Screening of Human Gut Bacterial Culture Collection Identifies Species That Biotransform Quercetin into Metabolites with Anticancer Properties
Abstract
:1. Introduction
2. Results
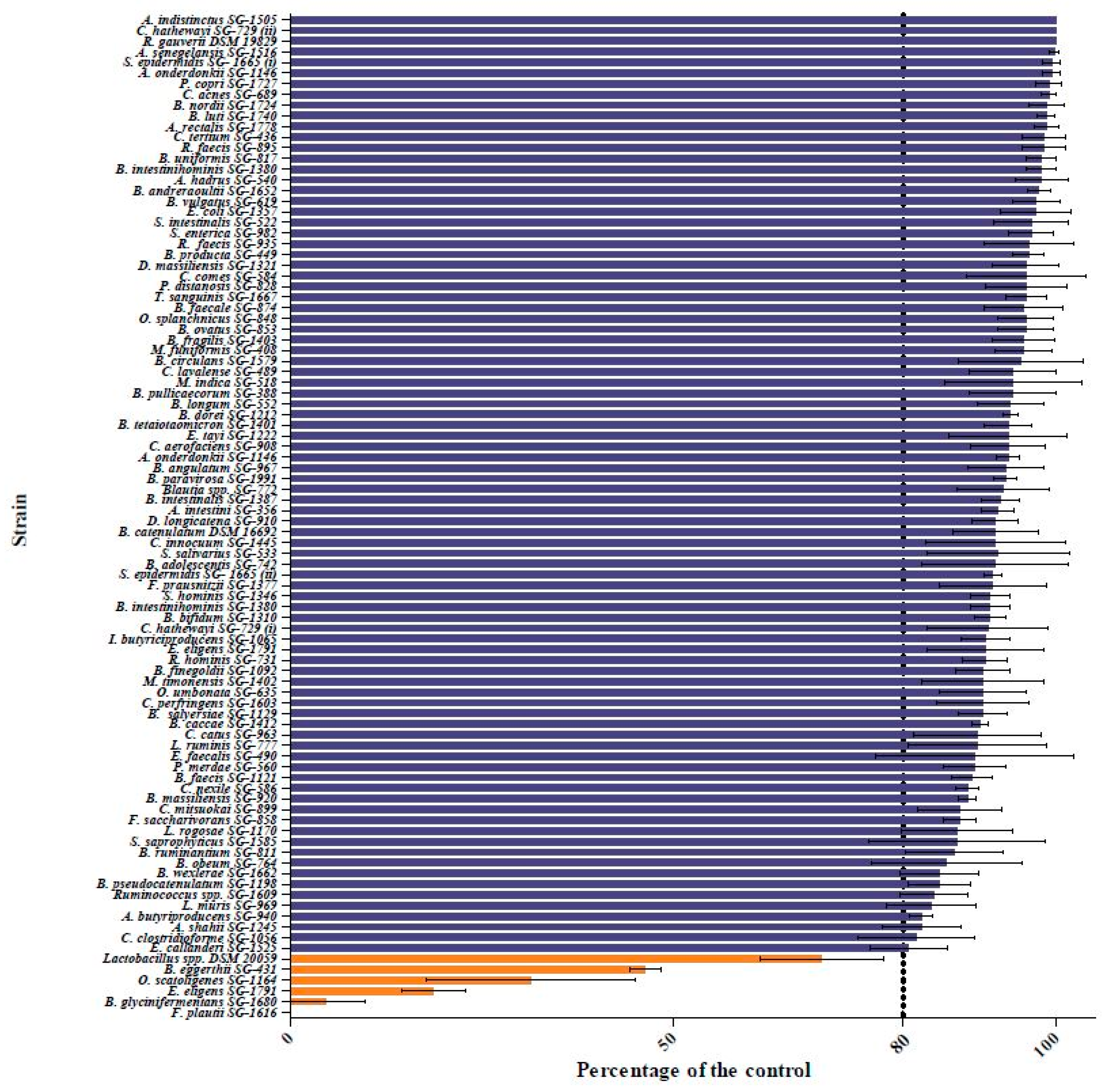
2.1. Screening the Human Gut Bacterial Library for Quercetin-Degrading Strains
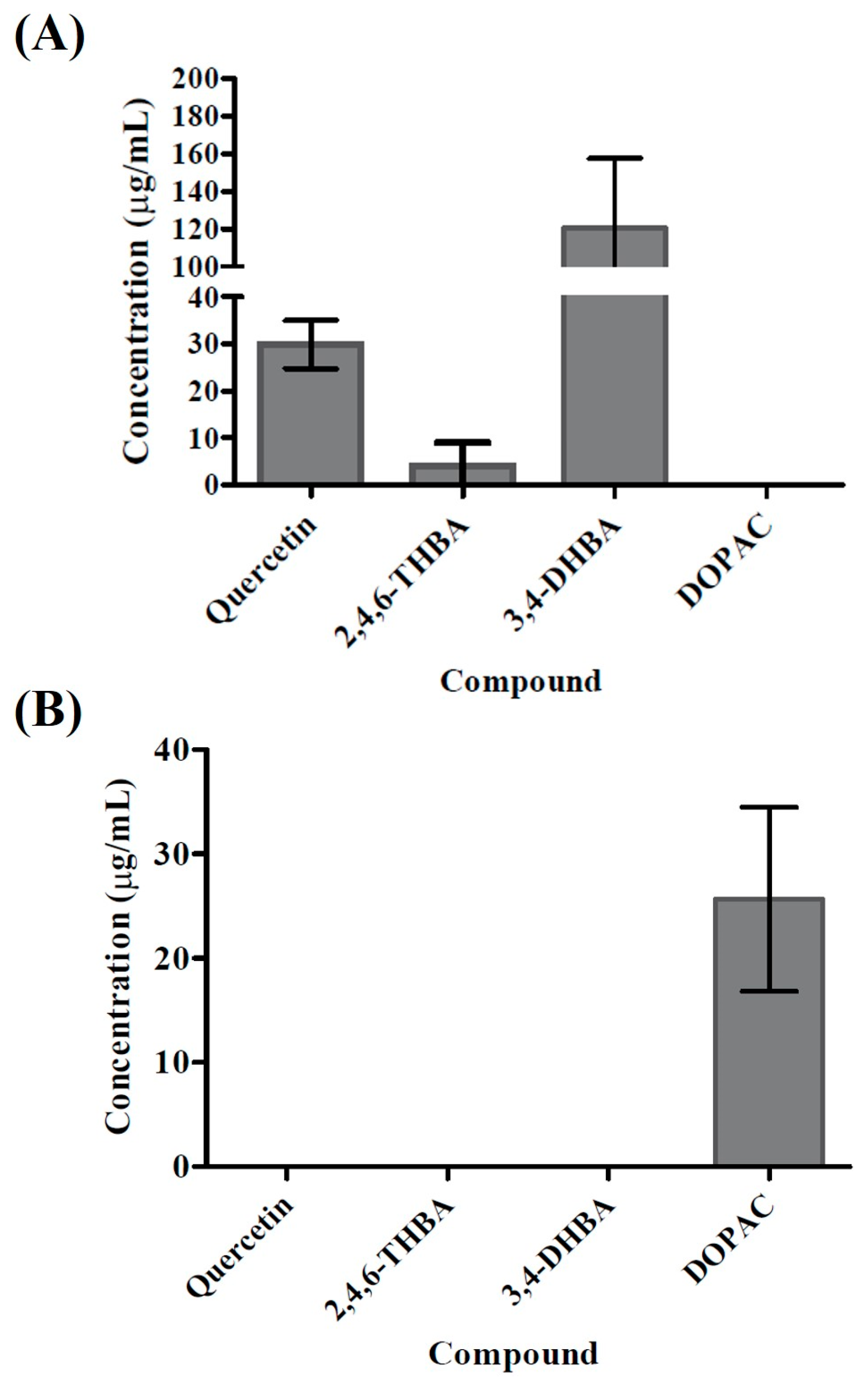
2.2. HPLC Analysis of Bacillus Glycinifermentans and Flavonifractor Plautii Revealed Different Metabolites Produced Following Quercetin Degradation
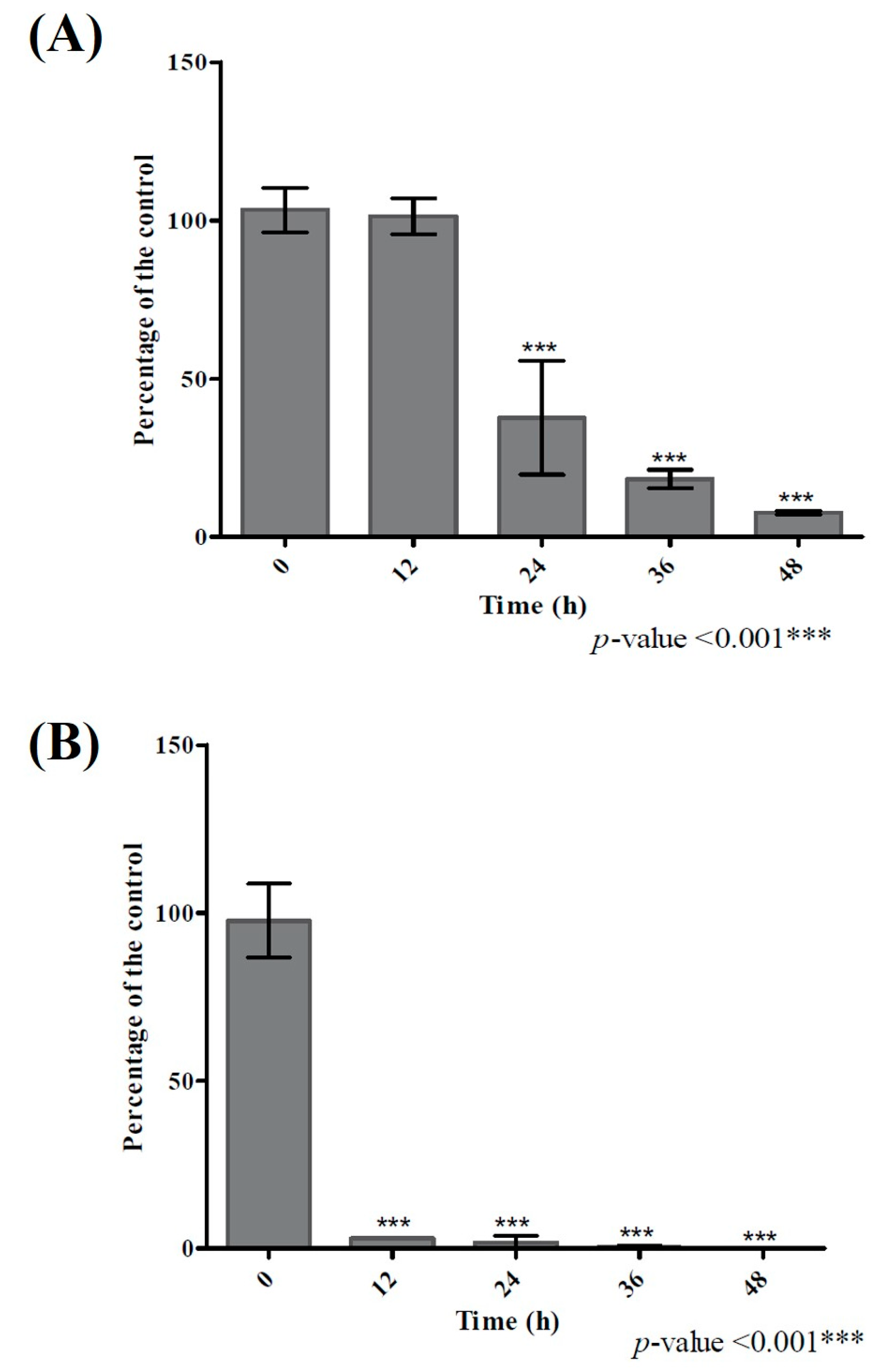
2.3. Degradation Kinetics and Different End Products Suggested Different Mechanisms of Quercetin Degradation by B. glycinifermentans and F. plautii
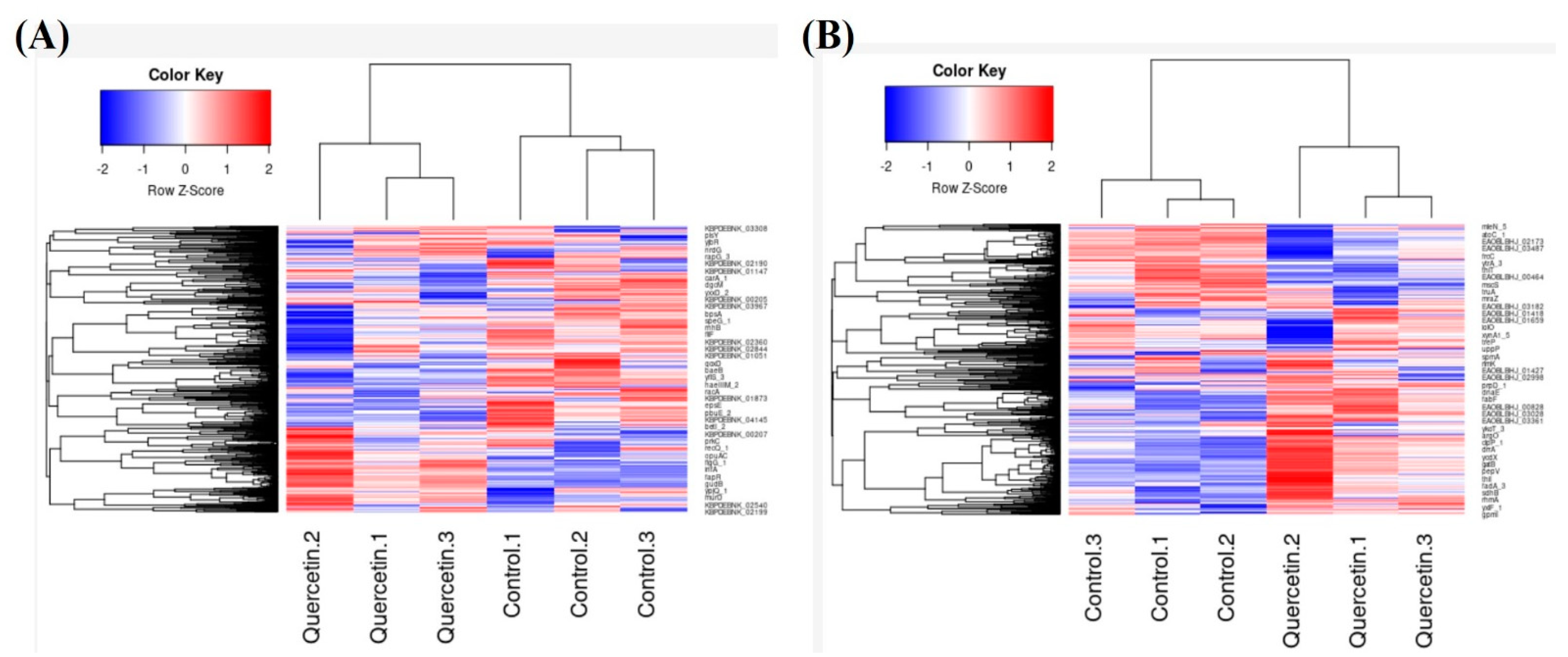
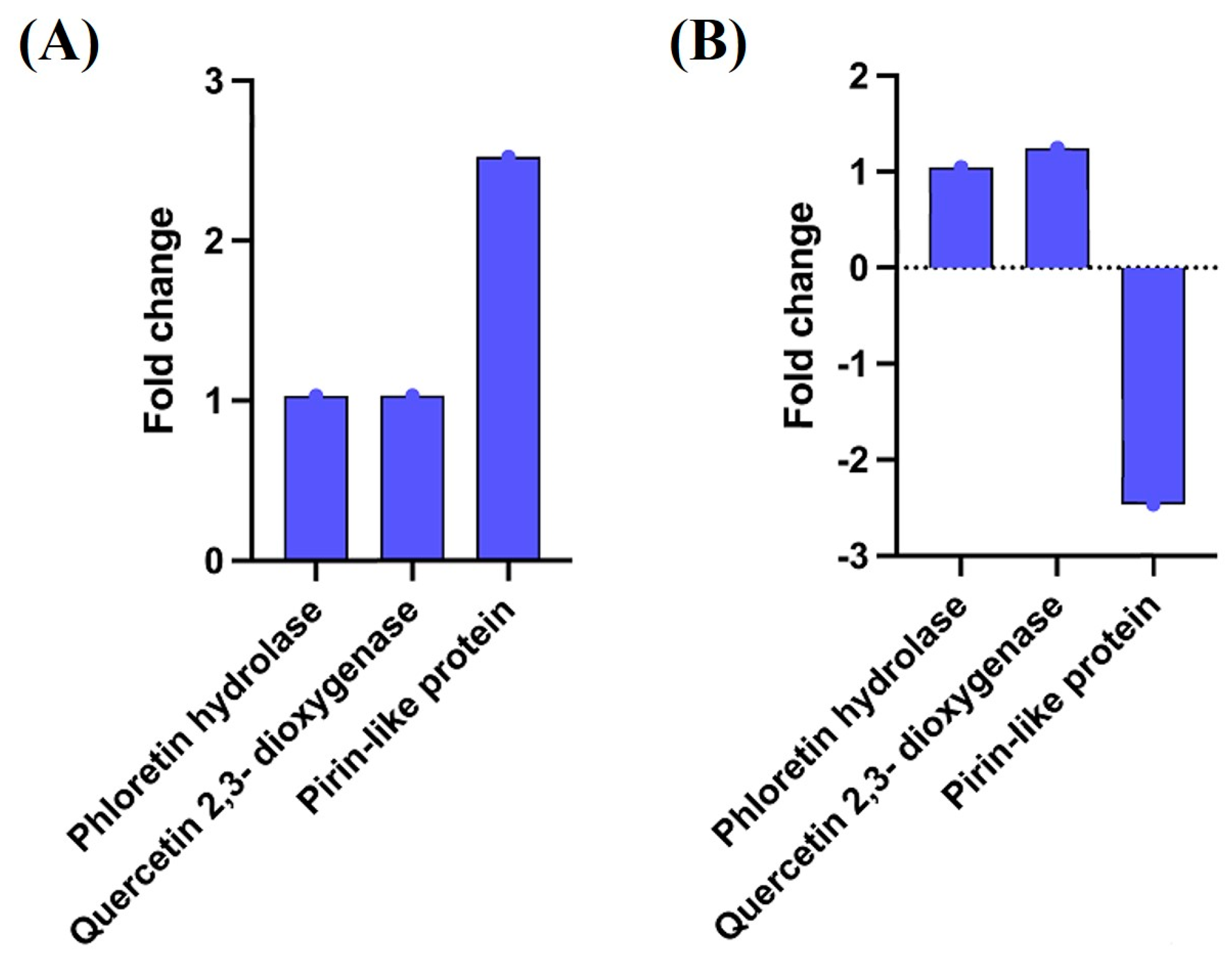
2.4. Genetic and Transcriptomic Analysis Revealed the Differential Expression of Genes in B. glycinifermentans and F. plautii upon Exposure to Quercetin
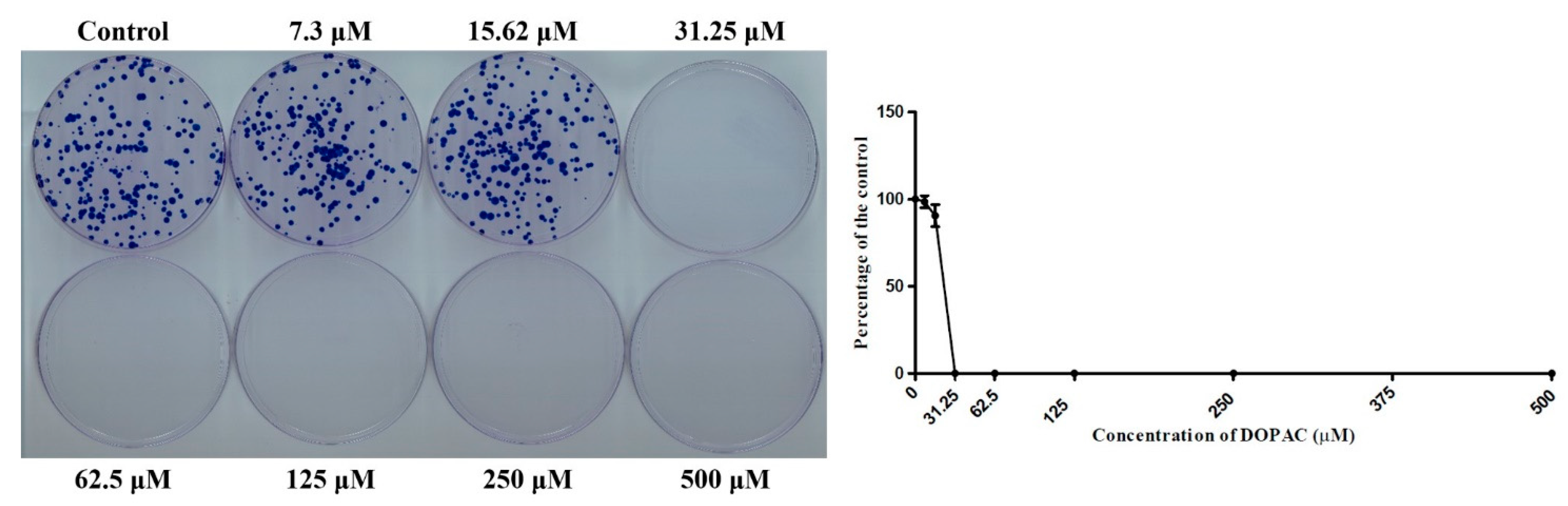
2.5. DOPAC Inhibited Cancer Cell Colony Formation
2.6. Clonogenic Assay Using the HCT-116 Cells with the Spent Bacterial Culture Supernatants
3. Discussion
4. Materials and Methods
4.1. Bacterial Culture Medium
4.2. Screening for Bacterial Species Capable of Metabolizing Quercetin
4.3. DPH Assay
4.4. UV-Visible Absorbance Spectroscopy
4.5. HPLC Analysis for the Detection of Quercetin/Metabolites in Bacterial Culture Supernatants
4.6. Lysis of Bacteria and Assay for Determination of Quercetin Degrading Enzyme Activity
4.7. Bacterial Whole Genome Sequencing
4.8. RNA Isolation and Transcriptomic Analysis
4.9. Synthesis and Sequencing of cDNA
