The Effect of Functional Fiber on Microbiota Composition in Different Intestinal Segments of Obese Mice
Abstract
1. Introduction
2. Results
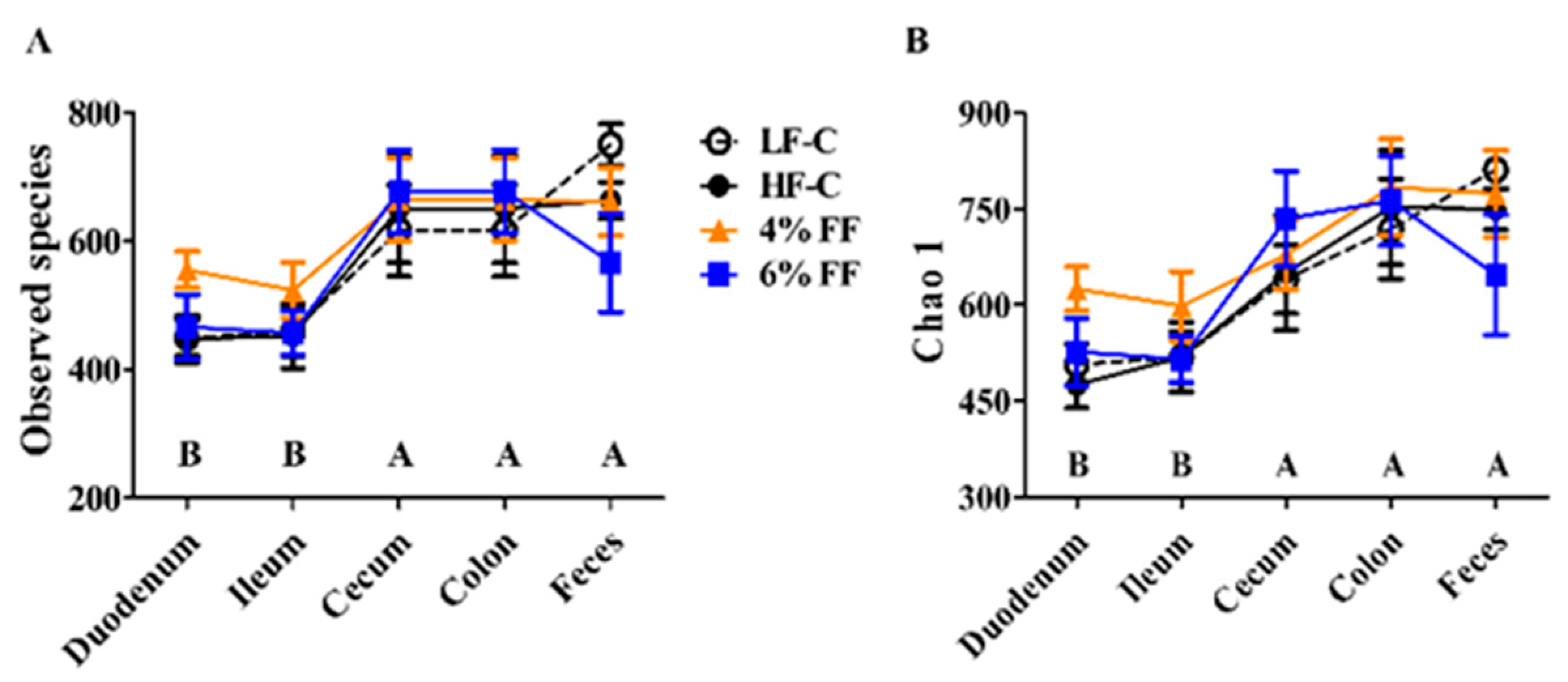
2.1. α-Diversity
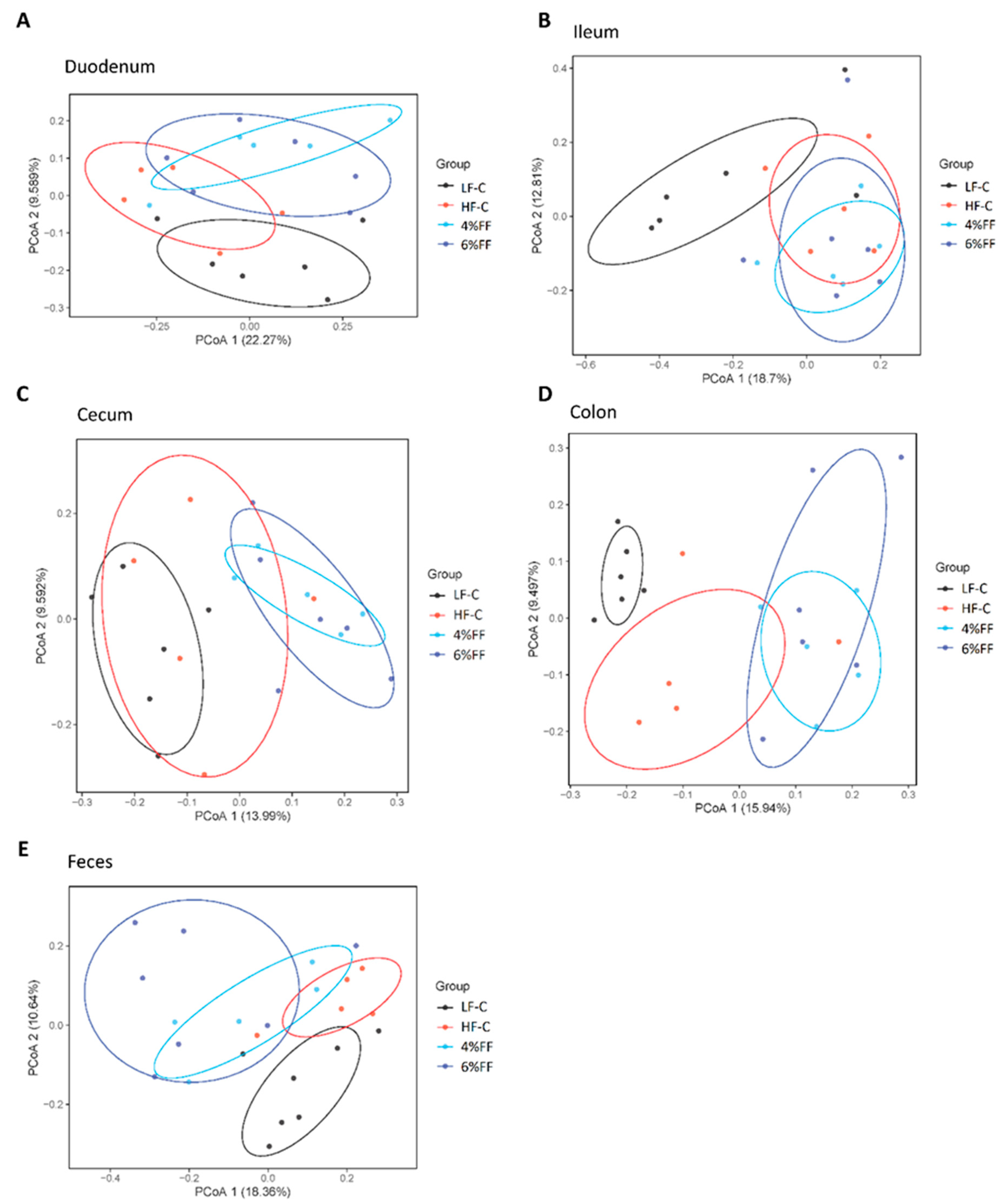
2.2. β-Diversity
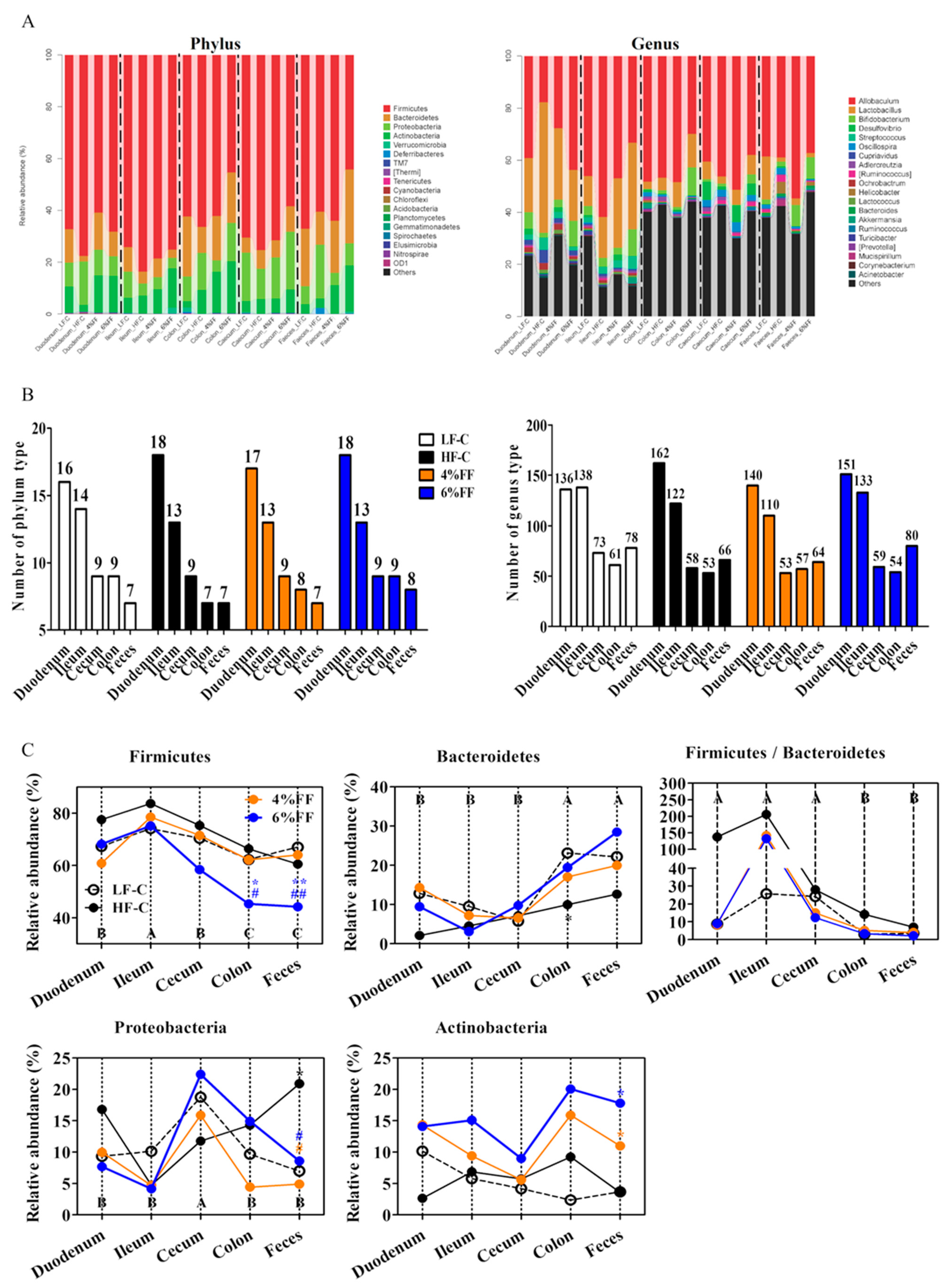
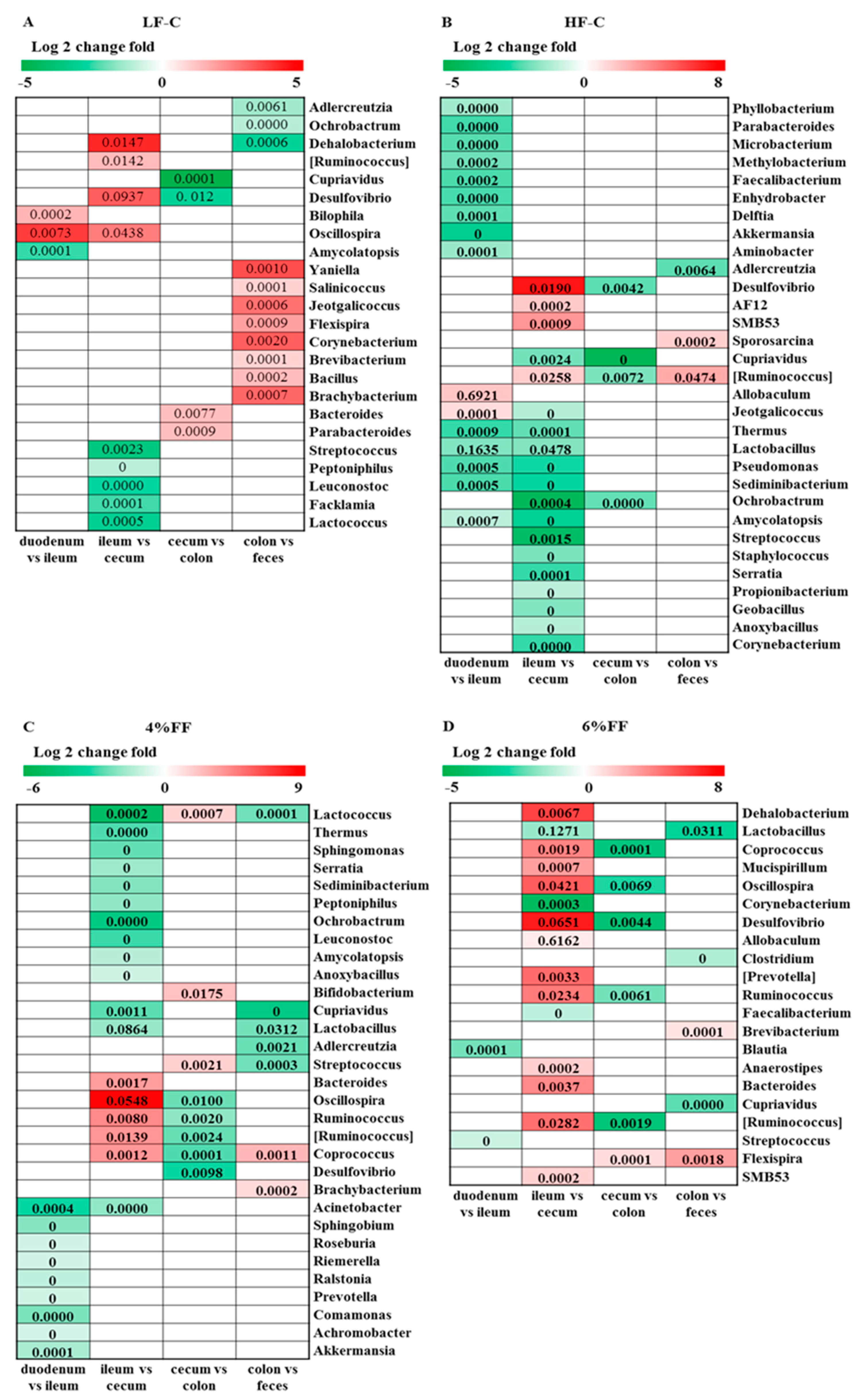
2.3. Changes in Microbial Composition of Different Intestinal Segments
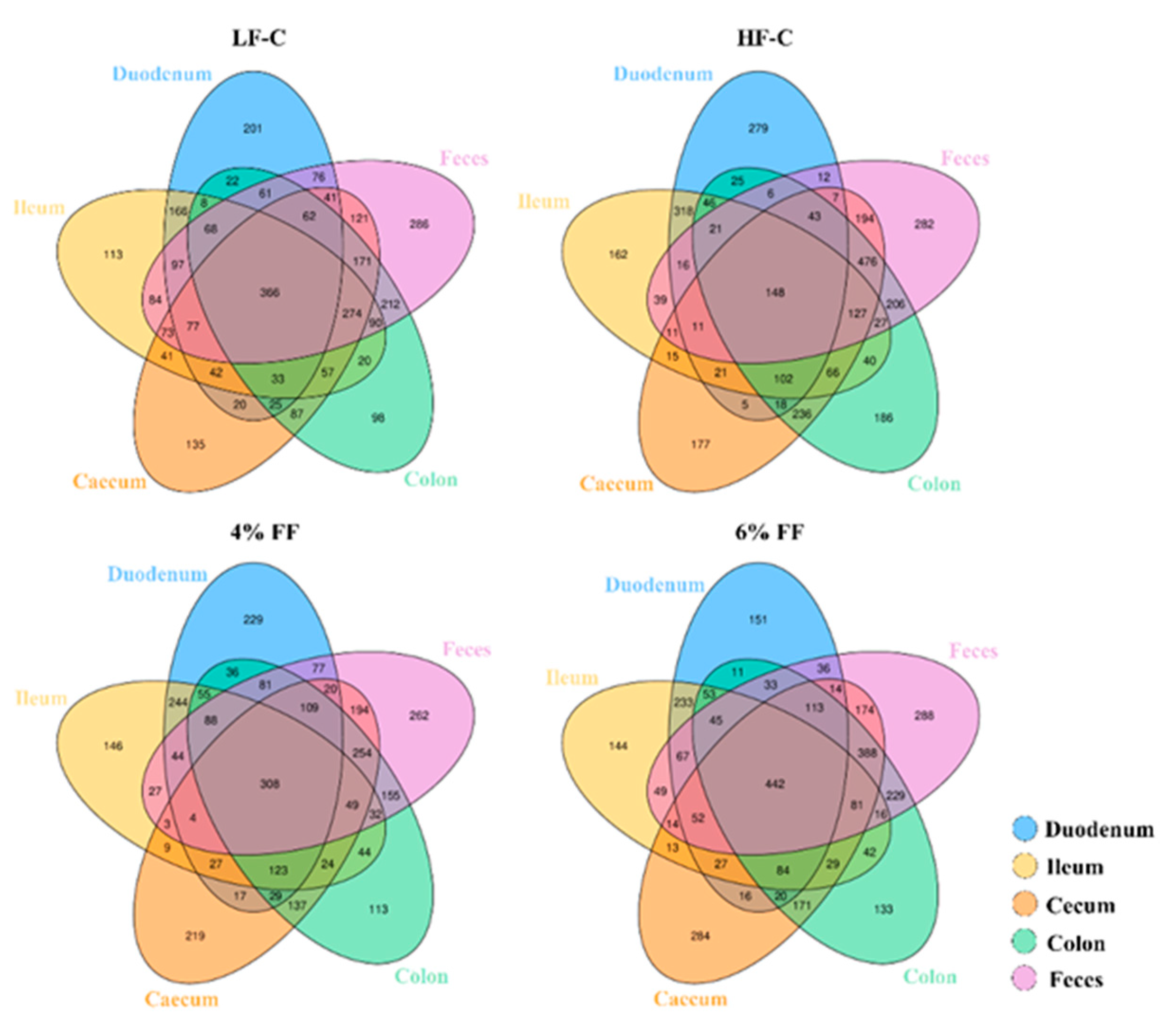
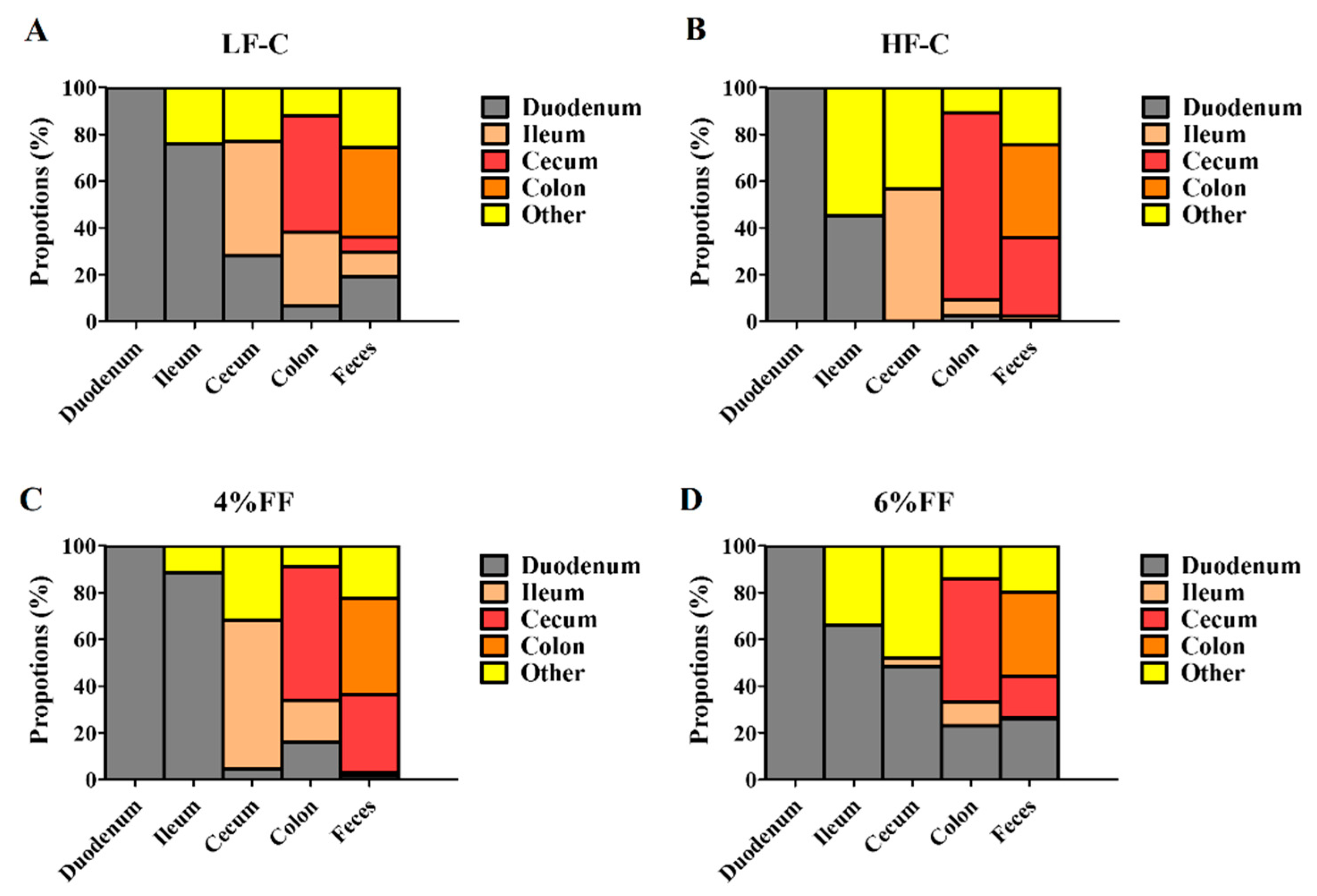
2.4. Microbial Traceability Analysis
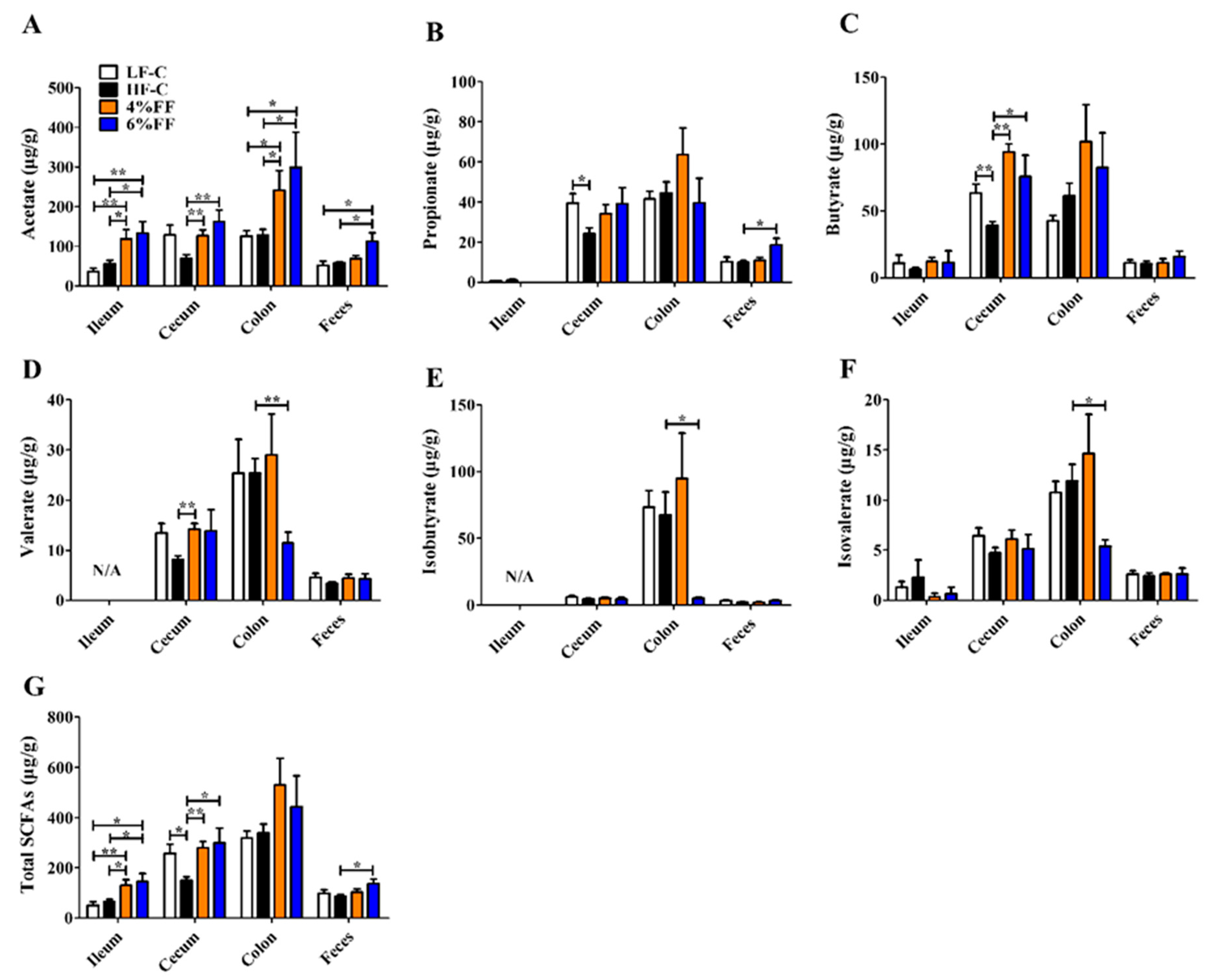
2.5. Changes in Microbial Metabolite Short-Chain Fatty Acids (SCFAs)
3. Discussion
4. Materials and Methods
4.1. Animal Diets
