Abstract
Elevated concentrations of heavy metals such as cadmium (Cd) have a negative impact on staple crop production due to their ability to elicit cytotoxic and genotoxic effects on plants. In order to understand the relationship between Cd stress and plants in an effort to improve Cd tolerance, studies have identified genetic mechanisms which could be important for conferring stress tolerance. In recent years epigenetic studies have garnered much attention and hold great potential in both improving the understanding of Cd stress in plants as well as revealing candidate mechanisms for future work. This review describes some of the main epigenetic mechanisms involved in Cd stress responses. We summarize recent literature and data pertaining to chromatin remodeling, DNA methylation, histone acetylation and miRNAs in order to understand the role these epigenetic traits play in cadmium tolerance. The review aims to provide the framework for future studies where these epigenetic traits may be used in plant breeding and molecular studies in order to improve Cd tolerance.
1. Introduction
The relationship between plants and their environment is highly complex. Plants are exposed to a multitude of stresses on a daily basis and therefore they have developed various strategies to combat these stresses [1]. These strategies include altering one or more of the following factors: morphological, biochemical and molecular factors [2]. One of the molecular factors that has been receiving increased attention in plants is the alteration of epigenetic factors [3]. It has been documented that epigenetic changes can be affected by both biotic and abiotic stresses [4,5].
Cadmium (Cd) is a toxic heavy metal which has detrimental effects on the environment which include the deterioration of soil quality, a decrease in plant health and a reduction in plant growth [6,7,8]. Due to mining, the use of chemical pesticides, improper watering practices and the non-ferrous metallurgy, the environment has become increasingly polluted with heavy metals such as Cd [9,10]. Due to Cd being nonbiodegradable, its high water solubility and its ability to change plant mineral uptake, it is often termed one of the most hazardous contaminants of soil [9,11]. To cope with adverse conditions such as high Cd concentrations some plants have evolved protective mechanisms such as the prevention of Cd entry in the roots, immobilization of Cd in vacuoles and the production of Cd phytochelatins [12,13,14]. These processes are dependent on the gene expression of certain genes and thus are controlled by genetic programming. DNA methylation is one of the more common epigenetic traits and it has been found to be important as it is involved in processes such as the induction of phenotypic alterations and maintaining genome stability when plants are under abiotic stress [15,16]. At times these alterations have been observed to improve the tolerance of the plants to specific abiotic stresses. Another important epigenetic trait includes chromatin remodeling, which has been reported to be active in many organisms when they are under stress. Certain authors have theorized that chromatin remodeling may be part of the mechanisms for stress memory [17]. Another important epigenetic trait, histone acetylation, has been shown to regulate and improve abiotic stress tolerance, such as salt tolerance in T. aestivum. The literature has displayed the importance of epigenetic modification and its role in the complex rearrangement of gene expression and the role it plays in crucial biological processes related to the plant’s response to environmental factors. To date, only a few studies have focused on the relationship between Cd and epigenetic changes in plants [18]. Therefore, this review describes the different epigenetic mechanisms which are involved in crop plants’ responses to Cd toxicity.
2. Plant Epigenetic Mechanisms for Cadmium Tolerance
2.1. Chromatin Remodeling and the Effect of Cadmium Stress in Plants
Chromatin is the repeating unit of nucleosomes, and it is composed of a complex of DNA and proteins assembled within the nucleus of cells [19]. Chromatin formation is carried out by a tightly condensed DNA wrapped around nuclear proteins called histones [20]. One hundred and forty-six base pairs of double-stranded DNA wrapped around eight histone proteins in a repeating unit are termed nucleosomes [21]. The more condensed a chromatin is, the harder it is for transcription factors and DNA-binding proteins to access DNA to carry out their functions [22]. Chromatin remodeling, therefore, is the rearrangement of chromatin from a condensed state to a transcriptionally accessible form, allowing transcription factors or other DNA binding proteins to access DNA and control gene expression [23]. Chromatin that is tightly packed and not actively being transcribed is referred to as heterochromatin. When the chromatin is packed loosely to allow transcription to occur, it is called euchromatin. Chromatin remodeling is highly implicated in the epigenetic modification of histone proteins such as demethylation/methylation, acetylation/deacetylation, which can change chromatin structure, causing transcriptional activation or repression [24].
This section’s focal point is to enumerate the effect of Cd on epigenetic modulation using chromatin remodeling. Understanding Cd utilization during chromatin remodeling involves two significant pathways which regulate DNA replication and nucleosome stability of chromatin structure [25]. One of these pathways is dependent on the histone-gene repressor (HIRA), and the other requires chromatin assembly factor 1 (CAF-1), which can be altered in response to Cd stress [26]. The variation of metals’ chemical properties and their reactive toxicities indicates that a uniform mechanism of action for all toxic metals is unlikely [27]. Heavy metals, including Cd, exert their cytotoxic and genotoxic effects by disrupting histone and other protein structures and functions by attacking the thiol groups in them, thus causing conformational changes to these proteins [28]. The overall effect of these conformational changes is the inhibition of DNA replication, gene expression, and cell division.
Few studies have investigated the effects of Cd on chromatin remodeling epigenetic modification in plants through the expression of stress-responsive genes. However, it is established that during Cd stress in plants, various forms of chromatin modification are possible, which include acetylation, methylation, phosphorylation, and ubiquitination that affect gene expression by altering chromatin structure and accessibility of transcription factors [29]. For example, histone’s hyperacetylation is generally associated with transcriptionally active chromatin, whereas deacetylated histone is correlated with inactive chromatin regions [30]. The heterotrimeric CAF-1 chaperone complex in A. thaliana, for instance, targets acetylated histone H3/H4 onto nascent DNA strand for the de novo assembly of nucleosomes [31]. Shafiq et al. [32] studied the interplay and coordination of histone acetylation and DNA methylation using metal stress tolerance dynamics as the focal point in Z. mays. The research found that Zn, Cd, and Pb differentially regulated DNA methyltransferase expression with varied histone deacetylases expression and dissuaded the excessive use of zinc fertilizers.
