Immortalization and Characterization of Rat Lingual Keratinocytes in a High-Calcium and Feeder-Free Culture System Using ROCK Inhibitor Y-27632
Abstract
:1. Introduction
2. Results
2.1. Isolation of RLKs and Screening of Culture Media
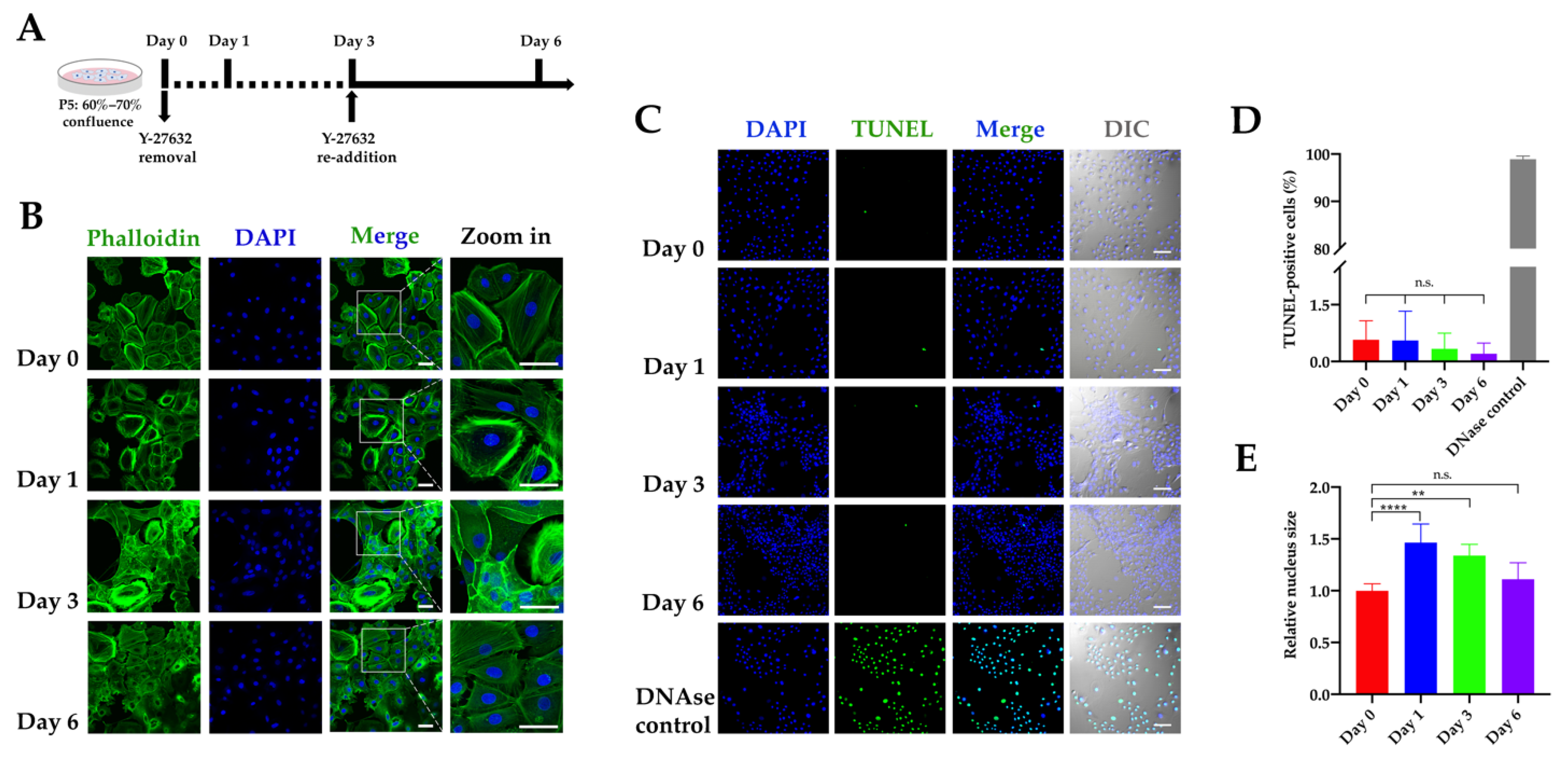
2.2. Conditional and Reversible Immortalization of RLKs by Y-27632
2.3. Proteome Profiling of RLKs in the Presence and Absence of Y-27632
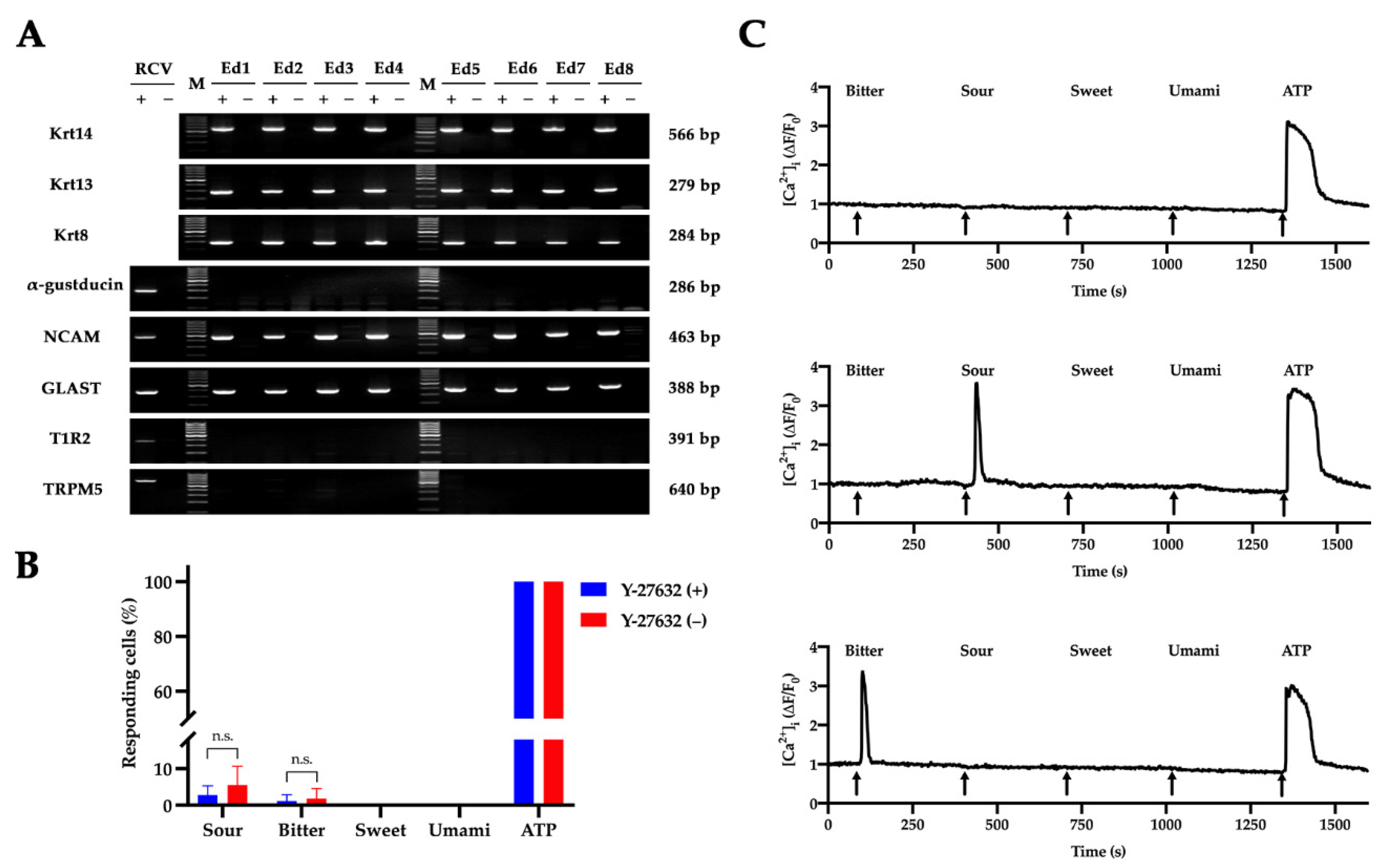
2.4. Characterization of RLKs
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Primary Culture and Maintenance of RLKs
4.3. Phalloidin Staining
4.4. TUNEL Assay
4.5. Sample Preparation for Mass Spectrometry
4.6. Nano-UPLC-MS/MS and Comparative Proteomic Analysis
4.7. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.8. Live-Cell Calcium Imaging
4.9. Cytogenetic Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Breidahl, A.F.; Juoson, R.T.; Clunie, G.J.A. Review of keratinocyte culture techniques: Problems of growing skin. Aust. N. Z. J. Surg. 1989, 59, 485–497. [Google Scholar] [CrossRef]
- Barrandon, Y.; Grasset, N.; Zaffalon, A.; Gorostidi, F.; Claudinot, S.; Droz-Georget, S.L.; Nanba, D.; Rochat, A. Capturing epidermal stemness for regenerative medicine. Semin. Cell Dev. Biol. 2012, 23, 937–944. [Google Scholar] [CrossRef]
- Choi, M.; Lee, C. Immortalization of primary keratinocytes and its application to skin research. Biomol. Ther. (Seoul) 2015, 23, 391–399. [Google Scholar] [CrossRef] [Green Version]
- Hammiller, B.O.; El-Abaseri, T.B.; Dlugosz, A.A.; Hansen, L.A. A method for the immortalization of newborn mouse skin keratinocytes. Front. Oncol. 2015, 5. [Google Scholar] [CrossRef] [Green Version]
- Hager, B.; Fleckman, P.; Bickenbach, J.R. Long-term culture of murine epidermal keratinocytes. J. Invest. Dermatol. 1999, 112, 971–976. [Google Scholar] [CrossRef] [Green Version]
- Reichelt, J.; Haase, I. Establishment of spontaneously immortalized keratinocyte lines from wild-type and mutant mice. In Epidermal Cells. Methods in Molecular Biology (Methods and Protocols), 2nd ed.; Turksen, K., Ed.; Humana Press: Totowa, NJ, USA, 2010; Volume 585, pp. 59–69. [Google Scholar]
