Expression of the E5 Oncoprotein of HPV16 Impacts on the Molecular Profiles of EMT-Related and Differentiation Genes in Ectocervical Low-Grade Lesions
Abstract
1. Introduction
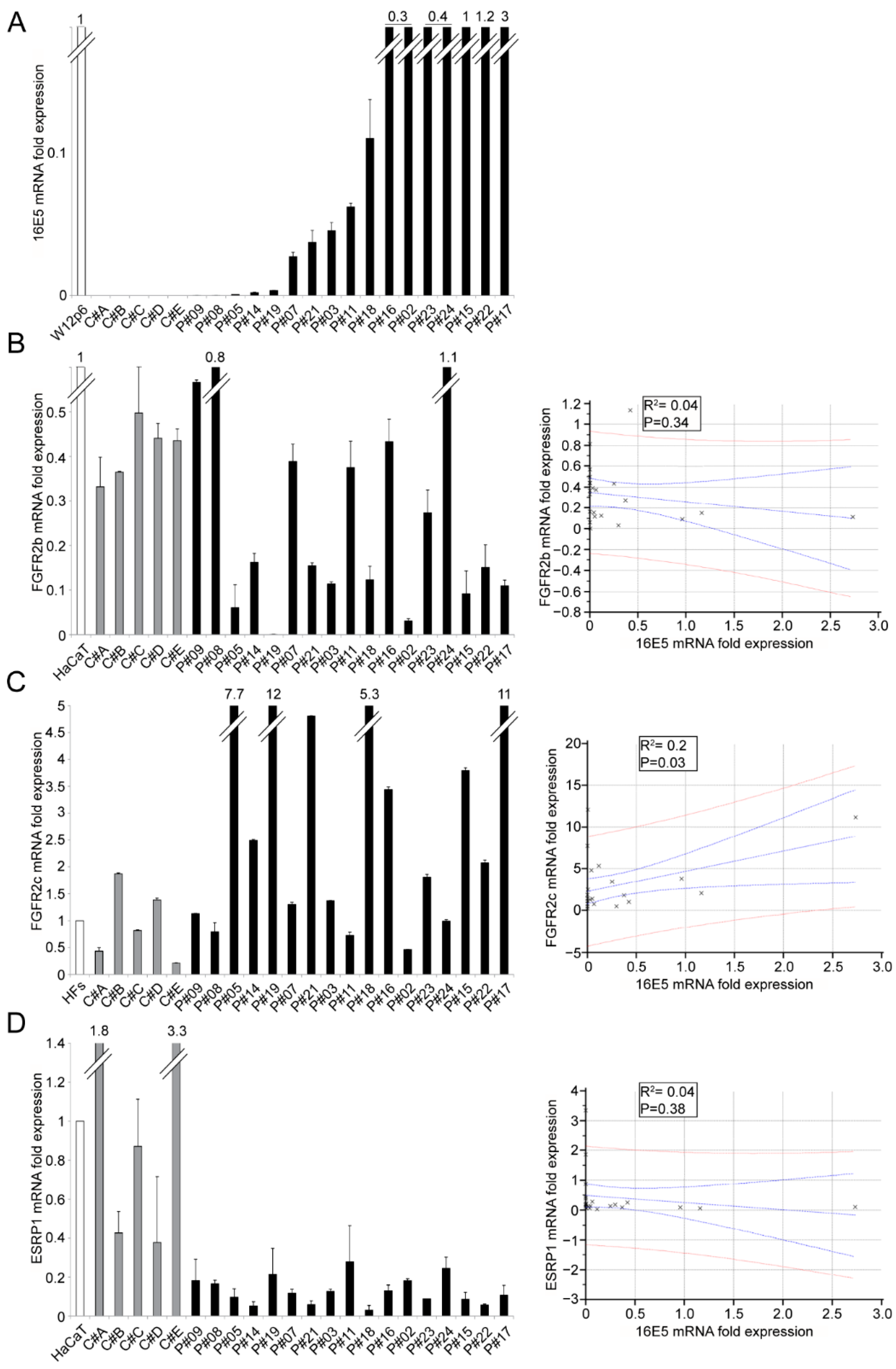
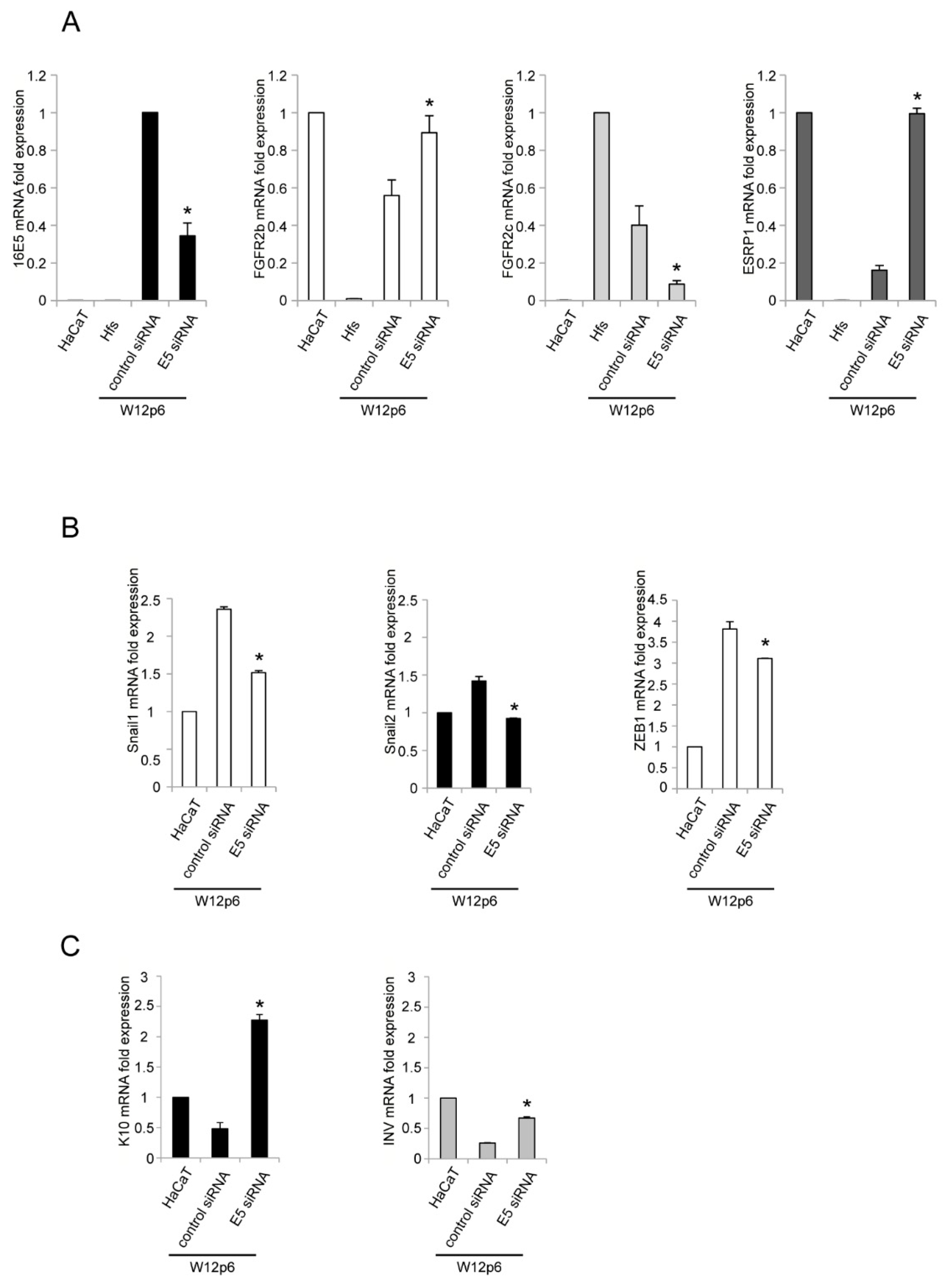
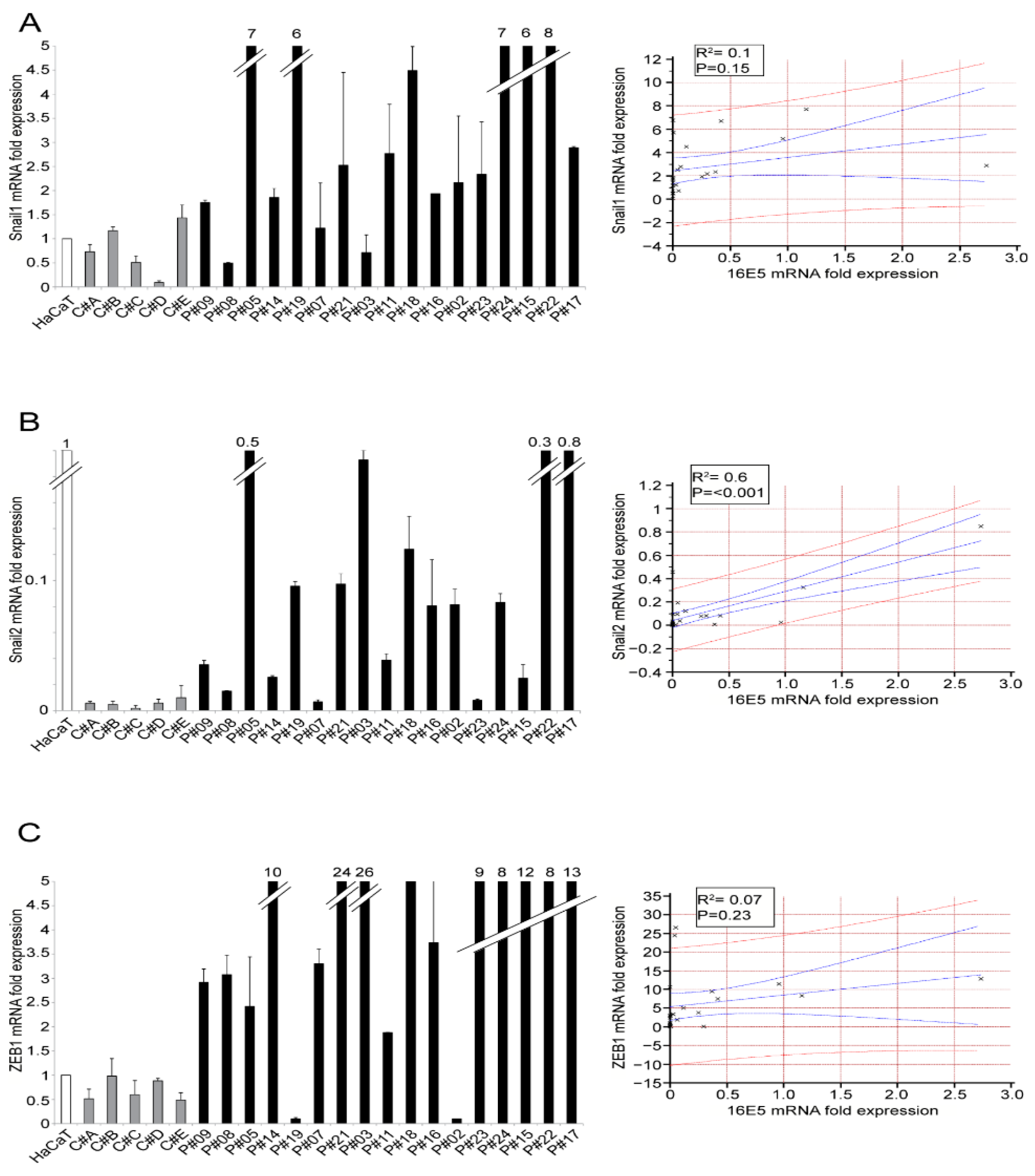
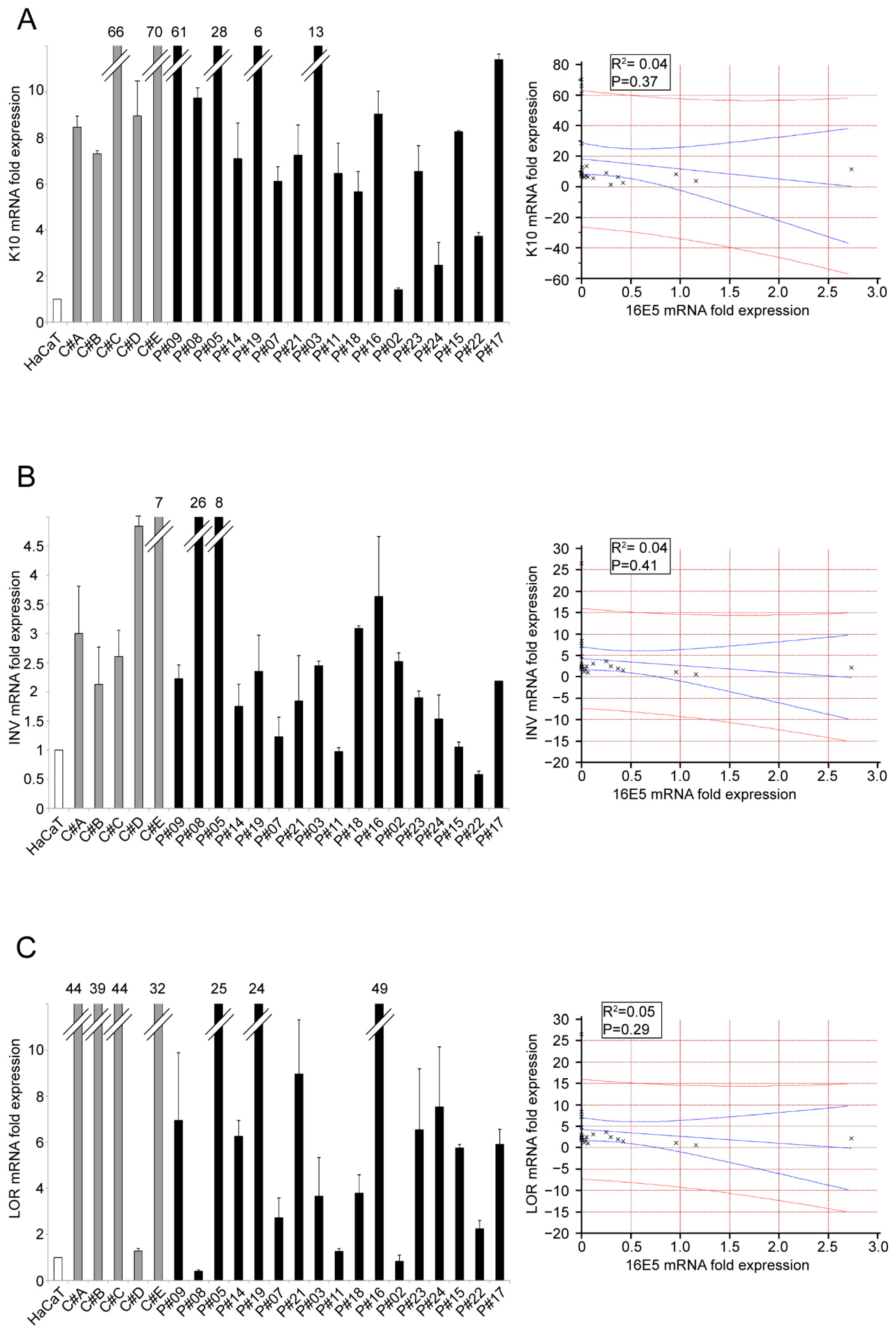
2. Results and Discussion
3. Materials and Methods
3.1. Cytological Samples
3.2. Cells and Treatments
4. Primers
4.1. RNA Extraction and cDNA Synthesis
4.2. PCR Amplification and Real-Time Quantitation
4.3. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crosbie, E.J.; Einstein, M.H.; Franceschi, S.; Kitchener, H.C. Human papillomavirus and cervical cancer. Lancet 2013, 382, 889–899. [Google Scholar] [CrossRef]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The biology and life-cycle of human papillomaviruses. Vaccine 2012, 30 (Suppl. S5), F55–F70. [Google Scholar] [CrossRef]
- Gray, E.; Pett, M.R.; Ward, D.; Winder, D.M.; Stanley, M.A.; Roberts, I.; Scarpini, C.G.; Coleman, N. In vitro progression of human papillomavirus 16 episome-associated cervical neoplasia displays fundamental similarities to integrant-associated carcinogenesis. Cancer Res. 2010, 70, 4081–4091. [Google Scholar] [CrossRef]