In another study by Gadhia et al. [33], the mitotic effects of inherited histone modification pathways in mouse embryonic stem cells (MES cells) were investigated, and the authors found that Cd exerted differential toxicity through selective disruption of chromatin on MES cells by targeting total histone protein (THP) production early in stem cell development with H3K27-monomethylation (H3K27me(1)) in later stages of differentiation. However, such a model study has not been replicated in plants and such a knowledge gap demands exploration by crop scientists and plant biotechnologists for future perspectives. Additionally, Zabka et al. [34] used apical root meristem in V. faba seedlings exposed to CdCl2 to monitor the epigenetic changes in transcription nucleosome assembly during the S-phase of the cell cycle. The study revealed cellular responses to Cd toxicity are interlinked with biochemical reactions through the generation of ROS and DNA-damage-induced replication stress.
2.2. The Role of DNA Methylation during Cadmium Stress
DNA methylation is a process involving the transfer of a methyl group to the C-5 position of the nucleotide cytosine and is an epigenetic modification which affects nuclear gene expression and genome stability [35]. DNA methylation/demethylation is a reversible reaction which allows this epigenetic trait to be associated with many diverse biological functions [18]. DNA methylation has been observed in all major taxonomic groups, which include mosses, ferns and angiosperms [36]. Given the functions that DNA methylation is involved in, as well as its conservation, it is quite evident that is required for proper plant development. DNA methylation of promoter regions has been shown to have an impact on gene expression and therefore affect the development of plants [37]. A study by Shafiq et al. [7] observed the effect Cd had on the DNA methylation levels in T. aestivum and how it affected metal tolerance. In the study the authors observed two T. aestivum cultivars, namely cv. Pirsabak 2004 (resistant) and cv. Fakhar-e-sarhad (sensitive), and tried to elucidate the mechanisms which could account for the metal tolerance observed in cv. Pirsabak 2004. The sensitive cultivar, cv. Fakhar-e-sarhad, was observed to have a higher DNA methylation level at the promoter regions of the transporters TaABCCs and TaHMA2 when compared to the resistant cultivar, cv. Pirsabak 2004, once exposed to Cd. It should be noted that TaABCCs and TaHMA2 are transporters which are required for metal transport [38,39]. The proposed theory was that the different levels of DNA methylation at the promoter sequences were due to Cd toxicity and resulted in altered expression of TaABCCs and TaHMA2 in the two cultivars and thus a difference in their metal transport capabilities was observed. A study by Fan et al. [9] observed the effect of inhibiting DNA demethylation on plant tolerance to Cd toxicity and iron nutrition. Cd exposure resulted in an increase in DNA methylation which was consistent with what was observed in the T. aestivum cultivar cv. Fakhar-e-sarhad in the study by Shafiq et al. [7,9]. The DNA methylation levels in Cd treated plants were 15% higher when compared to the control plants. The authors hypothesized that the demethylase genes (ROS1, DML2 and DML3) were important in the Cd-induced DNA methylation changes. This was investigated by using the A. thaliana rdd (ROS1/DML2/DML3) triple mutant. Rdd mutants exposed to Cd concentrations exceeding 20 µM displayed increased growth parameters when compared to the controls. This result confirmed that rdd mutants were more tolerant to Cd stress: the authors suggested that the improvement in tolerance was due to elevated DNA methylation in rdd mutants coupled with improved iron nutrition. The authors finally suggested that by inhibiting the RDD DNA, demethylation in the roots, which led to increased Cd tolerance, could be conferred through increasing DNA methylation and improving iron nutrition through a feedback mechanism. From the studies discussed it can be seen that epigenetic changes such as DNA methylation in plants when exposed to Cd are a complex system and may be different for different species of plants (Table 1). More studies are required to fully grasp the processes in which DNA methylation play a pivotal role, in order to improve plants against abiotic stresses such as Cd toxicity.

Table 1.
The relationship between Cd stress and DNA methylation in plants.
2.3. The Effects of Cadmium on Histone Acetylation
Histone acetylation was one of the earliest studied epigenetic mechanisms of transcriptional regulation. Histone acetylation is known to be involved in numerous and diverse cellular processes, including cell-cycle progression, DNA repair, gene silencing, growth and development, flowering, and seed development, and to deal with biotic stress and abiotic stress including salt, cold, and drought stresses [44]. Histone acetylation and deacetylation is a dynamic, reversible process catalyzed by two classes of enzymes, histone acetyltransferase (HAT) and histone deacetylase (HDAC), at lysine residues along the histone tail [45]. In general, histone acetylation correlates with transcriptional activation, and histone deacetylation correlates with transcriptional repression. Emerging evidence revealed that plant HATs and HDACs play essential roles in the regulation of gene expression in plant development and plant responses to environmental stresses. Furthermore, HATs and HDACs were shown to interact with various chromatin-remodeling factors and transcription factors involved in the transcriptional regulation of multiple developmental processes [46].