4.2. Treatment
4.3. Sample Collection
4.4. DNA Extraction, 16S rDNA Amplification, and Illumina MiSeq Sequencing
4.5. Sequence Filtering, OTU Clustering, and Sequence Analyses
4.6. Short-Chain Fatty Acids Determination
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tropini, C.; Earle, K.A.; Huang, K.C.; Sonnenburg, J.L. The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host Microbe 2017, 21, 433–442. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Leone, V.; Chang, E.B. Regional Diversity of the Gastrointestinal Microbiome. Cell Host Microbe 2019, 26, 314–324. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Gerard, P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013, 3, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Hegade, V.S.; Speight, R.A.; Etherington, R.E.; Jones, D.E.J. Novel bile acid therapeutics for the treatment of chronic liver diseases. Ther. Adv. Gastroenter. 2016, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Ji, Y.; Ma, N.; Zhang, J.; Wang, H.; Tao, T.; Pei, F.; Hu, Q. Dietary intake of mixture coarse cereals prevents obesity by altering the gut microbiota in high-fat diet fed mice. Food Chem. Toxicol. 2020, 147, 111901. [Google Scholar] [CrossRef]
- Wei, Y.; Liang, J.; Su, Y.; Wang, J.; Amakye, W.K.; Pan, J.; Chu, X.; Ma, B.; Song, Y.; Li, Y.; et al. The associations of the gut microbiome composition and short-chain