4.10. Clonogenic Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bischoff, S.C. Quercetin: Potentials in the prevention and therapy of disease. Curr. Opin. Clin. Nutr. Metab. Care 2008, 11, 733–740. [Google Scholar] [CrossRef]
- Blaut; Schoefer; Braune Transformation of Flavonoids by Intestinal Microorganisms. Int. J. Vitam. Nutr. Res. 2003, 73, 79–87. [CrossRef] [PubMed]
- Le Marchand, L. Cancer preventive effects of flavonoids—A review. Biomed. Pharmacother. 2002, 56, 296–301. [Google Scholar] [CrossRef]
- Vargas, J.A.; Burd, R. Hormesis and synergy: Pathways and mechanisms of quercetin in cancer prevention and management. Nutr. Rev. 2010, 68, 418–428. [Google Scholar] [CrossRef]
- Murakami, A.; Ashida, H.; Terao, J. Multitargeted cancer prevention by quercetin. Cancer Lett. 2008, 269, 315–325. [Google Scholar] [CrossRef]
- Aura, A.-M.; O’Leary, K.A.; Williamson, G.; Ojala, M.; Bailey, M.; Puupponen-Pimiä, R.; Nuutila, A.M.; Oksman-Caldentey, K.-M.; Poutanen, K. Quercetin derivatives are deconjugated and converted to hydroxyphenylacetic acids but not methylated by human fecal flora In Vitro. J. Agric. Food Chem. 2002, 50, 1725–1730. [Google Scholar] [CrossRef] [PubMed]
- Harborne, J.B. Nature, distribution and function of plant flavonoids. Prog. Clin. Boil. Res. 1986, 213, 15–24. [Google Scholar]
- Erlund, I.; Freese, R.; Marniemi, J.; Hakala, P.; Alfthan, G. Bioavailability of quercetin from berries and the diet. Nutr. Cancer 2006, 54, 13–17. [Google Scholar] [CrossRef]
- Hanske, L.; Engst, W.; Loh, G.; Sczesny, S.; Blaut, M.; Braune, A. Contribution of gut bacteria to the metabolism of cyanidin 3-glucoside in human microbiota-associated rats. Br. J. Nutr. 2013, 109, 1433–1441. [Google Scholar] [CrossRef]
