Emerging Evidence for Pleiotropism of Eosinophils
Abstract
1. Introduction
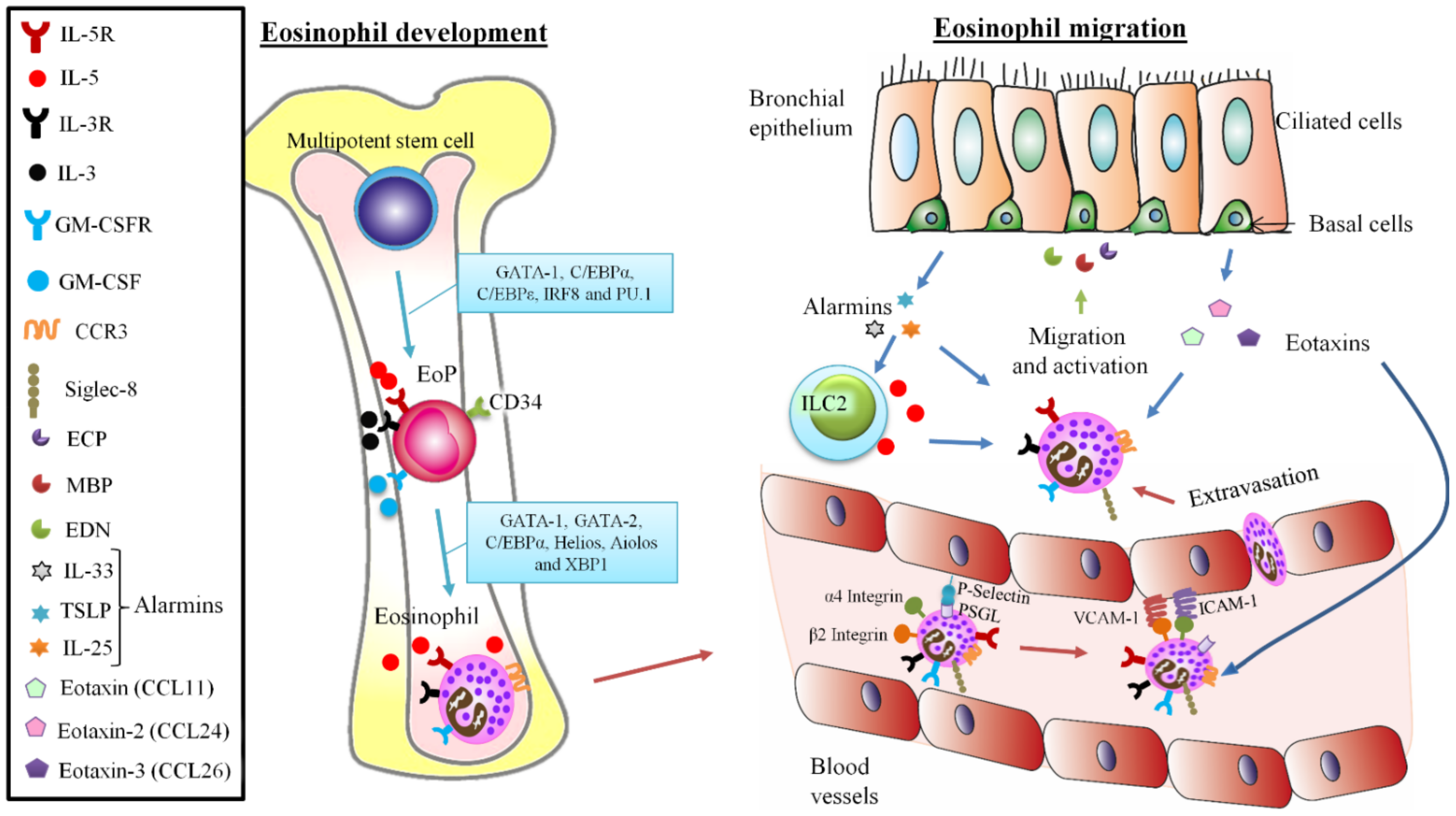
2. Unraveling the Complex Biology of Eosinophils: From Development to Traffic
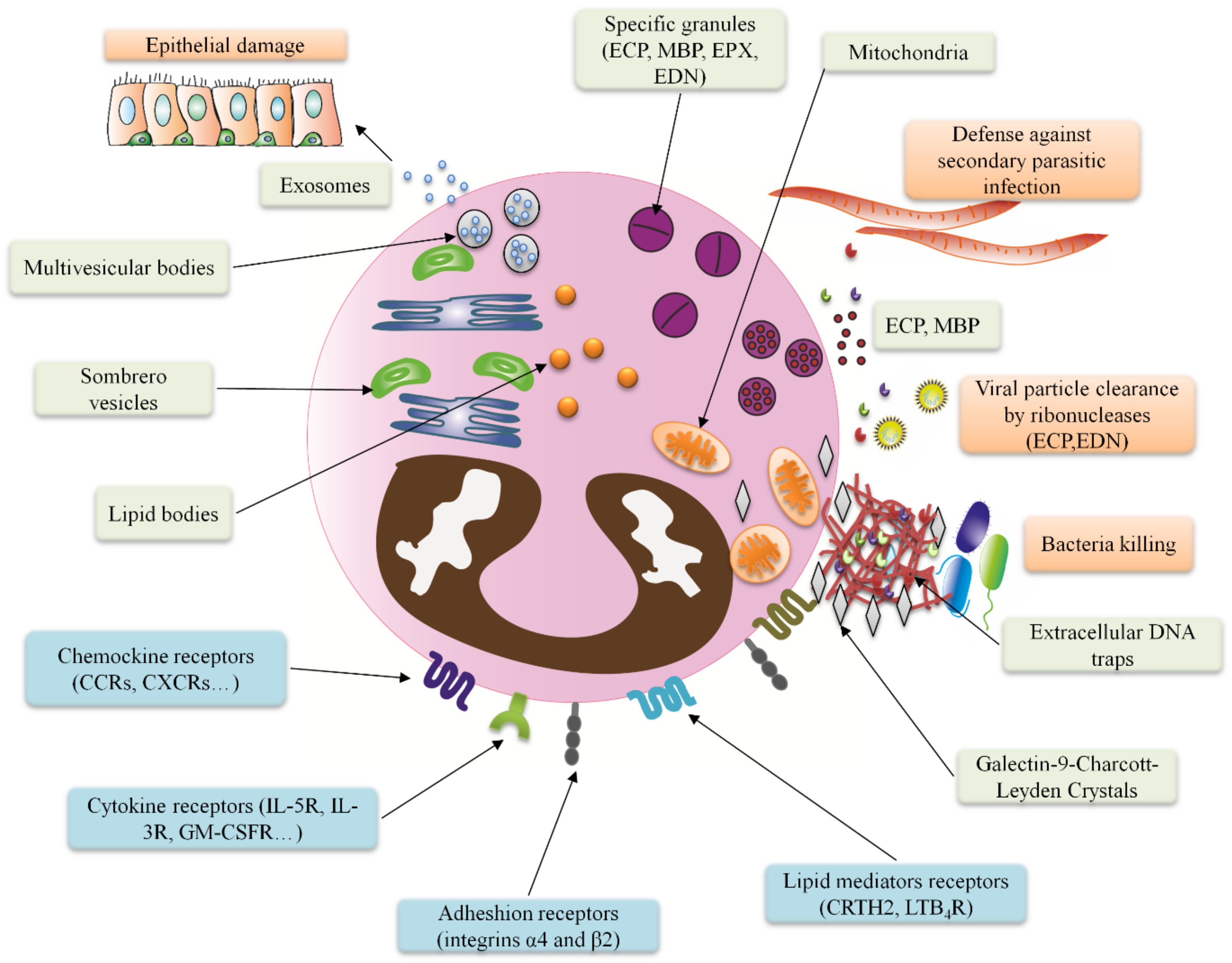
3. The Eosinophil Immune Response Mechanisms against Diverse Pathogens
3.1. Revisiting Eosinophil’s Role against Parasites
3.2. Eosinophil Responses against Bacteria, the Involvement of Extracellular Traps
3.3. Eosinophilic Responses against Virus and Current Knowledge about Eosinophils Involvement in COVID-19
| Main Findings | Number of Subjects | References |
|---|---|---|
| Blood eosinophilia is associated with good COVID-19 prognosis | 314 | [85] |
| 951 | [86] | |
| 9644 | [87] | |
| 10 | [88] | |
| 95 | [95] | |
| 4252 | [89] | |
| Higher eosinophil counts in severe COVID-19 | 135 | [97] |
| 15 | [98] | |
| 37 | [99] | |
| T2 diseases are not associated to COVID-19 | 189 | [108] |
| Blood eosinopenia is a marker of worst COVID-19 disease course | 324 | [90] |
| 95 | [101] | |
| 37 | [91] | |
| 96 | [93] | |
| 121 | [105] | |
| 40 | [94] | |
| 190 | [96] | |
| 294 | [118] | |
| 94 | [92] | |
| 429 | [102] | |
| 174 | [103] | |
| 37 | [104] | |
| ACE2 receptor is reduced in asthma | 365 | [109] |
| 66 | [110] | |
| Biological asthma treatment does not affect COVID-19 | 676 | [111] |
| 545 | [112] | |
| 1504 | [113] | |
| 2 cases | [115] | |
| 2 cases | [116] | |
| 1 case | [117] | |
| Biological asthma treatment might have worst COVID-19 outcome | 634 | [114] |
| ACE2 = Angiotensin-converting enzyme 2. COVID-19 = Coronavirus disease 2019. | ||
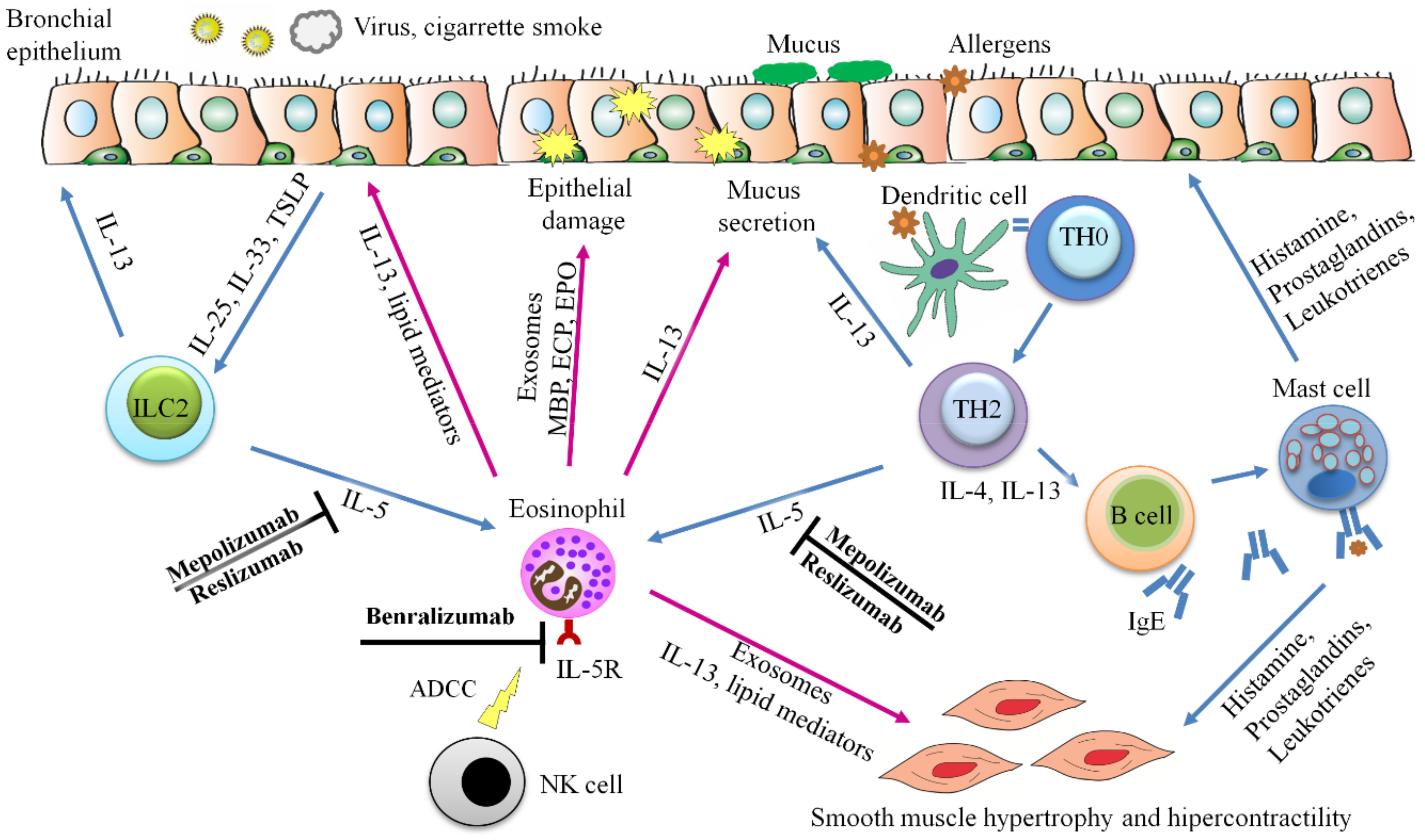
3.4. Exosomes from Eosinophils Contribute to Asthma Hallmarks
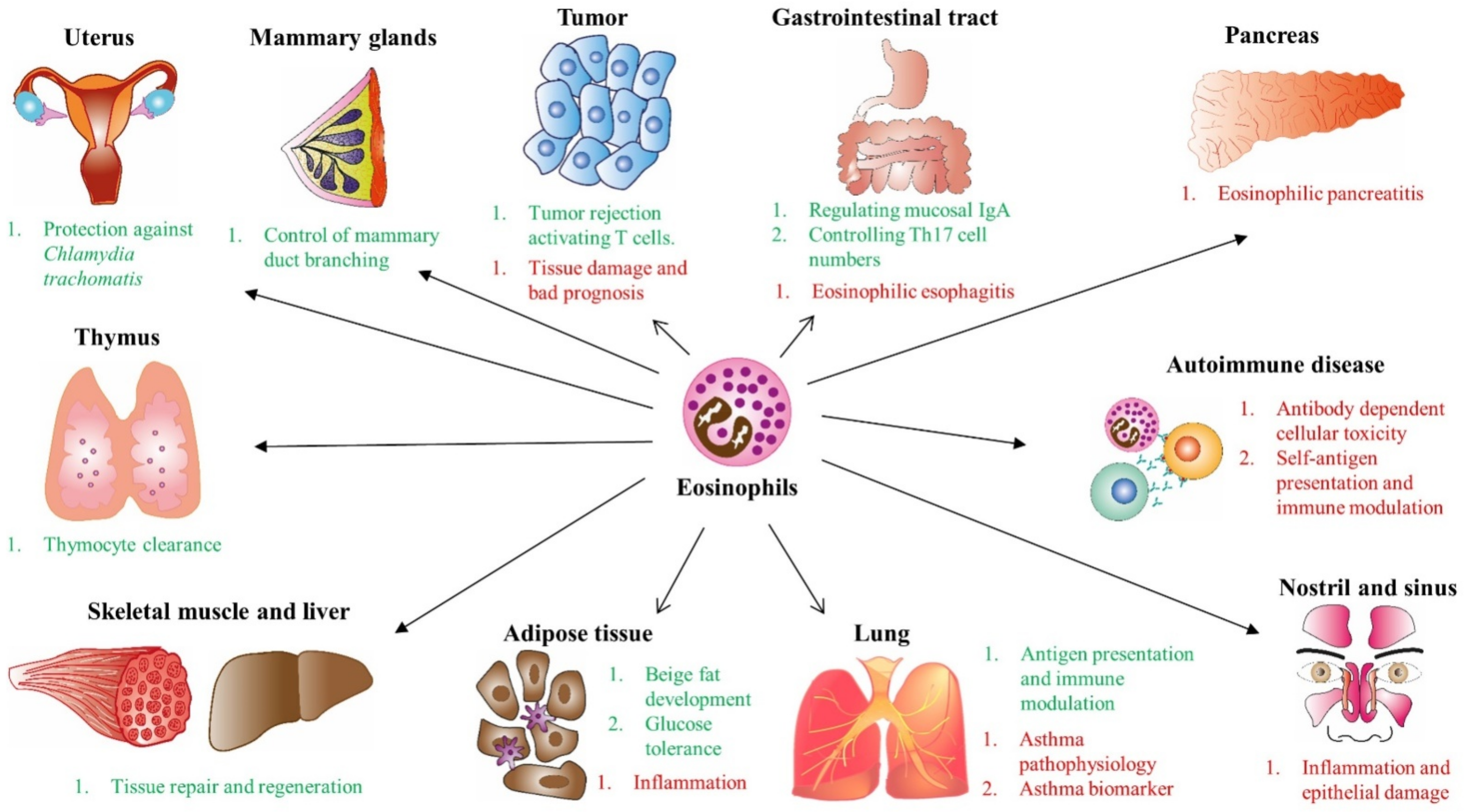