- Mendoza-Almanza, G.; Ortíz-Sánchez, E.; Rocha-Zavaleta, L.; Rivas-Santiago, C.; Esparza-Ibarra, E.; Olmos, J. Cervical cancer stem cells and other leading factors associated with cervical cancer development. Oncol. Lett. 2019, 18, 3423–3432. [Google Scholar] [CrossRef]
- Maufort, J.P.; Shai, A.; Pitot, H.C.; Lambert, P.F. A role for HPV16 E5 in cervical carcinogenesis. Cancer Res. 2010, 70, 2924–2931. [Google Scholar] [CrossRef]
- Paolini, F.; Curzio, G.; Cordeiro, M.N.; Massa, S.; Mariani, L.; Pimpinelli, F.; de Freitas, A.C.; Franconi, R.; Venuti, A. HPV 16 E5 oncoprotein is expressed in early stage carcinogenesis and can be a target of immunotherapy. Hum. Vaccines Immunother. 2017, 13, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Moody, C.A.; Laimins, L.A. Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer 2010, 10, 550–560. [Google Scholar] [CrossRef]
- Al Moustafa, A.-E. E5 and E6/E7 of high-risk HPVs cooperate to enhance cancer progression through EMT initiation. Cell Adh Migr. 2015, 9, 392–393. [Google Scholar] [CrossRef] [PubMed]
- de Freitas, A.C.; Araújo de Oliveira, T.H.; Rego Barros, M., Jr.; Venuti, A. hrHPV E5 oncoprotein: Immune evasion and related immunotherapies. J. Exp. Clin. Cancer Res. 2017, 36, 71. [Google Scholar] [CrossRef]
