Involvement of ABA Responsive SVB Genes in the Regulation of Trichome Formation in Arabidopsis
Abstract
:1. Introduction
2. Results
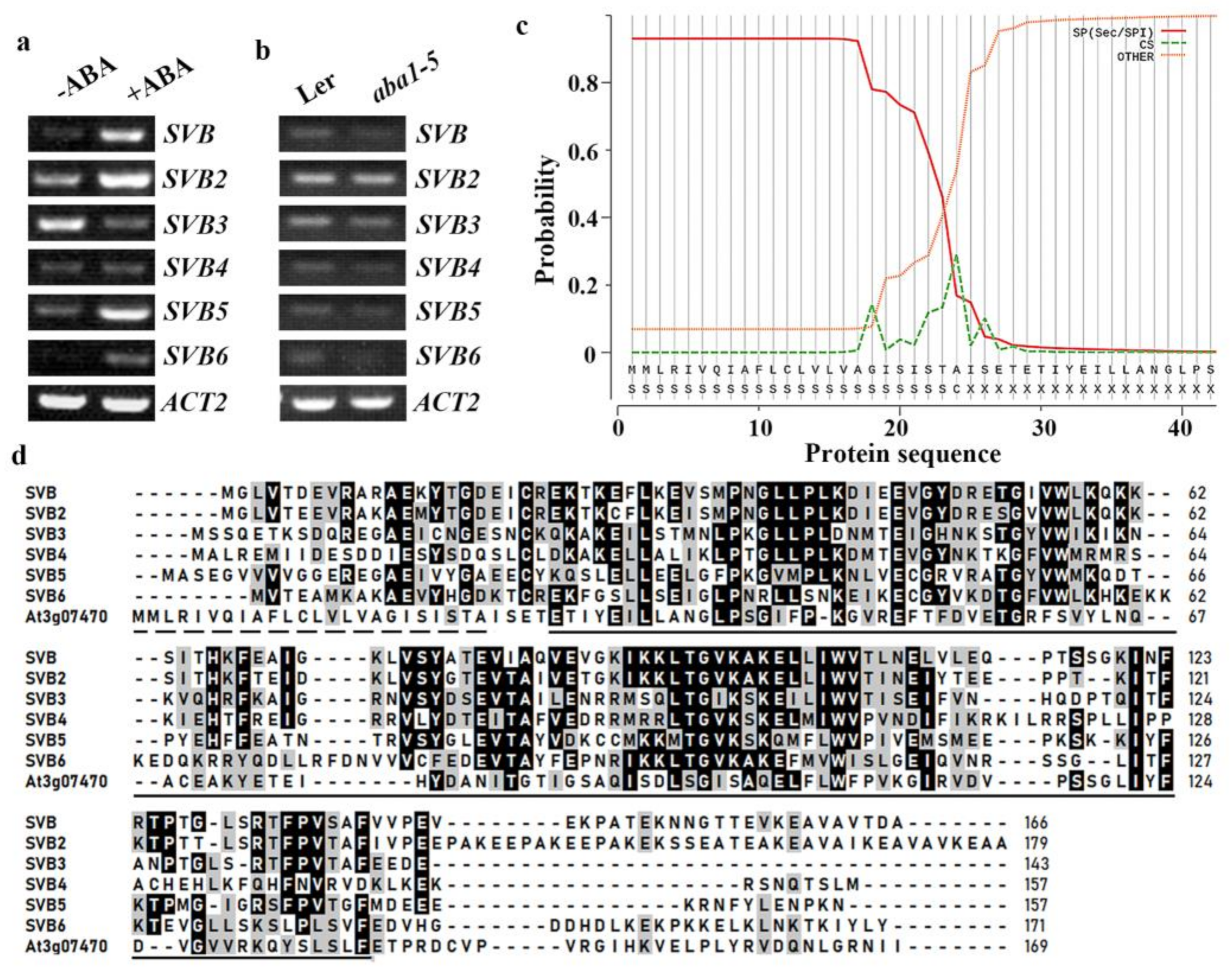
2.1. Expression of SVBs Are Regulated by ABA
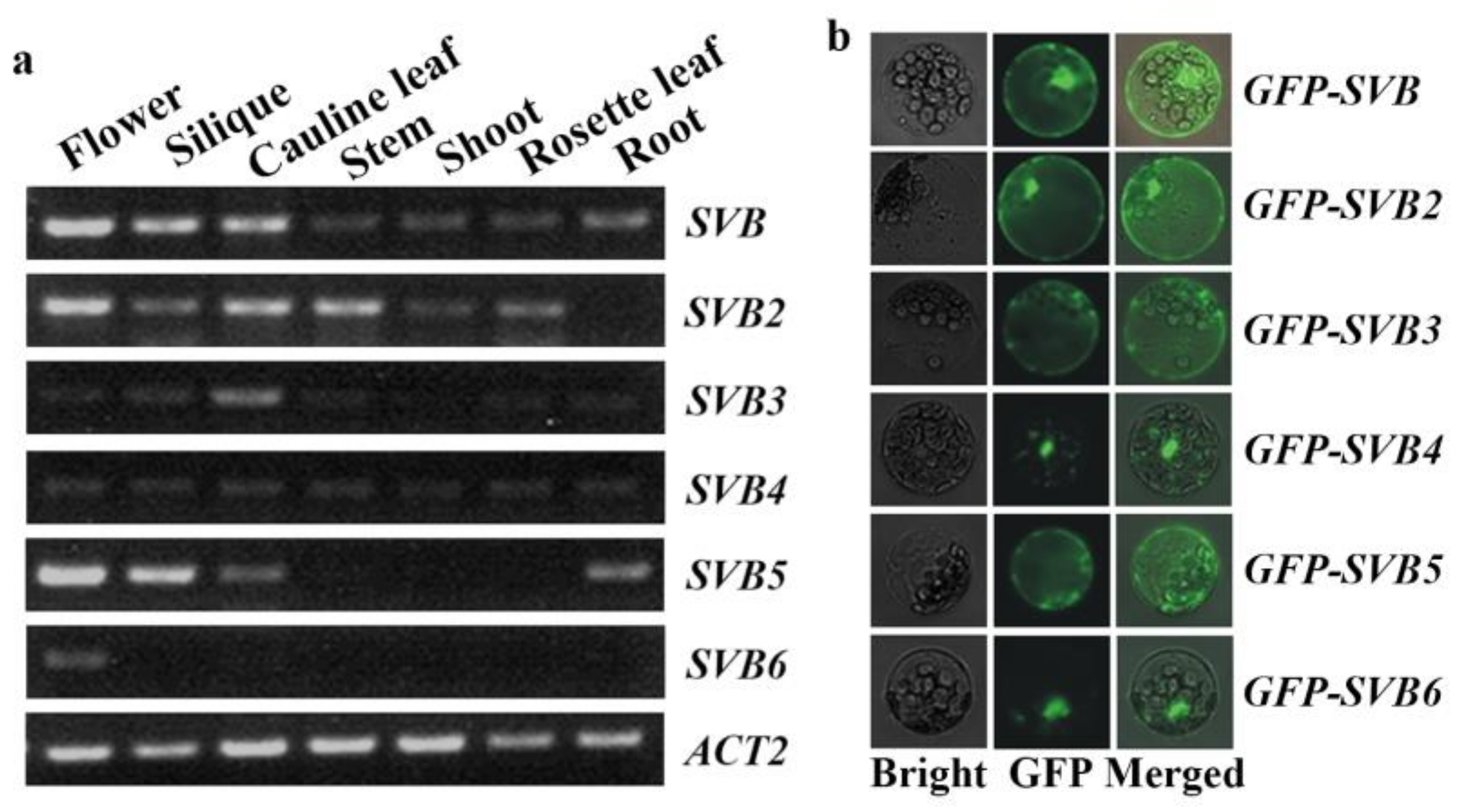
2.2. SVB and SVB2 Have Similar Expression Pattern and Similar Protein Subcellular Localization
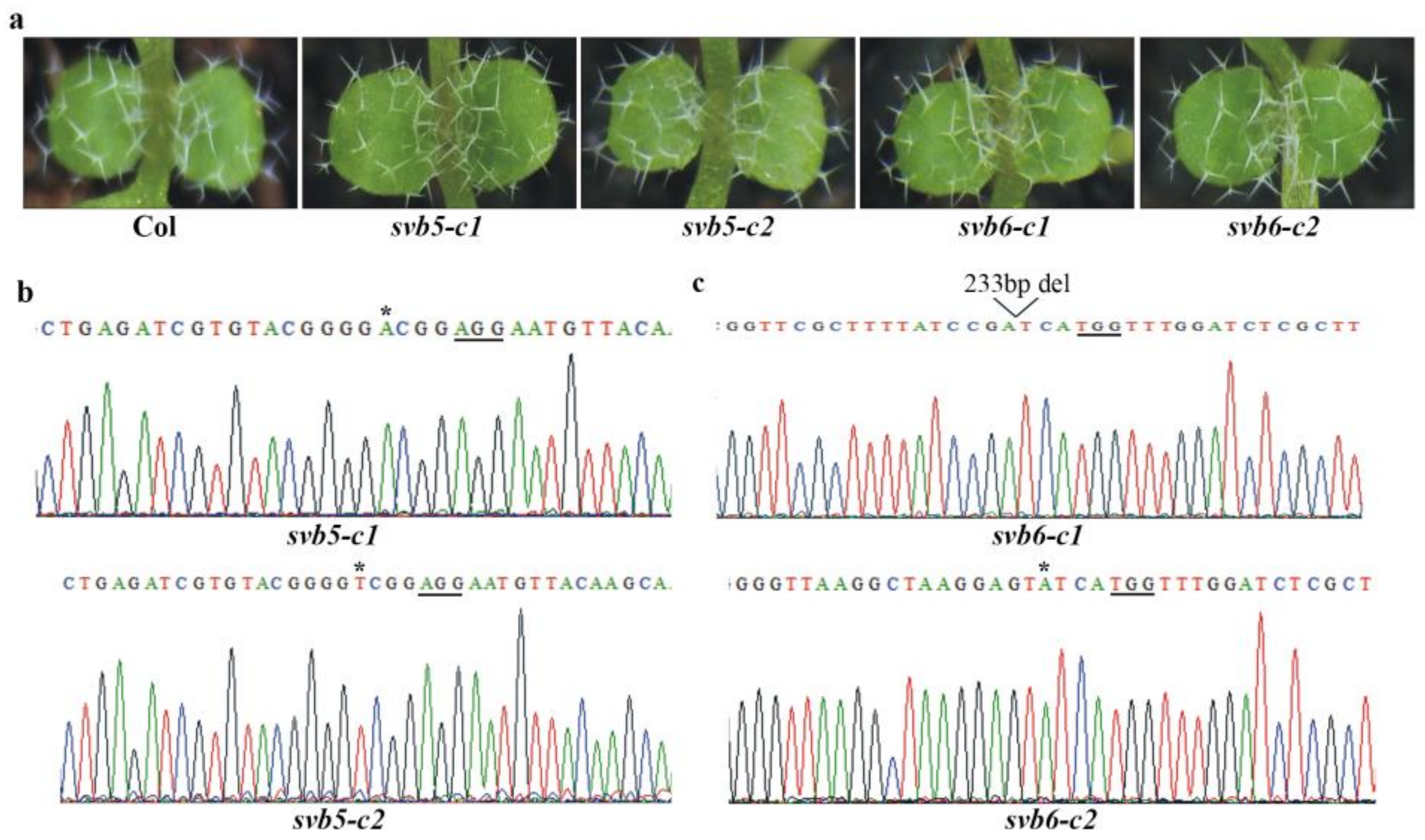
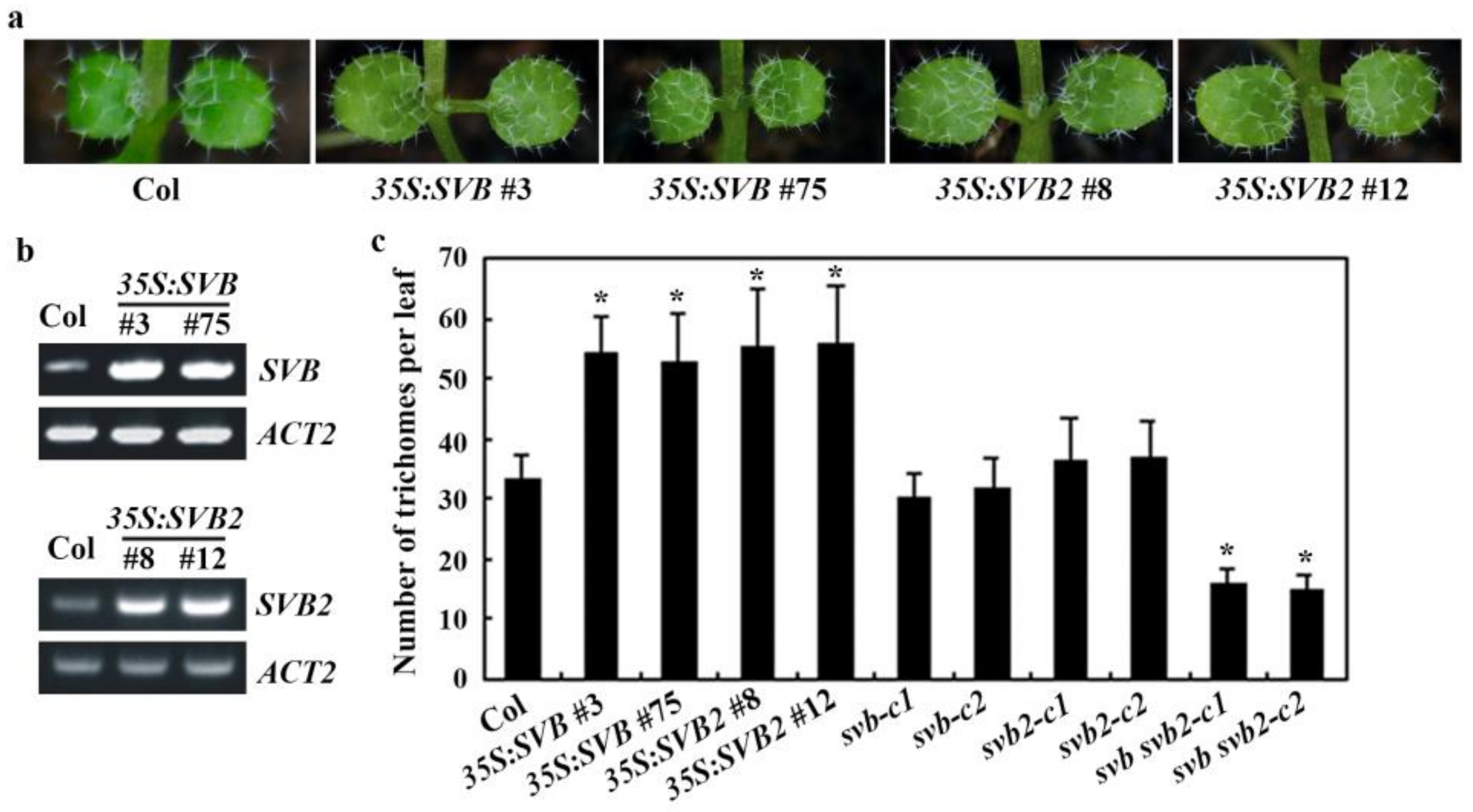
2.3. SVB but Not Other SVBs Affect Trichome Development
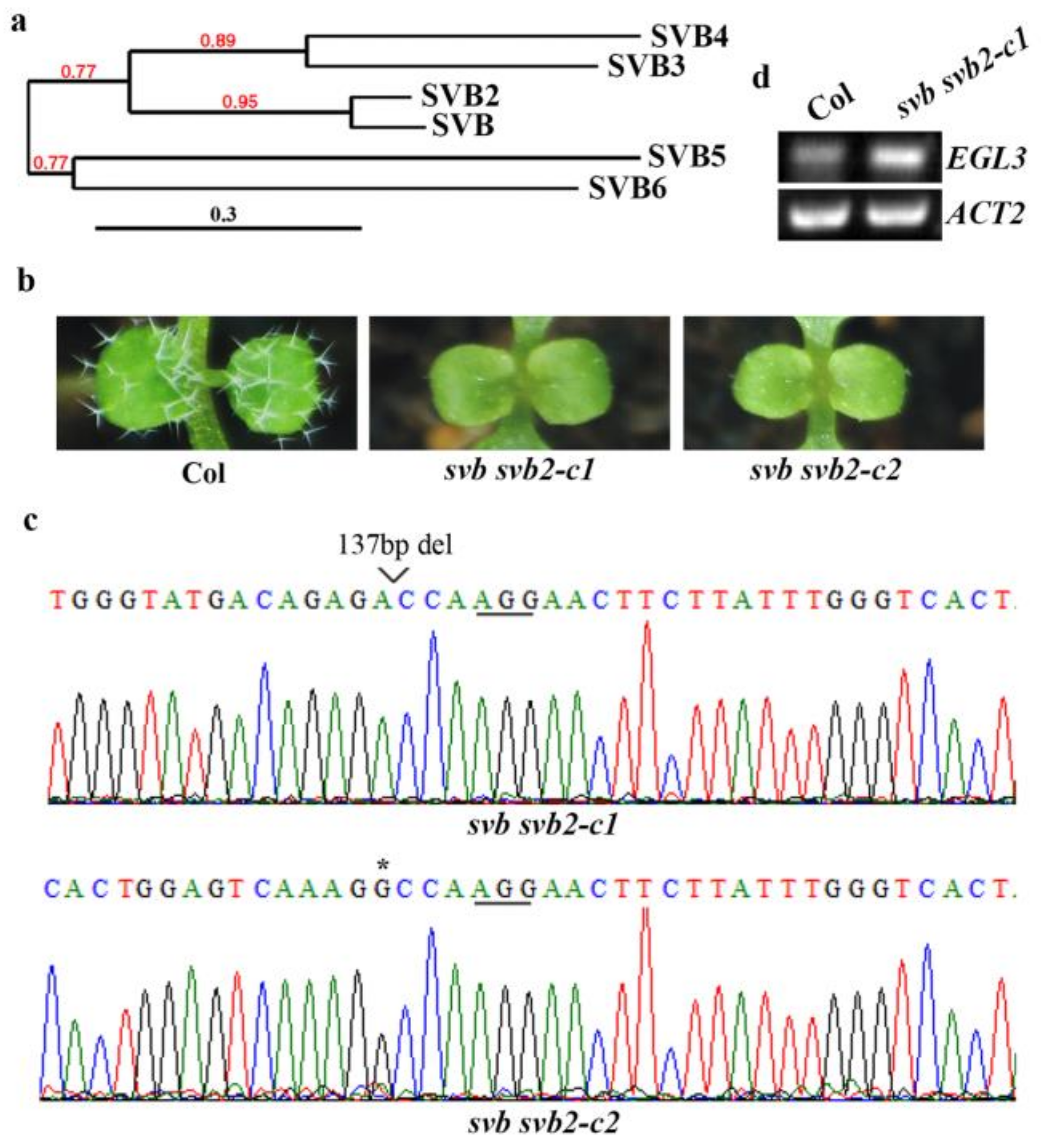
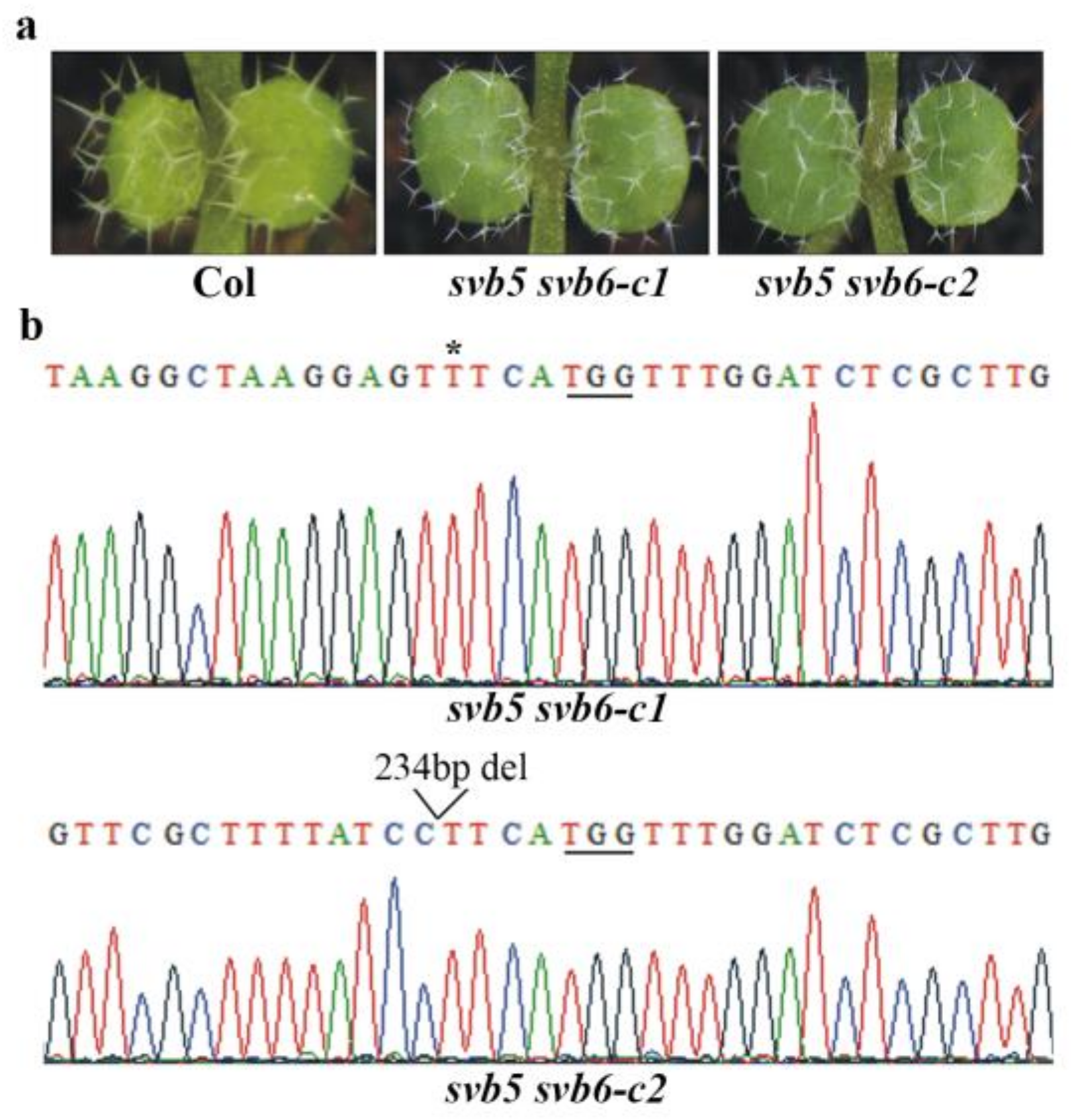
2.4. SVB and SVB2 Function Redundantly to Regulate Trichome Formation
2.5. SVB and SVB2 Function Redundantly to Regulate Plant Growth and Development
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Isolation and RT-PCR
4.3. Constructs
4.4. Plant Transformation and Transgenic Plants Selection
4.5. DNA Isolation and PCR
4.6. Plasmid DNA Isolation, Protoplast Isolation and Transfection
4.7. Morphological Assays
4.8. Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mauricio, R. Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. Am. Nat. 1998, 151, 20–28. [Google Scholar] [CrossRef]
- Szymanski, D.B.; Lloyd, A.M.; Marks, M.D. Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 2000, 5, 214–219. [Google Scholar] [CrossRef]
- Schiefelbein, J. Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 2003, 6, 74–78. [Google Scholar] [CrossRef]