4. Role of Eosinophils as Effector Cells in Homeostasis
5. Heterogeneity and Phenotypes of Eosinophils
6. Eosinophil Immune Dysfunction (EID)
6.1. Eosinophilic Gastrointestinal Diseases (EGID) and Pancreatic Disorders
6.2. Eosinophilia in Myeloid Neoplasms and Solid Tumors
6.3. Eosinophilia in Autoimmune Diseases
6.4. Eosinophilia in Lung Diseases: Atopic Diseases, Asthma and Interstitial Lung Disease
7. Treatments Focused on Eosinophils
| Drug | Disease | Mechanism | Ref. |
|---|---|---|---|
| Glucocorticoid | Systemic inflammation | Apoptotic effects and inhibition of cytokines implicated in eosinophil survival | [216] |
| Theophylline | Asthma | Anti-inflammatory effects | [217] |
| Antileukotrienes | Asthma | Anti-inflammatory effects | [217] |
| Cyclophosphamide | Lymphoma | Immunosuppressor | [218] |
| Alemtezumab | Severe T cell neoplasms or inflammatory diseases | Anti-CD52 | [218] |
| Levosimendan/destrosimendan | Commonly used to heart failure | Eosinophil proapoptotic effect in vitro | [219] |
| Bertilimumab | Pemphigus | Anti-eotaxin-1. Affect to eosinophil recruitment | [220] |
| Mepolizumab and reslizumab | Uncontrolled severe asthma | Anti-IL-5 | [221,222,223,224] |
| Benralizumab | Uncontrolled severe asthma | Anti-IL-5R. Inhibits the growth, maturation, activation and survival of eosinophils through an antibody-dependent cytotoxicity mechanism | [225,226] |
| Omalizumab | Persistent severe allergic asthma | Anti-IgE | [227] |
| Dupilumab | Moderate-to-severe atopic dermatitis | Anti-IL-13 | [228,229] |
| Tezepelumab | Severe asthma | Anti-TSLP | [230] |
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACE2 | Angiotensin-converting enzyme 2 |
| ADCC | Antibody-dependent cellular cytotoxicity |