- Purpura, V.; Belleudi, F.; Caputo, S.; Torrisi, M.R. HPV16 E5 and KGFR/FGFR2b interplay in differentiating epithelial cells. Oncotarget 2013, 4, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Rosato, B.; Ranieri, D.; Nanni, M.; Torrisi, M.R.; Belleudi, F. Role of FGFR2b expression and signaling in keratinocyte differentiation: Sequential involvement of PKCδ and PKCα. Cell Death Dis. 2018, 9, 565. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, D.; Belleudi, F.; Magenta, A.; Torrisi, M.R. HPV16 E5 expression induces switching from FGFR2b to FGFR2c and epithelial-mesenchymal transition. Int. J. Cancer 2015, 137, 61–72. [Google Scholar] [CrossRef]
- Ranieri, D.; Rosato, B.; Nanni, M.; Magenta, A.; Belleudi, F.; Torrisi, M.R. Expression of the FGFR2 mesenchymal splicing variant in epithelial cells drives epithelial-mesenchymal transition. Oncotarget 2016, 7, 5440–5460. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, D.; Rosato, B.; Nanni, M.; Belleudi, F.; Torrisi, M.R. Expression of the FGFR2c mesenchymal splicing variant in human keratinocytes inhibits differentiation and promotes invasion. Mol. Carcinog. 2018, 57, 272–283. [Google Scholar] [CrossRef]
- Scott, M.L.; Coleman, D.T.; Kelly, K.C.; Carroll, J.L.; Woodby, B.; Songock, W.K.; Cardelli, J.A.; Bodily, J.M. Human papillomavirus type 16 E5-mediated upregulation of Met in human keratinocytes. Virology 2018, 519, 1–11. [Google Scholar] [CrossRef]