- Rheinwald, J.G. Serial cultivation of normal human epidermal keratinocytes. In Methods in Cell Biology, 1st ed.; Harris, C.C., Trump, B.F., Stoner, G.D., Eds.; Academic Press: New York, NY, USA, 1980; Volume 21, pp. 229–254. [Google Scholar]
- Ścieżyńska, A.; Nogowska, A.; Sikorska, M.; Konys, J.; Karpińska, A.; Komorowski, M.; Ołdak, M.; Malejczyk, J. Isolation and culture of human primary keratinocytes—a methods review. Exp. Dermatol. 2019, 28, 107–112. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.-J. Isolation, culture, and characterization of primary mouse epidermal keratinocytes. In Mouse Cell Culture: Methods and Protocols, 1st ed.; Bertoncello, I., Ed.; Humana Press: New York, NY, USA, 2019; Volume 1940, pp. 205–215. [Google Scholar]
- Oku, H.; Kumamoto, C.; Miyagi, T.; Hiyane, T.; Nagata, J.; Chinen, I. Serum-free culture of rat keratinocytes. Vitr. Cell. Dev. Biol. Anim. 1994, 30, 496–503. [Google Scholar] [CrossRef]
- Hisha, H.; Tanaka, T.; Kanno, S.; Tokuyama, Y.; Komai, Y.; Ohe, S.; Yanai, H.; Omachi, T.; Ueno, H. Establishment of a novel lingual organoid culture system: Generation of organoids having mature keratinized epithelium from adult epithelial stem cells. Sci. Rep. 2013, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Simonson, L.; Vold, S.; Mowers, C.; Massey, R.J.; Ong, I.M.; Longley, B.J.; Chang, H. Keratin 13 deficiency causes white sponge nevus in mice. Dev. Biol. 2020, 468, 146–153. [Google Scholar] [CrossRef]
- Miura, H.; Kusakabe, Y.; Hashido, K.; Hino, A.; Ooki, M.; Harada, S. The glossopharyngeal nerve controls epithelial expression of Sprr2a and Krt13 around taste buds in the circumvallate papilla. Neurosci. Lett. 2014, 580, 147–152. [Google Scholar] [CrossRef]
- Iwasaki, S.-I.; Aoyagi, H.; Yoshizawa, H. Localization of keratins 13 and 14 in the lingual mucosa of rats during the morphogenesis of circumvallate papillae. Acta Histochem. 2011, 113, 395–401. [Google Scholar] [CrossRef]