A recent study by Shafiq et al. [32] investigated the interplay between Zn, Pb and Cd for their uptake and translocation in Z. mays. In the study the authors observed that in the presence of Zn facilitates the accumulation and transport of Pb and Cd in the aerial parts of the maize plants. Moreover, the Zn, Pb, and Cd interplay specifically interferes with the uptake and translocation of other divalent metals, such as calcium and magnesium. Zn, Pb, and Cd, individually and in combination, differentially regulate the expression of DNA methyltransferases, thus altering the DNA methylation levels at the promoter of Zinc-regulated transporters, Iron-regulated transporterlike Protein (ZIP) genes to regulate their expression. Furthermore, the expression of HDACs varies greatly in response to individual and combined metals, and HDACs’ expression showed a negative correlation with ZIP transporters. Moreover, the Pb, Cd and Zn caused different histone acetylation profiles. Interestingly, their Pearson correlation analysis showed a negative correlation between the expression of HDACs/DNA methyltransferases and the expression of IRT1, ZIP1, ZIP3, ZIP5, ZIP6, and ZIP7, suggesting that the altered DNA methylation and acetylation levels, alone or together, could regulate the ZIPs’ gene expression. However, the expression of ZIP2 and ZIP8 showed a positive correlation with the expression of HDACs and DNA methyltransferases, suggesting that the regulation of these ZIP transporters could be independent of HDACs and DNA methyltransferases. However, further studies are required to validate the crosstalk between histone acetylation and DNA methylation to regulate the ZIPs’ gene expression. Their study highlights the implication of DNA methylation and histone acetylation in regulating the metal stress tolerance dynamics through Zn transporters and warns against the excessive use of Zn fertilizers in Cd contaminated soils.
In 2017, Lee and Back [47] investigated the overexpression of OsSNAT1 in transgenic O. sativa. In the pharmaceutical industry Ovine SNAT is utilized over plant SNAT for the overproduction of melatonin in plants due to the higher catalytic activity of animal SNAT. In this study, Lee and Back [47] overexpressed O. sativa serotonin N-acetyltransferase 1 (SNAT1), which is naturally expressed in chloroplasts, in O. sativa. In contrast to transgenic O. sativa overexpressing ovine SNAT, OsSNAT1-overexpressing transgenic O. sativa plants with elevated levels of melatonin showed significant tolerance to Cd and senescence, phenotypes that were not observed in transgenic O. sativa plants overexpressing ovine SNAT. Further studies revealed that homozygous SNAT1-overexpressing transgenic O. sativa plants had higher grain yields than wild-type controls under field conditions. Their data suggest that plant SNAT gene overexpression provides an opportunity to uncover new functions of melatonin that were previously unrecognized from studies in transgenic plants expressing animal SNAT genes.
Zabka et al. [34], explored the possible relationships and linkages between Cd (II)-induced oxidative stress and the consequent damage at the genomic level of in the V. faba seedlings exposed to Cd treatment and to post stress recovery water incubations. Exposure to Cd toxicity therefore leads to two types of consequent Cd-induced secondary stress conditions, including oxidative stress, by which reactive oxygen species (ROS) played an important part in producing disorder at the genomic DNA level eventually leading to replication stress. The phosphorylation of histone H2AX on Ser-139 (γ-H2AX) is regarded as one of the earliest responses involved in cellular perception/signaling pathways activated by DNA double-strand breaks [48]. By using immunofluorescence labeling of γ-H2AX as a highly specific and sensitive molecular marker for monitoring oxidative stress associated DNA damage, they observed an increase in the number of visible γ-phosphorylation of histone H2AX (serine 139) in root apical meristem cells of V. faba exposed to Cd and phosphorylation of histone H3 (serine 10). Furthermore, Cd2+ ions induced considerable changes in cell-cycle-related acetylation of H4 histone at the N-terminal K5 residue, whereas earlier studies concentrated mainly on the rDNA genes in V. faba, and suggested that H4 acetylation is loosely associated with their transcriptional activity and inversely correlated with nucleolar rDNA replication during the early stages of the S-phase. In conclusion, cellular responses to Cd (II) toxicity give the impression of being composed of a series of interlinked biochemical reactions which via generation of ROS and DNA damage-induced replication stress, ultimately activate signal factors engaged in cell cycle control pathways, DNA repair systems, and epigenetic adaptations.
2.4. The Relationship between Cadmium Tolerance and microRNAs
In plants, there are small non-coding RNAs known as microRNAs (miRNAs), which serve the purpose of base-pairing to their specific target mRNA, resulting in the initiation of silencing or degradation of mRNA translation [38,49]. In addition, miRNAs are known epigenetic modulators which affect protein levels of the target mRNAs without modifying gene sequences. MicroRNA-guided gene regulation at the post-transcriptional level is one of the molecular mechanisms that respond to heavy metal stresses [49]. These roles are well established; however, limited knowledge exists about their response to Cd and whether they are involved in Cd reduction in plants. Previous studies revealed that heavy metal-related gene expression was regulated by an arrangement of miRNAs [50,51]. The following studies investigated the molecular mechanisms of miRNAs in Cd tolerance in various plants.
A study conducted by He et al. [52] investigated the molecular mechanisms of Cd tolerance in N. tabacum plants. Applying miRNA high-throughput sequencing technology, He et al. [52] conducted a genomewide identification and analysis of miRNA in the roots of two N. tabacum cultivars (Guiyan 1 (sensitive) and Yunyan 2 (resistant)). The authors employed qRT-PCR to validate the findings of the high-throughput sequencing, and obtained the validated miRNA profiles: they then utilized the miRBase database to classify tobacco miRNAs. He et al. [52] utilized RNAfold and Mireap to obtain novel miRNAs. Using the tobacco DFC gene index and psRNA target tool, the authors calculated putative targets. Cd stress caused significant alteration in miRNA length regardless of cultivar variation [52]. The authors suggested that alteration implicates miRNA involvement in extensive regulation of gene expression in response to the Cd stress in the N. tabacum roots. They found 72 and 14 differentially expressed miRNA, known and novel (respectively), of which 28 of the known and 5 of the novel miRNAs were deemed as Cd tolerance associated miRNA. Cadmium concentrations were then analyzed within plant organs, and it was noticed that the concentration was higher within the roots and shoots of both cultivars compared to their respective controls.