| CCR3 | CC-chemokine receptor 3 |
| COPD | Chronic obstructive pulmonary disease |
| cysLTR | Cysteinyl leukotriene receptor |
| CP | Chronic pancreatitis |
| CRSsNP | Chronic rhinosinusitis without nasal polyps |
| CRSwNP | Chronic rhinosinusitis with nasal polyps |
| CRTH2 | Prostaglandin D2 receptor 2 |
| DC | Dendritic cell |
| DNA | Deoxyribonucleic acid |
| ECP | Eosinophil cationic protein |
| EDN | Eosinophil derived neurotoxin |
| EETs | Eosinophil extracellular traps |
| EGF | Epidermal growth factor |
| EGID | Eosinophilic gastrointestinal disease |
| EID | Eosinophil immune dysfunction |
| EoE | Eosinophilic esophagitis |
| EoP | Eosinophil progenitors |
| Eos | Eosinophils |
| EPX | Eosinophil peroxidase |
| EVs | Extracellular vesicles |
| FeNO | Fraction of exhaled nitric oxide |
| GI | Gastrointestinal |
| GM-CSF | Granulocyte-macrophage colony stimulating factor |
| hEos | Homeostatic eosinophils |
| hpf | High-power field |
| ICAM1 | Intercellular adhesion molecule 1 |
| ICS | Inhaled corticosteroids |
| ICU | Intensive care unit |
| iEos | Inflammatory eosinophils |
| IFN | Interferon |
| IL-5 | Interleukin-5 |
| ILC2 | Type 2 innate lymphoid cells |
| LABA | Long-acting beta agonists |
| MBP | Major basic protein |
| MHC | Major histocompatibility complex |
| miRNA | MicroRNA |
| NO | Nitric oxide |
| pDCs | Plasmacytoid dendritic cells |
| PDGFR | Platelet-derived growth factor receptor |
| RCTs | Randomized clinical trials |
| RNA | Ribonucleic acid |
| ROS | Reactive oxygen species |
| RSV | Respiratory syncytial virus |
| RT-PCR | Reverse transcription polymerase chain reaction |
| RTC | Randomized clinical trial |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| Siglec-8 | Sialic acid-binding immunoglobulin-like lectin 8 |