- Gaillard, D.; Xu, M.; Liu, F.; Millar, S.E.; Barlow, L.A. β-catenin signaling biases multipotent lingual epithelial progenitors to differentiate and acquire specific taste cell fates. PLoS Genet. 2015, 11, e1005208. [Google Scholar] [CrossRef]
- Bierbaumer, L.; Schwarze, U.Y.; Gruber, R.; Neuhaus, W. Cell culture models of oral mucosal barriers: A review with a focus on applications, culture conditions and barrier properties. Tissue Barriers 2018, 6, 1479568. [Google Scholar] [CrossRef]
- Luo, X.; Okubo, T.; Randell, S.; Hogan, B.L. Culture of endodermal stem/progenitor cells of the mouse tongue. Vitr. Cell. Dev. Biol. Anim. 2009, 45, 44–54. [Google Scholar] [CrossRef]
- Nakamura, S.-I.; Kamakura, T.; Ookura, T. Tongue epithelial KT-1 cell-cycle arrest by TGF-β associated with induction of p21 Cip1 and p15 Ink4b. Cytotechnology 2009, 61, 109–116. [Google Scholar] [CrossRef] [Green Version]
- Ookura, T.; Kawamoto, K.; Tsuzaki, H.; Mikami, Y.; Ito, Y.; Oh, S.-H.; Hino, A. Fibroblast and epidermal growth factors modulate proliferation and neural cell adhesion molecule expression in epithelial cells derived from the adult mouse tongue. Vitr. Cell. Dev. Biol. Anim. 2002, 38, 365–372. [Google Scholar] [CrossRef]
- van den Bogaard, E.H.; Rodijk-Olthuis, D.; Jansen, P.A.; van Vlijmen-Willems, I.M.; van Erp, P.E.; Joosten, I.; Zeeuwen, P.L.; Schalkwijk, J. Rho kinase inhibitor Y-27632 prolongs the life span of adult human keratinocytes, enhances skin equivalent development, and facilitates lentiviral transduction. Tissue Eng. Part A 2012, 18, 1827–1836. [Google Scholar] [CrossRef] [Green Version]
- Chapman, S.; Liu, X.; Meyers, C.; Schlegel, R.; McBride, A.A. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Invest. 2010, 120, 2619–2626. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.-C.; Chiu, H.-T.; Lin, Y.-F.; Lee, K.-Y.; Pang, J.-H.S. Y-27632, a ROCK inhibitor, promoted limbal epithelial cell proliferation and corneal wound healing. PLoS ONE 2015, 10, e0144571. [Google Scholar] [CrossRef]
- Chapman, S.; McDermott, D.H.; Shen, K.; Jang, M.K.; McBride, A.A. The effect of Rho kinase inhibition on long-term keratinocyte proliferation is rapid and conditional. Stem Cell. Res. Ther. 2014, 5, 60. [Google Scholar] [CrossRef] [Green Version]
