1, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, M.; Beven, A.; Donovan, N.; Neill, S.J.; Peart, J.; Roberts, K.; Knox, J.P. Localization of cell wall proteins in relation to the developmental anatomy of the carrot root apex. Plant J. 1994, 5, 237–246. [Google Scholar] [CrossRef]


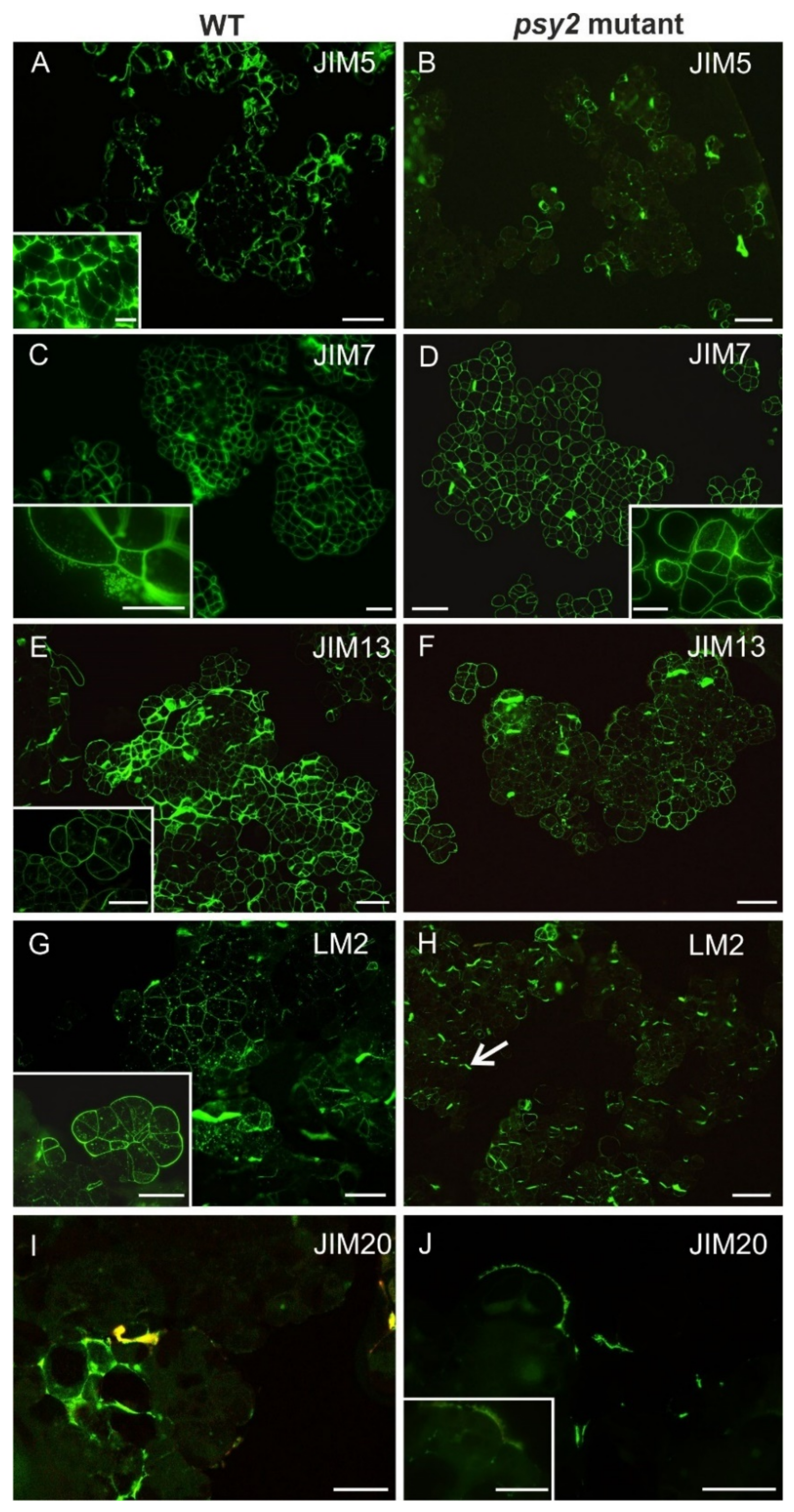
| Wall Constituents | Antibody | Epitope | References |
|---|---|---|---|
| Pectins | JIM5 | Low methyl-esterified HG | [102] |
| JIM7 | Highly methyl-esterified HG | [102] | |
| AGPs | LM2 | β-d-GlcpA | [90] [103] |
| JIM13 | β-d-GlcpA-(1→3)-α-d-GalpA-(1→2)-l-Rha | [104] [103] | |
| Extensins | JIM20 | Extensin/HRGP glycoprotein | [105] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oleszkiewicz, T.; Klimek-Chodacka, M.; Kruczek, M.; Godel-Jędrychowska, K.; Sala, K.; Milewska-Hendel, A.; Zubko, M.; Kurczyńska, E.; Qi, Y.; Baranski, R. Inhibition of Carotenoid Biosynthesis by CRISPR/Cas9 Triggers Cell Wall Remodelling in Carrot. Int. J. Mol. Sci. 2021, 22, 6516. https://doi.org/10.3390/ijms22126516
Oleszkiewicz T, Klimek-Chodacka M, Kruczek M, Godel-Jędrychowska K, Sala K, Milewska-Hendel A, Zubko M, Kurczyńska E, Qi Y, Baranski R. Inhibition of Carotenoid Biosynthesis by CRISPR/Cas9 Triggers Cell Wall Remodelling in Carrot. International Journal of Molecular Sciences. 2021; 22(12):6516. https://doi.org/10.3390/ijms22126516
Chicago/Turabian StyleOleszkiewicz, Tomasz, Magdalena Klimek-Chodacka, Michał Kruczek, Kamila Godel-Jędrychowska, Katarzyna Sala, Anna Milewska-Hendel, Maciej Zubko, Ewa Kurczyńska, Yiping Qi, and Rafal Baranski. 2021. "Inhibition of Carotenoid Biosynthesis by CRISPR/Cas9 Triggers Cell Wall Remodelling in Carrot" International Journal of Molecular Sciences 22, no. 12: 6516. https://doi.org/10.3390/ijms22126516
APA StyleOleszkiewicz, T., Klimek-Chodacka, M., Kruczek, M., Godel-Jędrychowska, K., Sala, K., Milewska-Hendel, A., Zubko, M., Kurczyńska, E., Qi, Y., & Baranski, R. (2021). Inhibition of Carotenoid Biosynthesis by CRISPR/Cas9 Triggers Cell Wall Remodelling in Carrot. International Journal of Molecular Sciences, 22(12), 6516. https://doi.org/10.3390/ijms22126516