| SNPs | Single nucleotide polymorphisms |
| TGF | Transforming growth factor |
| TNF-α | Tumor necrosis factor α |
| TLR | Toll-like receptor |
| TSLP | Thymic stromal lymphopoietin |
References
- Kay, A.B. The early history of the eosinophil. Clin. Exp. Allergy 2015, 45, 575–582. [Google Scholar] [CrossRef]
- Gleich, G.J.; Adolphson, C.R. The Eosinophilic Leukocyte: Structure and Function. Adv. Immunol. 1986, 39, 177–253. [Google Scholar] [CrossRef] [PubMed]
- Lacy, P.; Rosenberg, H.F.; Walsh, G.M. Eosinophil overview: Structure, biological properties, and key functions. Methods Mol. Biol. 2014, 1178, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dorosz, A.; Grosicki, M.; Dybas, J.; Matuszyk, E.; Rodewald, M.; Meyer, T.; Popp, J.; Malek, K.; Baranska, M. Eosinophils and Neutrophils-Molecular Differences Revealed by Spontaneous Raman, CARS and Fluorescence Microscopy. Cells 2020, 9, 2041. [Google Scholar] [CrossRef] [PubMed]
- Klion, A.D.; Ackerman, S.J.; Bochner, B.S. Contributions of Eosinophils to Human Health and Disease. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 179–209. [Google Scholar] [CrossRef] [PubMed]
- Weller, P.F.; Spencer, L.A. Functions of tissue-resident eosinophils. Nat. Rev. Immunol. 2017, 17, 746–760. [Google Scholar] [CrossRef]
- Acharya, K.R.; Ackerman, S.J. Eosinophil granule proteins: Form and function. J. Biol. Chem. 2014, 289, 17406–17415. [Google Scholar] [CrossRef]
- Plager, D.A.; Loegering, D.A.; Checkel, J.L.; Tang, J.; Kephart, G.M.; Caffes, P.L.; Adolphson, C.R.; Ohnuki, L.E.; Gleich, G.J. Major Basic Protein Homolog (MBP2): A Specific Human Eosinophil Marker. J. Immunol. 2006, 177, 7340–7345. [Google Scholar] [CrossRef] [PubMed]