- Schiefelbein, J.; Huang, L.; Zheng, X. Regulation of epidermal cell fate in Arabidopsis roots: The importance of multiple feedback loops. Front. Plant Sci. 2013, 5, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Chen, J.G. Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front. Plant Sci. 2014, 5, 133. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Wang, S. GLABRA2, a common regulator for epidermal cell fate determination and anthocyanin biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 4997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, H.; Wang, S. TRANSPARENT TESTA GLABRA1, a key regulator in plants with multiple roles and multiple function mechanisms. Int. J. Mol. Sci. 2020, 21, 4881. [Google Scholar] [CrossRef]
- Walker, A.R.; Davison, P.A.; Bolognesi-Winfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 1999, 11, 1337–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oppenheimer, D.G.; Herman, P.L.; Shan, S.; Esch, J.; Marks, M.D. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 1991, 67, 483–493. [Google Scholar] [CrossRef]
- Payne, C.T.; Zhang, F.; Lloyd, A.M. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 2000, 156, 1349–1362. [Google Scholar] [CrossRef]
- Zhang, F.; Gonzalez, A.; Zhao, M.Z.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef] [Green Version]
- Rerie, W.G.; Feldmann, K.A.; Marks, M.D. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994, 8, 1388–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wada, T.; Tachibana, T.; Shimura, Y.; Okada, K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 1997, 277, 1113–1116. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, A.; Folkers, U.; Schwab, B.; Jürgens, G.; Hülskamp, M. Generation of a spacing pattern: The role of triptychon in trichome patterning in Arabidopsis. Plant Cell 1999, 11, 1105–1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esch, J.J.; Chen, M.A.; Hillestad, M.; Marks, M.D. Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 2004, 40, 860–869. [Google Scholar] [CrossRef]
- Kirik, V.; Simon, M.; Huelskamp, M.; Schiefelbein, J. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 2004, 268, 506–513. [Google Scholar] [CrossRef] [Green Version]
- Simon, M.; Lee, M.; Lin, Y.; Gish, L.; Schiefelbein, J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 2007, 311, 566–578. [Google Scholar] [CrossRef]
- Tominaga, R.; Iwata, M.; Sano, R.; Inoue, K.; Okada, K.; Wada, T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 2008, 135, 1335–1345. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Kwak, S.H.; Zeng, Q.; Ellis, B.E.; Chen, X.Y.; Schiefelbein, J.; Chen, J.G. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 2007, 134, 3873–3882. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Hubbard, L.; Chang, Y.; Guo, J.; Schiefelbein, J.; Chen, J.G. Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 2008, 8, 81. [Google Scholar] [CrossRef] [Green Version]