During the study it was recorded that Yunyan 2 accumulated more cadmium (exhibited higher cadmium concentration in both roots and shoots organs) than Guiyan 1 under cadmium conditions; however, Yunyan 2 still exhibited higher cadmium tolerance when compared to Guiyan 1. Yunyan 2 was considered less sensitive to cadmium due to only indicating a 16.19% and 54.44% reduction in plant height and SPAD value (measures leaf chlorophyll concentration), respectively, when compared to its control. Guiyan 1 cultivar, on the other hand, displayed a 27.75% and 60.22% reduction in plant height and SPAD value, respectively, when compared to its control. Zhou et al., 2019 ultimately graded each with cultivar an integrated score of 43.173 (Guiyan 1) and 35.077 (Yunyan 2).
The study then provided the addition of new novel mRNA to the tobacco microRnome, and He et al. [52] concluded that 28 known and 5 novel miRNAs were linked to Cd tolerance in Yunyan 2 cultivar. The Cd tolerance displayed by Yunyan 2 was suggested to be due to the miRNA profile, indicating repression of certain miRNAs that accounted for efficient scavenging of Cd-induced ROS, thereby reducing Cd toxicity in tobacco. The profile also indicated that the regulation of processes involved in cell growth, ion homeostasis, stress defense, redox maintenance and hormone signaling contributed to better growth performance in Yunyan 2 than in the Guiyan 1 cultivar.
The plant miRNA miR390 is extremely vital in Cd stress responses [49] and this miRNA is conserved in a few plants (O. sativa, A. thaliana and Z. mays). In the O. sativa genome, miR390 is encoded to regulate the expression of the O. sativa stress-responsive leucine-rich repeat receptorlike kinase by cleaving the OsSRK mRNA [49].
Ding et al. [49] employed a transgenic approach to investigate miRNAs role in the O. sativa response to Cd stress. O. sativa was transformed with a MiR390 overexpression plasmid, followed by validation of transgenic plants by analyzing their transcript levels of mature MiR390 and OsSRK. [49]. Transgenic plants exhibited a significant increase in miR390 transcripts (Figure 1), confirming overexpression and the OsSRK transcripts were much lower than the controls. Ding et al. [49] recorded an increase in OsHMA2, known to transport Zn/Cd from the roots to shoots, and OsNRAMP5, known to transport Mn and Cd into the roots in the transgenic plants under Cd stress [53,54]. The study recorded that in the presence of Cd the transgenic lines accumulated more Cd, which resulted in Cd toxicity. The conclusion was that these transcripts (OsHMA2 and OsNRAMP5) were positively regulated by miR390, indicating their importance in Cd accumulation and translocation in O. sativa plants. The seedling growth of transgenic lines was retarded, the chlorophyll content decreased, and MDA and H2O2 increased by 40% and 2-folds, respectively, when compared to the control plants [49]. The outcomes of the study by Ding et al. [49] correlated with a study conducted by Marin et al. [55] which concluded that in response to Cd stress, miR390 exerted a negative effect on the lateral root formation of A. thaliana plants (regulated the pathway involved in auxin signaling and lateral root development). Considering all of the results obtained, it can be concluded that miR390 acts as a negative regulator involved in Cd tolerance in O. sativa plants and that the expression of miR390 alters the expression pattern of several metal transporter genes, resulting in Cd accumulation and toxicity in O. sativa plants.
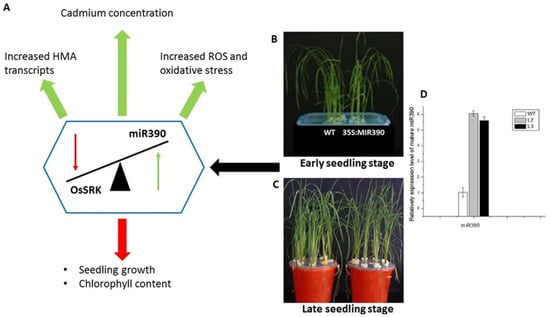
Figure 1.
Adapted from Ding et al., 2016. Represents the transgenic O. sativa (35S:MIR390) and the effects of the overexpression of miR390 under cadmium stress. (A) Indicates how the overexpression influenced the OsSRK expression and how this expression relationship altered the biochemistry of O. sativa seedlings. (B) Represents the physiology of the Wild type (WT) and transgenic O. sativa plant in their early seedling stage and (C) represents the physiology of the Wild type (WT) and transgenic O. sativa plants in their late seedling stage. (D) A graphical representation of the relative expression of miR390 in both WT and transgenic lines, confirming the overexpression of miR390. The green arrows indicate an increase and the red arrow indicates a decrease.