- Hollman, P.C.; Van Trijp, J.; Buysman, M.N.; Gaag, M.S.V.; Mengelers, M.J.; De Vries, J.H.; Katan, M.B. Relative bioavailability of the antioxidant flavonoid quercetin from various foods in man. FEBS Lett. 1997, 418, 152–156. [Google Scholar] [CrossRef] [Green Version]
- Amaretti, A.; Raimondi, S.; Leonardi, A.; Quartieri, A.; Rossi, M. Hydrolysis of the Rutinose-Conjugates Flavonoids Rutin and Hesperidin by the Gut Microbiota and Bifidobacteria. Nutrients 2015, 7, 2788–2800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdes, A.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-B. Impacts of Gut Bacteria on Human Health and Diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Possemiers, S.; Bolca, S.; Verstraete, W.; Heyerick, A. The intestinal microbiome: A separate organ inside the body with the metabolic potential to influence the bioactivity of botanicals. Fitoterapia 2011, 82, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Dahmus, J.D.; Kotler, D.L.; Kastenberg, D.M.; Kistler, C.A. The gut microbiome and colorectal cancer: A review of bacterial pathogenesis. J. Gastrointest. Oncol. 2018, 9, 769–777. [Google Scholar] [CrossRef]
- O’Keefe, S.J.D. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef]
- Ozdal, T.; Sela, D.A.; Xiao, J.; Boyacioglu, D.; Chen, F.; Capanoglu, E. The Reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients 2016, 8, 78. [Google Scholar] [CrossRef]
- Sofia, O.; Alberto, M. Insights into the role of the intestinal microbiota in colon cancer. Ther. Adv. Gastroenterol. 2017, 10, 417–428. [Google Scholar]