- Strudwick, X.L.; Lang, D.L.; Smith, L.E.; Cowin, A.J. Combination of low calcium with Y-27632 rock inhibitor increases the proliferative capacity, expansion potential and lifespan of primary human keratinocytes while retaining their capacity to differentiate into stratified epidermis in a 3D skin model. PLoS ONE 2015, 10, e0123651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Adase, C.A.; Zhang, L.-J. Isolation and culture of primary mouse keratinocytes from neonatal and adult mouse skin. J. Vis. Exp. 2017, 125, 56027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, G.; Liao, P.Y.; Cai, X.L.; Pi, X.X.; Zhang, M.F.; Li, S.J.; Quan, J.H.; Fan, Y.M. FoxO1 enhances differentiation and apoptosis in human primary keratinocytes. Exp. Dermatol. 2018, 27, 1254–1260. [Google Scholar] [CrossRef] [PubMed]
- Ozdener, H.; Yee, K.K.; Cao, J.; Brand, J.G.; Teeter, J.H.; Rawson, N.E. Characterization and long-term maintenance of rat taste cells in culture. Chem. Senses 2006, 31, 279–290. [Google Scholar] [CrossRef] [Green Version]
- Halbert, C.L.; Demers, G.W.; Galloway, D.A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 1991, 65, 473–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palechor-Ceron, N.; Suprynowicz, F.A.; Upadhyay, G.; Dakic, A.; Minas, T.; Simic, V.; Johnson, M.; Albanese, C.; Schlegel, R.; Liu, X. Radiation induces diffusible feeder cell factor (s) that cooperate with ROCK inhibitor to conditionally reprogram and immortalize epithelial cells. Am. J. Pathol. 2013, 183, 1862–1870. [Google Scholar] [CrossRef] [Green Version]
- Riento, K.; Ridley, A.J. Rocks: Multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 2003, 4, 446–456. [Google Scholar] [CrossRef]