In a study conducted by Qiu et al. [56], Real-Time PCR was utilized to obtain the expression profiles of T. aestivum microRNA. The webtool, psRNATarget, was employed to recognize the miRNA targets (mRNA). Qiu et al. [56] recorded different miRNA expression profiles under Cd stress, in the roots and shoots of T. aestivum seedlings. Physiological variations, such as reduction in shoot and root elongation, were observed between the control seedling and the Cd treated seedlings. The relative water content of the leaves of the Cd treated seedlings was reduced (67%) when compared to the control seedlings (94–96%) after 48 h of exposure to Cd. The chlorophyll content of the treated seedlings was reduced, and the lipid peroxidation of the membranes was increased under Cd stress [53]. Figure 2 exemplifies the five miRNAs differentially expressed between the roots and shoots of Cd treated seedings [56]. miR398 was observed to target CSD, a Cu/Zn Superoxide Dismutase (a ROS scavenger). A reduction in miR398 expression led to an increase in CSD expression [56]. However, this decreased expression in miR398 indirectly led to the increase in hydrogen peroxide production (correlates to the increase in lipid peroxidation) due to the role of CSD in the dismutation of superoxide radicals to produce more hydrogen peroxide [57]. Thus, miR398 was deemed a main constituent for the detoxification of ROS, by the control of the target gene, CDS [56]. To conclude, Qiu et al. [56] suggested wheat miRNAs are involved in the facilitation of Cd stress signaling responses and, furthermore, suggested that the differential expression patterns of miRNA observed in T. aestivum roots and leaves suggest that there are variations in the functional roles of miRNA in particular organs of T. aestivum plants with regard to the regulation of plant tolerance [53]. Another study by Zhou et al. [58] took a different approach in which they focused on the regulatory mechanisms between heavy metal ATPases (HMAs) and microRNAs in T. aestivum plants. The aim was to comprehend the global transcriptional response of T. aestivum plants to Cd stress and how this response was controlled by miRNAs. Two wheat cultivars (low-Cd accumulation (L17) and High-Cd accumulation (H17)) were analyzed using next generation transcriptomic and miRNA sequencing to the miRNA profiles of each cultivar in response to Cd stress. qRT-PCR was employed to obtain and observe the differentially expressed miRNA profiles and the TaHMA profiles in the root sections [58]. Figure 3 exemplifies the differential expression between the two Cd treated cultivars. The results obtained in this study noted two particular miRNAs (miR9664-3p and tea-miR159a) which were upregulated in L17Cd (Cd treated), compared to its control L17CK, but downregulated in H17Cd [58]. Further analysis demonstrated that the putative targets of both miR9664-3p and tea-miR159a were downregulated in L17Cd plants. Han et al. [59] previously recorded that tea-miR159a expression in leaf and developing seeds of wheat plants play a positive role in the response to Puccinia striiformis, through the regulation of TaMyb3 expression [59,60]. Zhou et al. [58] also identified 32 TaHMA genes in T. aestivum plants. Zhou et al. [58] deduced that miRNA can regulate TaHMAs; however, further validation was required for future works. Concluding, both studies [56,58] depict miRNAs as potentially playing an essential role in Cd responses in T. aestivum plants, either through the regulation of the miRNAs responsible in ROS scavengers or in the regulation of the TaHMA2;1 gene.
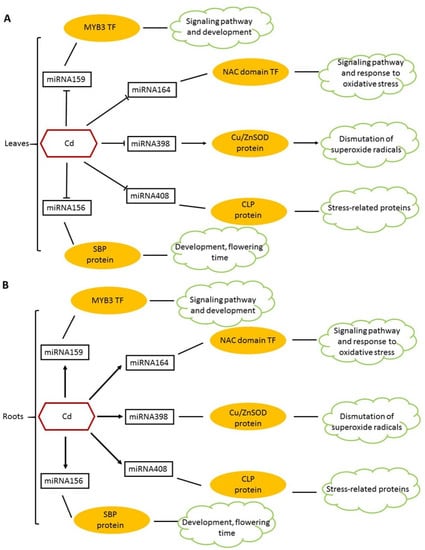
Figure 2.
Represents the miRNA expression profiles in T. aestivum plants under CD stress. (A) Represents the 5 miRNAs that were downregulated in the leaves at 12 h after exposure to Cd stress. The miRNA159 (targets MYB3 transcription factor), miRNA164 (targets NAC domain Transcription factor), miRNA398 (targets CSD) and miRNA408 (targets CLP). Besides the other 4 miRNAs, miR156 remained downregulated at all time points within the study. (B) Represents the 5 miRNAs that were upregulated in the roots from 12 h until 24 h after exposure. miRNA408 remained upregulated at all time points in this study. The arrows indicate the expression of the different micro RNAs, yellow ovals represent the targets of the miRNAs and the green clouds represent the function of the proteins. Negative correlations in the transcript expression were experienced between miR398 and CSD in the roots, miR159 and MYB3 in the roots and leaves, miR408 and CLP in the roots and leaves and between miR164 and NAC in the leaves. There was a positive correlation between miR156 and SBP in the roots and leaves. Adapted from Qiu et al. [56].
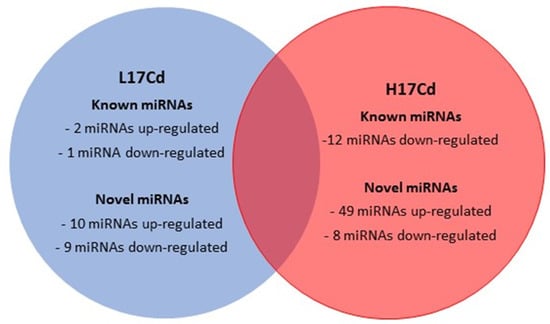
Figure 3.
Adapted from Zhou et al., 2019. Represents a Venn diagram of the differential expression patterns (DEP) between L17Cd and H17Cd wheat cultivars, compared to their controls, under cadmium stress. The DEP was obtained by conducting next generation transcriptomic and miRNA sequencing. Zhou et al., 2019 noted that miR9664-3p and tea-miR159a (known positive role in P. striiformis infection) in seed were both upregulated in the L17Cd cultivar when compared to its control L17CK, however, these two miRNAs were downregulated in H17Cd.
3. Conclusions
Knowledge about Cd tolerance in plants has been extensively documented and updated with regard to physiological and biochemical effects and responses to the toxic Cd conditions. An increase in Cd uptake in plant cells mainly leads to a decrease in plant growth, development and yield. In order to improve plant growth under high Cd concentrations, researchers identified various key genes and proteins which regulate Cd tolerance in various plants. In addition to these molecular mechanisms, epigenetic mechanisms have emerged as an important complex factor in plant responses to heavy metal stresses. However, very few studies have focused primarily on epigenetic mechanisms to improve plant performance under Cd stress. Epigenetic mechanisms could protect plants from Cd-induced DNA damage through random DNA methylation as well as through direct epigenetic regulation of Cd stress responsive genes. The interplay between chromatin remodeling, DNA methylation, histone modification, and microRNAs provides plants with a multilayered and robust epigenetic mechanism to improve survival under Cd stress. Therefore, there is a need to develop more robust bioinformatic pipelines for epigenetic analysis in plants under Cd stress. Furthermore, these studies will be crucial in future plant breeding studies where more stable epigenetic plant genotypes with improved Cd tolerance will be developed.