- Wasson, C.W.; Morgan, E.L.; Müller, M.; Ross, R.L.; Hartley, M.; Roberts, S.; Macdonald, A. Human papillomavirus type 18 E5 oncogene supports cell cycle progression and impairs epithelial differentiation by modulating growth factor receptor signalling during the virus life cycle. Oncotarget 2017, 8, 103581–103600. [Google Scholar] [CrossRef]
- Stanley, M.A.; Browne, H.M.; Appleby, M.; Minson, A.C. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int. J. Cancer 1989, 43, 672–676. [Google Scholar] [CrossRef]
- French, D.; Belleudi, F.; Mauro, M.V.; Mazzetta, F.; Raffa, S.; Fabiano, V.; Frega, A.; Torrisi, M.R. Expression of HPV16 E5 down-modulates the TGFbeta signaling pathway. Mol. Cancer 2013, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Lorenzon, L.; Mazzetta, F.; Venuti, A.; Frega, A.; Torrisi, M.R.; French, D. In vivo HPV 16 E5 mRNA: Expression pattern in patients with squamous intra-epithelial lesions of the cervix. J. Clin. Virol. 2011, 52, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Cheunim, T.; Zhang, J.; Milligan, S.G.; McPhillips, M.G.; Graham, S.V. The alternative splicing factor hnRNP A1 is up-regulated during virus-infected epithelial cell differentiation and binds the human papillomavirus type 16 late regulatory element. Virus Res. 2008, 131, 189–198. [Google Scholar] [CrossRef]