- Dakic, A.; DiVito, K.; Fang, S.; Suprynowicz, F.; Gaur, A.; Li, X.; Palechor-Ceron, N.; Simic, V.; Choudhury, S.; Yu, S.; et al. ROCK inhibitor reduces Myc-induced apoptosis and mediates immortalization of human keratinocytes. Oncotarget 2016, 7, 66740–66753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Darzynkiewicz, Z. Biomarkers of cell senescence assessed by imaging cytometry. In Cell Senescence: Methods and Protocols, 1st ed.; Galluzzi, L., Vitale, I., Kepp, O., Kroemer, G., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 965, pp. 83–92. [Google Scholar]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Vong, K.I.; Ma, T.C.; Li, B.; Leung, T.C.N.; Nong, W.; Ngai, S.M.; Hui, J.H.L.; Jiang, L.; Kwan, K.M. SOX9-COL9A3–dependent regulation of choroid plexus epithelial polarity governs blood-cerebrospinal fluid barrier integrity. Proc. Natl. Acad. Sci. USA 2021, 118, e2009568118. [Google Scholar] [CrossRef]
- Roper, S.D.; Chaudhari, N. Taste buds: Cells, signals and synapses. Nat. Rev. Neurosci. 2017, 18, 485–497. [Google Scholar] [CrossRef]
- Ren, W.; Lewandowski, B.C.; Watson, J.; Aihara, E.; Iwatsuki, K.; Bachmanov, A.A.; Margolskee, R.F.; Jiang, P. Single Lgr5-or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 16401–16406. [Google Scholar] [CrossRef] [Green Version]
- Butler, C.R.; Hynds, R.E.; Gowers, K.H.; Lee, D.D.H.; Brown, J.M.; Crowley, C.; Teixeira, V.H.; Smith, C.M.; Urbani, L.; Hamilton, N.J.; et al. Rapid expansion of human epithelial stem cells suitable for airway tissue engineering. Am. J. Respir. Crit. Care Med. 2016, 194, 156–168. [Google Scholar] [CrossRef] [Green Version]
- Alkhilaiwi, F.; Wang, L.; Zhou, D.; Raudsepp, T.; Ghosh, S.; Paul, S.; Palechor-Ceron, N.; Brandt, S.; Luff, J.; Liu, X.; et al. Long-term expansion of primary equine keratinocytes that maintain the ability to differentiate into stratified epidermis. Stem Cell. Res. Ther. 2018, 9, 181. [Google Scholar] [CrossRef]
- Mou, H.; Vinarsky, V.; Tata, P.R.; Brazauskas, K.; Choi, S.H.; Crooke, A.K.; Zhang, B.; Solomon, G.M.; Turner, B.; Bihler, H.; et al. Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell 2016, 19, 217–231. [Google Scholar] [CrossRef] [Green Version]
- Dale, T.P.; Borg D’anastasi, E.; Haris, M.; Forsyth, N.R. Rock Inhibitor Y-27632 enables feeder-free, unlimited expansion of sus scrofa domesticus swine airway stem cells to facilitate respiratory research. Stem Cells Int. 2019, 2019, 3010656. [Google Scholar] [CrossRef]