Author Contributions
A.G., M.F.C., O.O.B., L.-A.N., V.M. and M.K. wrote the manuscript. A.G., V.M. and M.K. edited and checked the manuscript. All authors contributed to the article. All authors have read and agreed to the published version of the manuscript.
Funding
L.-A.N. and M.F.C. were funded by the National Research Foundation of South Africa (NRF). O.O.B. is a recipient of a NRF Post-Doctoral Fellowship. This research was financially supported by the NRF to M.K. (Grant numbers: 116346 and 109083) and A.G. (Grant number: 129493). Part of the research was funded by a DST-NRF Centre of Excellence in Food Security award (Project ID: 170202) as well as GrainSA (GB0200066).
Acknowledgments
The authors would like to thank the various institutions, namely, the University of the Western Cape and the University of Free State for infrastructure and administrative support.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the review; in the collection and interpretation of literature; in the writing of the manuscript, or in the decision to publish the review.
References
- Abhinandan, K.; Skori, L.; Stanic, M.; Hickerson, N.; Jamshed, M.; Samuel, M.A. Abiotic Stress Signaling in Wheat—An Inclusive Overview of Hormonal Interactions during Abiotic Stress Responses in Wheat. Front. Plant Sci. 2018, 9, 734. [Google Scholar] [CrossRef] [PubMed]
- Lamalakshmi Devi, E.; Kumar, S.; Basanta Singh, T.; Sharma, S.K.; Beemrote, A.; Devi, C.P.; Chongtham, S.K.; Singh, C.H.; Yumlembam, R.A.; Haribhushan, A.; et al. Adaptation strategies and defence mechanisms of plants during environmental stress. In Medicinal Plants and Environmental Challenges; Springer International Publishing: Basel, Switzerland, 2017; pp. 359–413. ISBN 9783319687179. [Google Scholar]
- Gallo-Franco, J.J.; Sosa, C.C.; Ghneim-Herrera, T.; Quimbaya, M. Epigenetic Control of Plant Response to Heavy Metal Stress: A New View on Aluminum Tolerance. Front. Plant Sci. 2020, 11, 2025. [Google Scholar] [CrossRef] [PubMed]
- Annacondia, M.L.; Magerøy, M.H.; Martinez, G. Stress response regulation by epigenetic mechanisms: Changing of the guards. Physiol. Plant. 2018, 162, 239–250. [Google Scholar] [CrossRef]
- Begcy, K.; Dresselhaus, T. Epigenetic responses to abiotic stresses during reproductive development in cereals. Plant Reprod. 2018, 31, 343–355. [Google Scholar] [CrossRef] [Green Version]
- Bali, A.S.; Sidhu, G.P.S.; Kumar, V. Root exudates ameliorate cadmium tolerance in plants: A review. Environ. Chem. Lett. 2020, 18, 1243–1275. [Google Scholar] [CrossRef]
- Shafiq, S.; Zeb, Q.; Ali, A.; Sajjad, Y.; Nazir, R.; Widemann, E.; Liu, L. Lead, Cadmium and Zinc phytotoxicity alter DNA methylation levels to confer heavy metal tolerance in Wheat. Int. J. Mol. Sci. 2019, 20, 4676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.K.; Archana, G. Cadmium minimization in food crops by cadmium resistant plant growth promoting rhizobacteria. Appl. Soil Ecol. 2016, 107, 66–78. [Google Scholar] [CrossRef]
- Fan, S.K.; Ye, J.Y.; Zhang, L.L.; Chen, H.S.; Zhang, H.H.; Zhu, Y.X.; Liu, X.X.; Jin, C.W. Inhibition of DNA demethylation enhances plant tolerance to cadmium toxicity by improving iron nutrition. Plant Cell Environ. 2020, 43, 275–291. [Google Scholar] [CrossRef]
- Guan, D.L.; Ding, R.R.; Hu, X.Y.; Yang, X.R.; Xu, S.Q.; Gu, W.; Zhang, M. Cadmium-induced genome-wide DNA methylation changes in growth and oxidative metabolism in Drosophila melanogaster. BMC Genom. 2019, 20, 356. [Google Scholar] [CrossRef] [Green Version]
- Mourato, M.; Pinto, F.; Moreira, I.; Sales, J.; Leitão, I.; Martins, L.L. The Effect of Cd Stress in Mineral Nutrient Uptake in Plants. In Cadmium Toxicity and Tolerance in Plants: From Physiology to Remediation; Academic Press: Lisbon, Portugal, 2019; pp. 327–348. ISBN 9780128148655. [Google Scholar]
- Huang, X.; Duan, S.; Wu, Q.; Yu, M.; Shabala, S. Reducing cadmium accumulation in plants: Structure–function relations and tissue-specific operation of transporters in the spotlight. Plants 2020, 9, 223. [Google Scholar] [CrossRef] [Green Version]
- Guan, M.Y.; Zhang, H.H.; Pan, W.; Jin, C.W.; Lin, X.Y. Sulfide alleviates cadmium toxicity in Arabidopsis plants by altering the chemical form and the subcellular distribution of cadmium. Sci. Total Environ. 2018, 627, 663–670. [Google Scholar] [CrossRef]
- Sebastian, A.; Prasad, M.N.V. Exogenous citrate and malate alleviate cadmium stress in Oryza sativa L.: Probing role of cadmium localization and iron nutrition. Ecotoxicol. Environ. Saf. 2018, 166, 215–222. [Google Scholar] [CrossRef]
- Liu, J.; He, Z. Small DNA Methylation, Big Player in Plant Abiotic Stress Responses and Memory. Front. Plant Sci. 2020, 11, 595603. [Google Scholar] [CrossRef] [PubMed]