- Gan, L.; Xia, K.; Chen, J.G.; Wang, S. Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol. 2011, 11, 176. [Google Scholar] [CrossRef] [Green Version]
- Hajdukiewicz, P.; Svab, Z.; Maliga, P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994, 25, 989–994. [Google Scholar] [CrossRef]
- Pesch, M.; Hülskamp, M. Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 2004, 14, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kurata, T.; Okada, K.; Wada, T. A genetic regulatory network in the development of trichomes and root hairs. Plant Biol. 2008, 59, 365–386. [Google Scholar] [CrossRef]
- Gan, Y.; Kumimoto, R.; Liu, C.; Ratcliffe, O.; Yu, H.; Broun, P. GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 2006, 18, 1383–1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, Y.; Liu, C.; Yu, H.; Broun, P. Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 2007, 134, 2073–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; An, L.; Sun, L.; Zhu, S.; Xi, W.; Broun, P.; Yu, H.; Gan, Y. Zinc finger protein5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis. Plant Physiol. 2011, 157, 673–682. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Sun, L.; Zhao, Y.; An, L.; Yan, A.; Meng, X.; Gan, Y. Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol. 2013, 198, 699–708. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, A.; Zhou, Z.; Zhao, Y.; Yan, A.; Bao, S.; Yu, H.; Gan, Y. GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol. 2015, 206, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Cai, W.J.; Wang, S.; Shan, C.M.; Wang, L.J.; Chen, X.Y. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 2010, 22, 2322–2335. [Google Scholar] [CrossRef] [Green Version]