- Amin, E.; Dubey, B.N.; Zhang, S.-C.; Gremer, L.; Dvorsky, R.; Moll, J.M.; Taha, M.S.; Nagel-Steger, L.; Piekorz, R.P.; Somlyo, A.V.; et al. Rho-kinase: Regulation, (dys)function, and inhibition. Biol. Chem. 2013, 394, 1399–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, K.; Ueno, M.; Kamiya, D.; Nishiyama, A.; Matsumura, M.; Wataya, T.; Takahashi, J.B.; Nishikawa, S.; Nishikawa, S.-I.; Muguruma, K.; et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007, 25, 681–686. [Google Scholar] [CrossRef]
- Zhang, L.; Valdez, J.M.; Zhang, B.; Wei, L.; Chang, J.; Xin, L. ROCK inhibitor Y-27632 suppresses dissociation-induced apoptosis of murine prostate stem/progenitor cells and increases their cloning efficiency. PLoS ONE 2011, 6, e18271. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Miao, Y.; Fan, Z.; Zhang, J.; Guo, Y.; Dai, D.; Huang, J.; Liu, Z.; Chen, R.; Hu, Z. Establishment of an efficient primary culture system for human hair follicle stem cells using the rho-associated protein kinase inhibitor Y-27632. Front. Cell Dev. Biol. 2021, 9, 632882. [Google Scholar] [CrossRef]
- Zhang, C.; Lee, H.J.; Shrivastava, A.; Wang, R.; McQuiston, T.J.; Challberg, S.S.; Pollok, B.A.; Wang, T. Long-term in vitro expansion of epithelial stem cells enabled by pharmacological inhibition of PAK1-ROCK-Myosin II and TGF-β signaling. Cell Rep. 2018, 25, 598–610.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, Y.M.; Hong, J. Rho-associated protein kinase inhibitor, Y-27632, significantly enhances cell adhesion and induces a delay in G1 to S phase transition in rabbit corneal endothelial cells. Mol. Med. Rep. 2015, 12, 1951–1956. [Google Scholar] [CrossRef]