- Akhter, Z.; Bi, Z.; Ali, K.; Sun, C.; Fiaz, S.; Haider, F.U.; Bai, J. In Response to Abiotic Stress, DNA Methylation Confers EpiGenetic Changes in Plants. Plants 2021, 10, 1096. [Google Scholar] [CrossRef] [PubMed]
- Bhadouriya, S.L.; Mehrotra, S.; Basantani, M.K.; Loake, G.J.; Mehrotra, R. Role of Chromatin Architecture in Plant Stress Responses: An Update. Front. Plant Sci. 2021, 11, 2131. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.J.; Liu, X.S.; Ma, L.Y.; Khan, I.U.; Rono, J.K.; Yang, Z.M. Identification of epigenetic mechanisms in paddy crop associated with lowering environmentally related cadmium risks to food safety. Environ. Pollut. 2020, 256, 113464. [Google Scholar] [CrossRef] [PubMed]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Chen, H.; Wang, J.P.; Liu, H.; Li, H.; Lin, Y.C.J.; Shi, R.; Yang, C.; Gao, J.; Zhou, C.; Li, Q.; et al. Hierarchical transcription factor and chromatin binding network for wood formation in populus trichocarpa. Plant Cell 2019, 31, 602–626. [Google Scholar] [CrossRef] [Green Version]
- Nagashima, R.; Hibino, K.; Ashwin, S.S.; Babokhov, M.; Fujishiro, S.; Imai, R.; Nozaki, T.; Tamura, S.; Tani, T.; Kimura, H.; et al. Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II. J. Cell Biol. 2019, 218, 1511–1530. [Google Scholar] [CrossRef] [Green Version]
- Damodaran, K.; Venkatachalapathy, S.; Alisafaei, F.; Radhakrishnan, A.V.; Jokhun, D.S.; Shenoy, V.B.; Shivashankar, G.V. Compressive force induces reversible chromatin condensation and cell geometry–dependent transcriptional response. Mol. Biol. Cell 2018, 29, 3039–3051. [Google Scholar] [CrossRef]
- Jing, Y.; Guo, Q.; Zha, P.; Lin, R. The chromatin-remodelling factor PICKLE interacts with CONSTANS to promote flowering in Arabidopsis. Plant Cell Environ. 2019, 42, 2495–2507. [Google Scholar] [CrossRef]
- Imai, K.; Ochiai, K. Role of histone modification on transcriptional regulation and HIV-1 gene expression: Possible mechanisms of periodontal diseases in AIDS progression. J. Oral Sci. 2011, 53, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Escobar, T.M.; Oksuz, O.; Saldaña-Meyer, R.; Descostes, N.; Bonasio, R.; Reinberg, D. Active and Repressed Chromatin Domains Exhibit Distinct Nucleosome Segregation during DNA Replication. Cell 2019, 179, 953–963.e11. [Google Scholar] [CrossRef]
- Mendiratta, S.; Gatto, A.; Almouzni, G. Histone supply: Multitiered regulation ensures chromatin dynamics throughout the cell cycle. J. Cell Biol. 2019, 218, 39–54. [Google Scholar] [CrossRef] [Green Version]
- Ageeva-Kieferle, A.; Rudolf, E.E.; Lindermayr, C. Redox-dependent chromatin remodeling: A new function of nitric oxide as architect of chromatin structure in plants. Front. Plant Sci. 2019, 10, 625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The effects of cadmium toxicity. Int. J. Environ. Res. Public Health 2020, 17, 3782. [Google Scholar] [CrossRef]
- Liu, Y.; Tavana, O.; Gu, W. P53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 2019, 11, 564–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, C.E.; English, D.M.; Cowley, S.M. Acetylation and Co: An expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem. 2019, 63, 97–107. [Google Scholar] [PubMed] [Green Version]
- Ng, C.; Aichinger, M.; Nguyen, T.; Au, C.; Najar, T.; Wu, L.; Mesa, K.R.; Liao, W.; Quivy, J.P.; Hubert, B.; et al. The histone chaperone CAF-1 cooperates with the DNA methyl transferases to maintain Cd4 silencing in cytotoxic T cells. Genes Dev. 2019, 33, 669–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafiq, S.; Ali, A.; Sajjad, Y.; Zeb, Q.; Shahzad, M.; Khan, A.R.; Nazir, R.; Widemann, E. The Interplay between Toxic and Essential Metals for Their Uptake and Translocation Is Likely Governed by DNA Methylation and Histone Deacetylation in Maize. Int. J. Mol. Sci. 2020, 21, 6959. [Google Scholar] [CrossRef]
- Gadhia, S.R.; O’Brien, D.; Barile, F.A. Cadmium affects mitotically inherited histone modification pathways in mouse embryonic stem cells. Toxicol. In Vitro 2015, 30, 583–592. [Google Scholar] [CrossRef]
- Żabka, A.; Winnicki, K.; Polit, J.T.; Wróblewski, M.; Maszewski, J. Cadmium (II)-Induced Oxidative Stress Results in Replication Stress and Epigenetic Modifications in Root Meristem Cell Nuclei of Vicia faba. Cells 2021, 10, 640. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Bräutigam, K.; Cronk, Q. DNA methylation and the evolution of developmental complexity in plants. Front. Plant Sci. 2018, 9, 1447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartels, A.; Han, Q.; Nair, P.; Stacey, L.; Gaynier, H.; Mosley, M.; Huang, Q.; Pearson, J.; Hsieh, T.-F.; An, Y.-Q.; et al. Dynamic DNA Methylation in Plant Growth and Development. Int. J. Mol. Sci. 2018, 19, 2144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, G.; Qin, L.; Tian, W.; Meng, L.; He, T.; Zhao, D. Heavy metal transporters-associated proteins in s. Tuberosum: Genome-wide identification, comprehensive gene feature, evolution and expression analysis. Genes 2020, 11, 1269. [Google Scholar] [CrossRef]