- Tarallo, S.; Ferrero, G.; Gallo, G.; Francavilla, A.; Clerico, G.; Luc, A.R.; Manghi, P.; Thomas, A.M.; Vineis, P.; Segata, N.; et al. Altered fecal small RNA profiles in colorectal cancer reflect gut microbiome composition in stool samples. mSystems 2019, 4, e00289-19. [Google Scholar] [CrossRef] [Green Version]
- Vipperla, K.; O’Keefe, S.J. Diet, microbiota, and dysbiosis: A ’recipe’ for colorectal cancer. Food Funct. 2016, 7, 1731–1740. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Xu, A.; Krul, C.; Venema, K.; Liu, Y.; Niu, Y.; Lu, J.; Bensoussan, L.; Seeram, N.P.; Heber, D.; et al. Of the major phenolic acids formed during human microbial fermentation of tea, citrus, and soy flavonoid supplements only 3,4-dihydroxyphenylacetic acid has antiproliferative activity. J. Nutr. 2006, 136, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Braune, A.; Blaut, M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes 2016, 7, 216–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sankaranarayanan, R.; Valiveti, C.; Kumar, D.; Van Slambrouck, S.; Kesharwani, S.; Seefeldt, T.; Scaria, J.; Tummala, H.; Bhat, G. The flavonoid metabolite 2,4,6-trihydroxybenzoic acid is a cdk inhibitor and an anti-proliferative agent: A potential role in cancer prevention. Cancers 2019, 11, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowater, L.; Fairhurst, S.A.; Just, V.J.; Bornemann, S. Bacillus subtilis YxaG is a novel Fe-containing quercetin 2,3-dioxygenase. FEBS Lett. 2003, 557, 45–48. [Google Scholar] [CrossRef]
- Tang, Y.; Nakashima, S.; Saiki, S.; Myoi, Y.; Abe, N.; Kuwazuru, S.; Zhu, B.; Ashida, H.; Murata, Y.; Nakamura, Y. 3,4-Dihydroxyphenylacetic acid is a predominant biologically-active catabolite of quercetin glycosides. Food Res. Int. 2016, 89, 716–723. [Google Scholar] [CrossRef]