- Solomon, D.; Davey, D.; Kurman, R.; Moriarty, A.; O’Connor, D.; Prey, M.; Raab, S.; Sherman, M.; Wilbur, D.; Wright, T., Jr.; et al. The 2001 Bethesda System: Terminology for reporting results of cervical cytology. JAMA 2002, 287, 2114–2119. [Google Scholar] [CrossRef] [PubMed]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human kera- tinocyte cell line. J. Cell Biol. 1988, 106, 761–771. [Google Scholar] [CrossRef]
- Pett, M.R.; Alazawi, W.O.F.; Roberts, I.; Dowen, S.; Smith, D.I.; Stanley, M.A.; Coleman, N. Acquisition of high-level chromosomal instability is associated with integration of human papillomavirus type 16 in cervical keratinocytes. Cancer Res. 2004, 64, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Persechino, F.; Ranieri, D.; Guttieri, L.; Nanni, M.; Torrisi, M.R.; Belleudi, F. Expression Profile of Fibroblast Growth Factor Receptors, Keratinocyte Differentiation Markers, and Epithelial Mesenchymal Transition-Related Genes in Actinic Keratosis: A Possible Predictive Factor for Malignant Progression? Biology 2021, 10, 331. [Google Scholar] [CrossRef]
- Oh, J.M.; Kim, S.H.; Lee, Y.I.; Seo, M.; Kim, S.Y.; Song, Y.S.; Kim, W.H.; Juhnn, Y.S. Human papillomavirus E5 protein induces expression of the EP4 subtype of prostaglandin E2 receptor in cyclic AMP response element-dependent pathways in cervical cancer cells. Carcinogenesis 2009, 30, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranieri, D.; French, D.; Raffa, S.; Guttieri, L.; Torrisi, M.R.; Belleudi, F. Expression of the E5 Oncoprotein of HPV16 Impacts on the Molecular Profiles of EMT-Related and Differentiation Genes in Ectocervical Low-Grade Lesions. Int. J. Mol. Sci. 2021, 22, 6534. https://doi.org/10.3390/ijms22126534
Ranieri D, French D, Raffa S, Guttieri L, Torrisi MR, Belleudi F. Expression of the E5 Oncoprotein of HPV16 Impacts on the Molecular Profiles of EMT-Related and Differentiation Genes in Ectocervical Low-Grade Lesions. International Journal of Molecular Sciences. 2021; 22(12):6534. https://doi.org/10.3390/ijms22126534
Chicago/Turabian StyleRanieri, Danilo, Deborah French, Salvatore Raffa, Luisa Guttieri, Maria Rosaria Torrisi, and Francesca Belleudi. 2021. "Expression of the E5 Oncoprotein of HPV16 Impacts on the Molecular Profiles of EMT-Related and Differentiation Genes in Ectocervical Low-Grade Lesions" International Journal of Molecular Sciences 22, no. 12: 6534. https://doi.org/10.3390/ijms22126534
APA StyleRanieri, D., French, D., Raffa, S., Guttieri, L., Torrisi, M. R., & Belleudi, F. (2021). Expression of the E5 Oncoprotein of HPV16 Impacts on the Molecular Profiles of EMT-Related and Differentiation Genes in Ectocervical Low-Grade Lesions. International Journal of Molecular Sciences, 22(12), 6534. https://doi.org/10.3390/ijms22126534