- Park, J.; Song, W.Y.; Ko, D.; Eom, Y.; Hansen, T.H.; Schiller, M.; Lee, T.G.; Martinoia, E.; Lee, Y. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 2012, 69, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.J.; Liu, X.S.; Tao, H.; Tan, S.K.; Chu, S.S.; Oono, Y.; Zhang, X.D.; Chen, J.; Yang, Z.M. Variation of DNA methylation patterns associated with gene expression in rice (Oryza sativa) exposed to cadmium. Plant Cell Environ. 2016, 39, 2629–2649. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 2012, 63, 695–709. [Google Scholar] [CrossRef] [Green Version]
- Greco, M.; Sáez, C.A.; Contreras, R.A.; Rodríguez-Rojas, F.; Bitonti, M.B.; Brown, M.T. Cadmium and/or copper excess induce interdependent metal accumulation, DNA methylation, induction of metal chelators and antioxidant defences in the seagrass Zostera marina. Chemosphere 2019, 224, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; He, L.; Song, J.; Cui, W.; Zhang, Y.; Jia, C.; Francis, D.; Rogers, H.J.; Sun, L.; Tai, P.; et al. Cadmium-induced genomic instability in Arabidopsis: Molecular toxicological biomarkers for early diagnosis of cadmium stress. Chemosphere 2016, 150, 258–265. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Ding, A.B.; Liu, F.; Zhong, X. Linking signaling pathways to histone acetylation dynamics in plants. J. Exp. Bot. 2020, 71, 5179–5190. [Google Scholar] [CrossRef]
- Gao, S.; Li, L.; Han, X.; Liu, T.; Jin, P.; Cai, L.; Xu, M.; Zhang, T.; Zhang, F.; Chen, J.; et al. Genome-wide identification of the histone acetyltransferase gene family in Triticum aestivum. BMC Genom. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, S.; Yu, C.W.; Chen, C.Y.; Wu, K. Histone Acetylation and Plant Development. Enzymes 2016, 40, 173–199. [Google Scholar] [PubMed]
- Lee, K.; Back, K. Overexpression of rice serotonin N-acetyltransferase 1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. Pineal Res. 2017, 62. [Google Scholar] [CrossRef] [PubMed]
- Rybaczek, D.; Bodys, A.; Maszewski, J. H2AX foci in late S/G2- and M-phase cells after hydroxyurea- and aphidicolin-induced DNA replication stress in Vicia. Histochem. Cell Biol. 2007, 128. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ye, Y.; Jiang, Z.; Wang, Y.; Zhu, C. MicroRNA390 is involved in cadmium tolerance and accumulation in rice. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.S.; Song, J.B.; Yang, Z.M. Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. J. Exp. Bot. 2012, 63. [Google Scholar] [CrossRef] [Green Version]
- Gielen, H.; Remans, T.; Vangronsveld, J.; Cuypers, A. MicroRNAs in metal stress: Specific roles or secondary responses? Int. J. Mol. Sci. 2012, 13, 15826–15847. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Zheng, W.; Cao, F.; Wu, F. Identification and comparative analysis of the microRNA transcriptome in roots of two contrasting tobacco genotypes in response to cadmium stress. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Satoh-Nagasawa, N.; Mori, M.; Nakazawa, N.; Kawamoto, T.; Nagato, Y.; Sakurai, K.; Takahashi, H.; Watanabe, A.; Akagi, H. Mutations in rice (oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012, 53. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Takahashi, R.; Bashir, K.; Shimo, H.; Senoura, T.; Sugimoto, K.; Ono, K.; Yano, M.; Ishikawa, S.; Arao, T.; et al. Characterizing the role of rice NRAMP5 in Manganese, Iron and Cadmium Transport. Sci. Rep. 2012, 2. [Google Scholar] [CrossRef]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. mir390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, Z.B.; Hai, B.Z.; Guo, J.L.; Li, Y.F.; Zhang, L. Characterization of wheat miRNAs and their target genes responsive to cadmium stress. Plant Physiol. Biochem. 2016, 101, 60–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Zheng, S.; Liu, R.; Lu, L.; Zhang, C.; Zhang, L.; Yant, L.; Wu, Y. The genome-wide impact of cadmium on microRNA and mRNA expression in contrasting Cd responsive wheat genotypes. BMC Genom. 2019, 20, 615. [Google Scholar] [CrossRef] [Green Version]
- Han, R.; Jian, C.; Lv, J.; Yan, Y.; Chi, Q.; Li, Z.; Wang, Q.; Zhang, J.; Liu, X.; Zhao, H. Identification and characterization of microRNAs in the flag leaf and developing seed of wheat (Triticum aestivum L.). BMC Genom. 2014, 15, 289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.; Zhang, Q.; Li, H.; Wang, X.; Wang, X.; Duan, X.; Wang, B.; Kang, Z. VsiRNAs derived from the miRNA-generating sites of pri-tae-miR159a based on the BSMV system play positive roles in the wheat response to Puccinia striiformis f. sp. tritici through the regulation of taMyb3 expression. Plant Physiol. Biochem. 2013, 68, 90–95. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).