- Braune, A.; Engst, W.; Elsinghorst, P.W.; Furtmann, N.; Bajorath, J.; Gütschow, M.; Blaut, M. Chalcone isomerase from eubacterium ramulus catalyzes the ring contraction of flavanonols. J. Bacteriol. 2016, 198, 2965–2974. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Hong, S.; Yang, P.; Sun, Y.; Wang, Y.; Zhang, P.; Jiang, W.; Gu, Y. Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria. Nat. Commun. 2021, 12, 1–15. [Google Scholar]
- Gall, M.; Thomsen, M.; Peters, C.; Pavlidis, I.V.; Jonczyk, P.; Grünert, P.P.; Beutel, S.; Scheper, T.; Gross, E.; Backes, M.; et al. Enzymatic conversion of flavonoids using bacterial chalcone isomerase and enoate reductase. Angew. Chem. Int. Ed. 2013, 53, 1439–1442. [Google Scholar] [CrossRef]
- Das, S.; Rosazza, J.P.N. Microbial and enzymatic transformations of flavonoids. J. Nat. Prod. 2006, 69, 499–508. [Google Scholar] [CrossRef]
- Ghimire, S.; Roy, C.; Wongkuna, S.; Antony, L.; Maji, A.; Keena, M.C.; Foley, A.; Scaria, J. Identification of clostridioides difficile-inhibiting gut commensals using culturomics, phenotyping, and combinatorial community assembly. mSystems 2020, 5, e00620-19. [Google Scholar] [CrossRef] [Green Version]
- Schoefer, L.; Braune, A.; Blaut, M. A fluorescence quenching test for the detection of flavonoid transformation. FEMS Microbiol. Lett. 2001, 204, 277–280. [Google Scholar] [CrossRef]
- Kim, S.-J.; Dunlap, C.; Kwon, S.-W.; Rooney, A. Bacillus glycinifermentans sp. nov., isolated from fermented soybean paste. Int. J. Syst. Evol. Microbiol. 2015, 65, 3586–3590. [Google Scholar] [CrossRef] [PubMed]