- Li, Z.; Han, S.; Wang, X.; Han, F.; Zhu, X.; Zheng, Z.; Wang, H.; Zhou, Q.; Wang, Y.; Su, L. Rho kinase inhibitor Y-27632 promotes the differentiation of human bone marrow mesenchymal stem cells into keratinocyte-like cells in xeno-free conditioned medium. Stem Cell. Res. Ther. 2015, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neilson, K.A.; Ali, N.A.; Muralidharan, S.; Mirzaei, M.; Mariani, M.; Assadourian, G.; Lee, A.; van Sluyter, S.C.; Haynes, P.A. Less label, more free: Approaches in label-free quantitative mass spectrometry. Proteomics 2011, 11, 535–553. [Google Scholar] [CrossRef] [PubMed]
- Rauniyar, N.; Yates, J.R., III. Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res. 2014, 13, 5293–5309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gevaert, K.; Impens, F.; Ghesquière, B.; van Damme, P.; Lambrechts, A.; Vandekerckhove, J. Stable isotopic labeling in proteomics. Proteomics 2008, 8, 4873–4885. [Google Scholar] [CrossRef] [Green Version]
- Feng, P.; Huang, L.; Wang, H. Taste bud homeostasis in health, disease, and aging. Chem. Senses 2014, 39, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Barlow, L.A. Progress and renewal in gustation: New insights into taste bud development. Development 2015, 142, 3620–3629. [Google Scholar] [CrossRef] [Green Version]
- Asano-Miyoshi, M.; Hamamichi, R.; Emori, Y. Cytokeratin 14 is expressed in immature cells in rat taste buds. J. Mol. Histol. 2008, 39, 193–199. [Google Scholar] [CrossRef]
- Barlow, L.A.; Klein, O.D. Developing and regenerating a sense of taste. Curr. Top. Dev. Biol. 2015, 111, 401–419. [Google Scholar] [PubMed] [Green Version]
- Okubo, T.; Clark, C.; Hogan, B.L. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells 2009, 27, 442–450. [Google Scholar] [CrossRef] [Green Version]
- Castillo, D.; Seidel, K.; Salcedo, E.; Ahn, C.; de Sauvage, F.J.; Klein, O.D.; Barlow, L.A. Induction of ectopic taste buds by SHH reveals the competency and plasticity of adult lingual epithelium. Development 2014, 141, 2993–3002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanenko, A.A.; Kavsan, V.M. Immortalization and malignant transformation of eukaryotic cells. Tsitol. Genet. 2012, 46, 96–129. [Google Scholar] [CrossRef]