- Davoine, F.; Lacy, P. Eosinophil cytokines, chemokines, and growth factors: Emerging roles in immunity. Front. Immunol. 2014, 5, 570. [Google Scholar] [CrossRef]
- Woschnagg, C.; Rubin, J.; Venge, P. Eosinophil Cationic Protein (ECP) Is Processed during Secretion. J. Immunol. 2009, 183, 3949–3954. [Google Scholar] [CrossRef]
- Melo, R.C.N.; Weller, P.F. Contemporary understanding of the secretory granules in human eosinophils. J. Leukoc. Biol. 2018, 104, 85–93. [Google Scholar] [CrossRef]
- Ueki, S.; Miyabe, Y.; Yohei, Y.; Fukuchi, M.; Hirokawa, M.; Spencer, L.A.; Weller, P.F. Charcot-Leyden Crystals in Eosinophilic Inflammation: Active Cytolysis Leads to Crystal Formation. Curr. Allergy Asthma Rep. 2019, 19, 35. [Google Scholar] [CrossRef] [PubMed]
- Melo, R.C.N.; Weller, P.F. Unraveling the complexity of lipid body organelles in human eosinophils. J. Leukoc. Biol. 2014, 96, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Mazzeo, C.; Cañas, J.A.; Zafra, M.P.; Rojas Marco, A.; Fernández-Nieto, M.; Sanz, V.; Mittelbrunn, M.; Izquierdo, M.; Baixaulli, F.; Sastre, J.; et al. Exosome secretion by eosinophils: A possible role in asthma pathogenesis. J. Allergy Clin. Immunol. 2015, 135, 1603–1613. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.A.; Bochner, B.S. Eosinophils and eosinophil-associated diseases: An update. J. Allergy Clin. Immunol. 2018, 141, 505–517. [Google Scholar] [CrossRef]
- Mori, Y.; Iwasaki, H.; Kohno, K.; Yoshimoto, G.; Kikushige, Y.; Okeda, A.; Uike, N.; Niiro, H.; Takenaka, K.; Nagafuji, K.; et al. Identification of the human eosinophil lineage-committed progenitor: Revision of phenotypic definition of the human common myeloid progenitor. J. Exp. Med. 2009, 206, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Bossios, A.; Rådinger, M. CD34+ eosinophil-lineage-committed cells in the mouse lung. Methods Mol. Biol. 2014, 1178, 29–43. [Google Scholar] [CrossRef]