fatty acid concentrations with body fat distribution in children. Clin. Nutr. 2020, 40, 3379–3390. [Google Scholar] [CrossRef]
- Cheng, C.; Wei, H.; Xu, C.; Xie, X.; Jiang, S.; Peng, J. Maternal soluble fiber diet during pregnancy changes the intestinal microbiota, improves growth performance, and reduces intestinal permeability in piglets. Appl. Environ. Microbiol. 2018, 84, 84. [Google Scholar] [CrossRef]
- Xu, C.; Liu, J.; Gao, J.; Wu, X.; Cui, C.; Wei, H.; Zheng, R.; Peng, J. Combined Soluble Fiber-Mediated Intestinal Microbiota Improve Insulin Sensitivity of Obese Mice. Nutrients 2020, 12, 351. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Raes, J.; van den Bogert, B.; Arumugam, M.; Booijink, C.C.; Troost, F.J.; Bork, P.; Wels, M.; de Vos, W.M.; Kleerebezem, M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012, 6, 1415–1426. [Google Scholar] [CrossRef]
- Islam, K.B.; Fukiya, S.; Hagio, M.; Fujii, N.; Ishizuka, S.; Ooka, T.; Ogura, Y.; Hayashi, T.; Yokota, A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011, 141, 1773–1781. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ. Microbiol. 2007, 9, 1101–1111. [Google Scholar] [CrossRef]
- Thomas, F.; Hehemann, J.H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut bacteroidetes: The food connection. Front. Microbiol. 2011, 2, 93. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458.e5–469.e5. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Tanabe, S.; Suzuki, T. High-fat Diet-induced Intestinal Hyperpermeability is Associated with Increased Bile Acids in the Large Intestine of Mice. J. Food Sci. 2016, 81, H216–H222. [Google Scholar] [CrossRef] [PubMed]
- Bortolotti, M.; Levorato, M.; Lugli, A.; Mazzero, G. Effect of a balanced mixture of dietary fibers on gastric emptying, intestinal transit and body weight. Ann. Nutr. Metab. 2008, 52, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Trowell, H. Definition of dietary fiber and hypotheses that it is a protective factor in certain diseases. Am. J. Clin. Nutr. 1976, 29, 417–427. [Google Scholar] [CrossRef]
- Al-Masaudi, S.; El Kaoutari, A.; Drula, E.; Redwan, E.M.; Lombard, V.; Henrissat, B. A metagenomics investigation of carbohydrate-active enzymes along the goat and camel intestinal tract. Int. Microbiol. 2019, 22, 429–435. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Liu, J.; Gao, J.; Wu, X.; Cui, C.; Wei, H.; Peng, J.; Zheng, R. The Effect of Functional Fiber on Microbiota Composition in Different Intestinal Segments of Obese Mice. Int. J. Mol. Sci. 2021, 22, 6525. https://doi.org/10.3390/ijms22126525
Xu C, Liu J, Gao J, Wu X, Cui C, Wei H, Peng J, Zheng R. The Effect of Functional Fiber on Microbiota Composition in Different Intestinal Segments of Obese Mice. International Journal of Molecular Sciences. 2021; 22(12):6525. https://doi.org/10.3390/ijms22126525
Chicago/Turabian StyleXu, Chuanhui, Jianhua Liu, Jianwei Gao, Xiaoyu Wu, Chenbin Cui, Hongkui Wei, Jian Peng, and Rong Zheng. 2021. "The Effect of Functional Fiber on Microbiota Composition in Different Intestinal Segments of Obese Mice" International Journal of Molecular Sciences 22, no. 12: 6525. https://doi.org/10.3390/ijms22126525
APA StyleXu, C., Liu, J., Gao, J., Wu, X., Cui, C., Wei, H., Peng, J., & Zheng, R. (2021). The Effect of Functional Fiber on Microbiota Composition in Different Intestinal Segments of Obese Mice. International Journal of Molecular Sciences, 22(12), 6525. https://doi.org/10.3390/ijms22126525