- Stadermann, K.B.; Blom, J.; Borgmeier, C.; Sciberras, N.; Herbold, S.; Kipker, M.; Meurer, G.; Molck, S.; Petri, D.; Pelzer, S.; et al. First complete genome sequence of Bacillus glycinifermentans B-27. J. Biotechnol. 2017, 257, 187–191. [Google Scholar] [CrossRef]
- Schoefer, L.; Mohan, R.; Schwiertz, A.; Braune, A.; Blaut, M. Anaerobic degradation of flavonoids by Clostridium orbiscindens. Appl. Environ. Microbiol. 2003, 69, 5849–5854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirooka, K.; Fujita, Y. Excess production ofBacillus subtilisQuercetin 2,3-dioxygenase affects cell viability in the presence of quercetin. Biosci. Biotechnol. Biochem. 2010, 74, 1030–1038. [Google Scholar] [CrossRef]
- Widiatningrum, T.; Maeda, S.; Kataoka, K.; Sakurai, T. A pirin-like protein from Pseudomonas stutzeri and its quercetinase activity. Biochem. Biophys. Rep. 2015, 3, 144–149. [Google Scholar] [CrossRef]
- Schoefer, L.; Braune, A.; Blaut, M. Cloning and expression of a phloretin hydrolase gene from Eubacterium ramulus and characterization of the recombinant enzyme. Appl. Environ. Microbiol. 2004, 70, 6131–6137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parasuraman, S.; David, A.V.A.; Arulmoli, R. Overviews of biological importance of quercetin: A bioactive flavonoid. Pharmacogn. Rev. 2016, 10, 84–89. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Peng, X.; Li, S.; Zhang, N.; Wang, Y.; Wei, H. Isolation and identification of quercetin degrading bacteria from human fecal microbes. PLoS ONE 2014, 9, e90531. [Google Scholar] [CrossRef] [Green Version]
- Rosa, L.D.S.; Jordão, N.A.; Soares, N.D.C.P.; De Mesquita, J.F.; Monteiro, M.; Teodoro, A.J. Pharmacokinetic, antiproliferative and apoptotic effects of phenolic acids in human colon adenocarcinoma cells using in vitro and in silico approaches. Molecules 2018, 23, 2569. [Google Scholar] [CrossRef] [Green Version]