- Tian, H.; Wang, X.; Guo, H.; Cheng, Y.; Hou, C.; Chen, J.G.; Wang, S. NTL8 Regulates Trichome Formation in Arabidopsis by Directly Activating R3 MYB Genes TRY and TCL1. Plant Physiol. 2017, 174, 2363–2375. [Google Scholar] [CrossRef] [Green Version]
- Vadde, B.V.L.; Challa, K.R.; Sunkara, P.; Hegde, A.S.; Nath, U. The TCP4 transcription factor directly activates TRICHOMELESS1 and 2 and suppresses trichome initiation. Plant Physiol. 2019, 181, 1587–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, J.; Zhang, J.; Yuan, R.; Yu, H.; An, F.; Sun, L.; Chen, H.; Zhou, Y.; Qian, W.; He, H.; et al. TCP Transcription Factors Suppress Cotyledon Trichomes by Impeding a Cell Differentiation-regulating Complex. Plant Physiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF COI1 complex during jasmonate signalling. Nature 2007, 448, 661. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Song, L.; Huang, S.C.; Wise, A.; Castanon, R.; Nery, J.R.; Chen, H.; Watanabe, M.; Thomas, J.; Bar-Joseph, Z.; Ecker, J.R. A transcription factor hierarchy defines an environmental stress response network. Science 2016, 354, 1550. [Google Scholar] [CrossRef] [Green Version]
- Tian, H.; Chen, S.; Yang, W.; Wang, T.; Zheng, K.; Wang, Y.; Cheng, Y.; Zhang, N.; Liu, S.; Li, D.; et al. A novel family of transcription factors conserved in angiosperms is required for ABA signalling. Plant Cell Environ. 2017, 40, 2958–2971. [Google Scholar] [CrossRef]