- Stirling, R.G.; Van Rensen, E.L.J.; Barnes, P.J.; Chung, K.F. Interleukin-5 induces CD34+ eosinophil progenitor mobilization and eosinophil CCR3 expression in asthma. Am. J. Respir. Crit. Care Med. 2001, 164, 1403–1409. [Google Scholar] [CrossRef]
- Robinson, D.S.; Damia, R.; Zeibecoglou, K.; Molet, S.; North, J.; Yamada, T.; Kay, A.B.; Hamid, Q. CD34+/interleukin-5Rα messenger RNA+ cells in the bronchial mucosa in asthma: Potential airway eosinophil progenitors. Am. J. Respir. Cell Mol. Biol. 1999, 20, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Fulkerson, P.C. Transcription factors in eosinophil development and as therapeutic targets. Front. Med. 2017, 4, 115. [Google Scholar] [CrossRef]
- Bouffi, C.; Kartashov, A.V.; Schollaert, K.L.; Chen, X.; Bacon, W.C.; Weirauch, M.T.; Barski, A.; Fulkerson, P.C. Transcription Factor Repertoire of Homeostatic Eosinophilopoiesis. J. Immunol. 2015, 195, 2683–2695. [Google Scholar] [CrossRef]
- Yu, C.; Cantor, A.B.; Yang, H.; Browne, C.; Wells, R.A.; Fujiwara, Y.; Orkin, S.H. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J. Exp. Med. 2002, 195, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Drissen, R.; Thongjuea, S.; Theilgaard-Mönch, K.; Nerlov, C. Identification of two distinct pathways of human myelopoiesis. Sci. Immunol. 2019, 4, eaau7148. [Google Scholar] [CrossRef] [PubMed]
- Pellin, D.; Loperfido, M.; Baricordi, C.; Wolock, S.L.; Montepeloso, A.; Weinberg, O.K.; Biffi, A.; Klein, A.M.; Biasco, L. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Weller, P.F. Cytokine regulation of eosinophil function. Clin. Immunol. Immunopathol. 1992, 62, S55–S59. [Google Scholar] [CrossRef]