- Guarner, F.; Malagelada, J.-R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Quigley, E.M. Gut bacteria in health and disease. Gastroenterol. Hepatol. 2013, 9, 560–569. [Google Scholar]
- Singh, R.K.; Chang, H.-W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [Green Version]
- Henning, S.M.; Wang, P.; Abgaryan, N.; Vicinanza, R.; De Oliveira, D.M.; Zhang, Y.; Lee, R.-P.; Carpenter, C.L.; Aronson, W.J.; Heber, D. Phenolic acid concentrations in plasma and urine from men consuming green or black tea and potential chemopreventive properties for colon cancer. Mol. Nutr. Food Res. 2013, 57, 483–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havlik, J.; Marinello, V.; Gardyne, A.; Hou, M.; Mullen, W.; Morrison, D.J.; Preston, T.; Combet, E.; Edwards, C.A. Dietary fibres differentially impact on the production of phenolic acids from rutin in an In Vitro fermentation model of the human gut microbiota. Nutrients 2020, 12, 1577. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Turroni, F.; Motherway, M.O.; Mac Sharry, J.; van Sinderen, D. Host–microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 2012, 20, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Castaño, G.P.; Dorris, M.R.; Liu, X.; Bolling, B.; Acosta-Gonzalez, A.; Rey, F.E. Bacteroides thetaiotaomicron starch utilization promotes quercetin degradation and butyrate production by eubacterium ramulus. Front. Microbiol. 2019, 10, 1145. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- CLC Genomics Work Bench 12.0. Aarhus, Denmark. Available online: http://www.clcbio.com/ (accessed on 1 June 2021).