- Thompson, S.L.; Compton, D.A. Chromosomes and cancer cells. Chromosome Res. 2011, 19, 433–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedrazzi, M.; Vercellone, S.; Barberis, E.; Capraro, M.; de Tullio, R.; Cresta, F.; Casciaro, R.; Castellani, C.; Patrone, M.; Marengo, E.; et al. Identification of potential leukocyte biomarkers related to drug recovery of cftr: Clinical applications in cystic fibrosis. Int. J. Mol. Sci. 2021, 22, 3928. [Google Scholar] [CrossRef] [PubMed]
- Gosselin, K.; Martien, S.; Pourtier, A.; Vercamer, C.; Ostoich, P.; Morat, L.; Sabatier, L.; Duprez, L.; de Roodenbeke, C.T.K.; Gilson, E.; et al. Senescence-associated oxidative DNA damage promotes the generation of neoplastic cells. Cancer Res. 2009, 69, 7917–7925. [Google Scholar] [CrossRef] [Green Version]
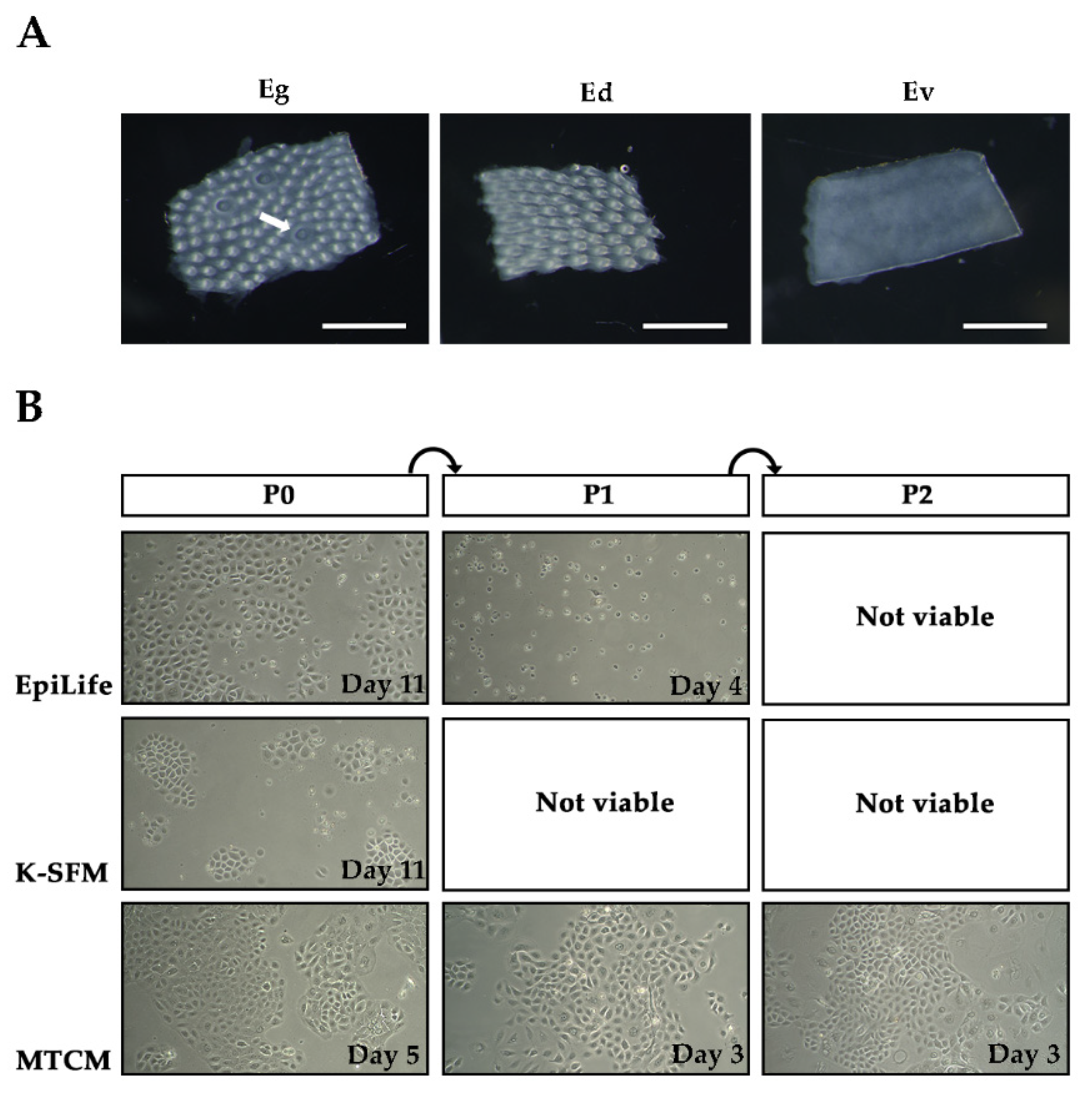



| EpiLife | K-SFM | MTCM | |
|---|---|---|---|
| Basal media | EpiLife™ medium | Defined keratinocytes SFM | IMDM + 10% FBS + 20% MCDB 153 medium |
| Supplements | EpiLife™ defined growth supplement, (10 μM Y-27632) 1 | Defined keratinocyte SFM growth supplement, (10 μM Y-27632) 1 | 10 ng/mL EGF, (10 μM Y-27632) 1 |
| Antibiotics | 1% antibiotic-antimycotic, 0.015% gentamicin | ||
| Calcium level | 60 µM | <0.1 mM | ~1.4 mM |
| References | [26] | [27] | [1,28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; He, W.; Leung, T.C.N.; Chung, H.Y. Immortalization and Characterization of Rat Lingual Keratinocytes in a High-Calcium and Feeder-Free Culture System Using ROCK Inhibitor Y-27632. Int. J. Mol. Sci. 2021, 22, 6782. https://doi.org/10.3390/ijms22136782
Chen Z, He W, Leung TCN, Chung HY. Immortalization and Characterization of Rat Lingual Keratinocytes in a High-Calcium and Feeder-Free Culture System Using ROCK Inhibitor Y-27632. International Journal of Molecular Sciences. 2021; 22(13):6782. https://doi.org/10.3390/ijms22136782
Chicago/Turabian StyleChen, Zixing, Wenmeng He, Thomas Chun Ning Leung, and Hau Yin Chung. 2021. "Immortalization and Characterization of Rat Lingual Keratinocytes in a High-Calcium and Feeder-Free Culture System Using ROCK Inhibitor Y-27632" International Journal of Molecular Sciences 22, no. 13: 6782. https://doi.org/10.3390/ijms22136782