- Rodriguez, P.L.; Leube, M.P.; Grill, E. Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol. Biol. 1998, 38, 879–883. [Google Scholar] [CrossRef]
- Gosti, F.; Beaudoin, N.; Serizet, C.; Webb, A.A.; Vartanian, N.; Giraudat, J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 1999, 11, 1897–1910. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Yang, X.; Weston, D.J.; Chen, J.G. Abscisic acid receptors: Past, present and future. J. Integr. Plant Biol. 2011, 53, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar]
- Marks, M.D.; Wenger, J.P.; Gilding, E.; Jilk, R.; Dixon, R.A. Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol. Plant 2009, 2, 803–822. [Google Scholar] [CrossRef]
- Yu, C.Y.; Kanehara, K. The Unfolded Protein Response Modulates a Phosphoinositide-Binding Protein through the IRE1-bZIP60 Pathway. Plant Physiol. 2020, 183, 221–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oxley, D.; Ktistakis, N.; Farmaki, T. Differential isolation and identification of PI(3)P and PI(3,5)P2 binding proteins from Arabidopsis thaliana using an agarose-phosphatidylinositol-phosphate affinity chromatography. J. Proteom. 2013, 91, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, S.; Valerius, O.; Hall, H.; Zeng, Q.; Li, J.F.; Weston, D.J.; Ellis, B.E.; Chen, J.G. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 2011, 155, 370–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ton, J.; Mauch-Mani, B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 2004, 38, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, J.G. Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol. 2008, 49, 1792–1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. AUXIN RESPONS EFACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 2005, 17, 1979–1993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hülskamp, M.; Misŕa, S.; Jürgens, G. Genetic dissection of trichome cell development in Arabidopsis. Cell 1994, 76, 555–566. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, N.; Hussain, S.; Ahmed, S.; Yang, W.; Wang, S. Integration of a FT expression cassette into CRISPR/Cas9 construct enables fast generation and easy identification of transgene-free mutants in Arabidopsis. PLoS ONE 2019, 14, e0218583. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhang, N.; Zhang, Q.; Zhou, G.; Tian, H.; Hussain, S.; Ahmed, S.; Wang, T.; Wang, S. Genome editing to integrate seed size and abiotic stress tolerance traits in Arabidopsis reveals a role for DPA4 and SOD7 in the regulation of inflorescence architecture. Int. J. Mol. Sci. 2019, 20, 2695. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Zhou, L.; Zhang, W.; Cai, L.; Guo, H.; Tian, H.; Schieflbein, J.; Wang, S. A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant Cell Environ. 2016, 39, 897–907. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, X.; Hu, Q.; Dai, X.; Tian, H.; Zheng, K.; Wang, X.; Mao, T.; Chen, J.G.; Wang, S. Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 2015, 83, 300–311. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, S.; Zhang, N.; Wang, W.; Ahmed, S.; Cheng, Y.; Chen, S.; Wang, X.; Wang, Y.; Hu, X.; Wang, T.; et al. Involvement of ABA Responsive SVB Genes in the Regulation of Trichome Formation in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 6790. https://doi.org/10.3390/ijms22136790
Hussain S, Zhang N, Wang W, Ahmed S, Cheng Y, Chen S, Wang X, Wang Y, Hu X, Wang T, et al. Involvement of ABA Responsive SVB Genes in the Regulation of Trichome Formation in Arabidopsis. International Journal of Molecular Sciences. 2021; 22(13):6790. https://doi.org/10.3390/ijms22136790
Chicago/Turabian StyleHussain, Saddam, Na Zhang, Wei Wang, Sajjad Ahmed, Yuxin Cheng, Siyu Chen, Xutong Wang, Yating Wang, Xiaojun Hu, Tianya Wang, and et al. 2021. "Involvement of ABA Responsive SVB Genes in the Regulation of Trichome Formation in Arabidopsis" International Journal of Molecular Sciences 22, no. 13: 6790. https://doi.org/10.3390/ijms22136790
APA StyleHussain, S., Zhang, N., Wang, W., Ahmed, S., Cheng, Y., Chen, S., Wang, X., Wang, Y., Hu, X., Wang, T., & Wang, S. (2021). Involvement of ABA Responsive SVB Genes in the Regulation of Trichome Formation in Arabidopsis. International Journal of Molecular Sciences, 22(13), 6790. https://doi.org/10.3390/ijms22136790