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2019. [Google Scholar]
- Zhou, S.; Xia, H.; Xu, H.; Tang, Q.; Nie, Y.; Yong Gong, Q.; Bi, F. ERRalpha suppression enhances the cytotoxicity of the MEK inhibitor trametinib against colon cancer cells. J. Exp. Clin. Cancer Res. 2018, 37, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Pathway ID | Pathway Name | Number of Differentially Expressed Genes |
|---|---|---|
| bgy01110 | Biosynthesis of secondary metabolites | 60 |
| bgy03010 | Ribosome | 45 |
| bgy01120 | Microbial metabolism in diverse environments | 30 |
| bgy01200 | Carbon metabolism | 25 |
| bgy01230 | Biosynthesis of amino acids | 23 |
| bgy00230 | Purine metabolism | 20 |
| bgy01240 | Biosynthesis of cofactors | 20 |
| bgy00190 | Oxidative phosphorylation | 13 |
| bgy00010 | Glycolysis/gluconeogenesis | 12 |
| bgy00250 | Alanine, aspartate and glutamate metabolism | 9 |
| bgy03018 | RNA degradation | 9 |
| bgy00680 | Methane metabolism | 8 |
| bgy00680 | Citrate cycle (TCA cycle) | 8 |
| bgy00030 | Pentose phosphate pathway | 7 |
| bgy02024 | Quorum sensing | 7 |
| bgy00630 | Glyoxylate and dicarboxylate metabolism | 6 |
| bgy00620 | Pyruvate metabolism | 6 |
| bgy00260 | Glycine, serine and threonine metabolism | 5 |
| bgy01210 | 2-oxocarboxylic acid metabolism | 5 |
| bgy00670 | One carbon pool by folate | 5 |
| bgy02020 | Two-component system | 4 |
| bgy02010 | ABC transporters | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sankaranarayanan, R.; Sekhon, P.K.; Ambat, A.; Nelson, J.; Jose, D.; Bhat, G.J.; Scaria, J. Screening of Human Gut Bacterial Culture Collection Identifies Species That Biotransform Quercetin into Metabolites with Anticancer Properties. Int. J. Mol. Sci. 2021, 22, 7045. https://doi.org/10.3390/ijms22137045
Sankaranarayanan R, Sekhon PK, Ambat A, Nelson J, Jose D, Bhat GJ, Scaria J. Screening of Human Gut Bacterial Culture Collection Identifies Species That Biotransform Quercetin into Metabolites with Anticancer Properties. International Journal of Molecular Sciences. 2021; 22(13):7045. https://doi.org/10.3390/ijms22137045
Chicago/Turabian StyleSankaranarayanan, Ranjini, Prabhjot Kaur Sekhon, Achuthan Ambat, Julia Nelson, Davis Jose, G. Jayarama Bhat, and Joy Scaria. 2021. "Screening of Human Gut Bacterial Culture Collection Identifies Species That Biotransform Quercetin into Metabolites with Anticancer Properties" International Journal of Molecular Sciences 22, no. 13: 7045. https://doi.org/10.3390/ijms22137045
APA StyleSankaranarayanan, R., Sekhon, P. K., Ambat, A., Nelson, J., Jose, D., Bhat, G. J., & Scaria, J. (2021). Screening of Human Gut Bacterial Culture Collection Identifies Species That Biotransform Quercetin into Metabolites with Anticancer Properties. International Journal of Molecular Sciences, 22(13), 7045. https://doi.org/10.3390/ijms22137045