- Dougan, M.; Dranoff, G.; Dougan, S.K. GM-CSF, IL-3, and IL-5 Family of Cytokines: Regulators of Inflammation. Immunity 2019, 50, 796–811. [Google Scholar] [CrossRef]
- Rothenberg, M.E.; Hogan, S.P. The eosinophil. Annu. Rev. Immunol. 2006, 24, 147–174. [Google Scholar] [CrossRef] [PubMed]
- Pelaia, C.; Paoletti, G.; Puggioni, F.; Racca, F.; Pelaia, G.; Canonica, G.W.; Heffler, E. Interleukin-5 in the Pathophysiology of Severe Asthma. Front. Physiol. 2019, 10, 1514. [Google Scholar] [CrossRef]
- Mortaz, E.; Amani, S.; Mumby, S.; Adcock, I.M.; Movassaghi, M.; Folkerts, J.; Garssen, J.; Folkerts, G. Role of mast cells and Type 2 Innate Lymphoid (ILC2) cells in lung transplantation. J. Immunol. Res. 2018, 2018, 2785971. [Google Scholar] [CrossRef]
- Kim, H.J.; Jung, Y. The Emerging Role of Eosinophils as Multifunctional Leukocytes in Health and Disease. Immune Netw. 2020, 20, e24. [Google Scholar] [CrossRef]
- Nakagome, K.; Nagata, M. Possible mechanisms of eosinophil accumulation in eosinophilic pneumonia. Biomolecules 2020, 10, 638. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.S.; Rokach, J. The eosinophil chemoattractant 5-oxo-ETE and the OXE receptor. Prog. Lipid Res. 2013, 52, 651–665. [Google Scholar] [CrossRef] [PubMed]
- Esnault, S.; Kelly, E.A. Essential mechanisms of differential activation of eosinophils by IL-3 compared to GM-CSF and IL-5. Crit. Rev. Immunol. 2016, 36, 429–444. [Google Scholar] [CrossRef]
- Schwartz, C.; Willebrand, R.; Huber, S.; Rupec, R.A.; Wu, D.; Locksley, R.; Voehringer, D. Eosinophil-specific deletion of IκBα in mice reveals a critical role of NF-κB-induced Bcl-xLfor inhibition of apoptosis. Blood 2015, 125, 3896–3904. [Google Scholar] [CrossRef]
- Mitchell, P.D.; O’Byrne, P.M. Epithelial-Derived Cytokines in Asthma. Chest 2017, 151, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- Johnston, L.K.; Hsu, C.-L.; Krier-Burris, R.A.; Chhiba, K.D.; Chien, K.B.; McKenzie, A.; Berdnikovs, S.; Bryce, P.J. IL-33 Precedes IL-5 in Regulating Eosinophil Commitment and Is Required for Eosinophil Homeostasis. J. Immunol. 2016, 197, 3445–3453. [Google Scholar] [CrossRef]
- Heffler, E.; Allegra, A.; Pioggia, G.; Picardi, G.; Musolino, C.; Gangemi, S. MicroRNA profiling in asthma: Potential biomarkers and therapeutic targets. Am. J. Respir. Cell Mol. Biol. 2017, 57, 642–650. [Google Scholar] [CrossRef]
- Bochner, B.S. Novel Therapies for Eosinophilic Disorders. Immunol. Allergy Clin. N. Am. 2015, 35, 577–598. [Google Scholar] [CrossRef]
- Takatsu, K. Interleukin-5 and IL-5 receptor in health and diseases. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 463–485. [Google Scholar] [CrossRef] [PubMed]
- Lamkhioued, B.; Abdelilah, S.G.; Hamid, Q.; Mansour, N.; Delespesse, G.; Renzi, P.M. The CCR3 Receptor Is Involved in Eosinophil Differentiation and Is Up-Regulated by Th2 Cytokines in CD34 + Progenitor Cells. J. Immunol. 2003, 170, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Youngblood, B.A.; Brock, E.C.; Leung, J.; Falahati, R.; Bochner, B.S.; Rasmussen, H.S.; Peterson, K.; Bebbington, C.; Tomasevic, N. Siglec-8 antibody reduces eosinophils and mast cells in a transgenic mouse model of eosinophilic gastroenteritis. JCI Insight 2019, 4, e126219. [Google Scholar] [CrossRef] [PubMed]
- Legrand, F.; Tomasevic, N.; Simakova, O.; Lee, C.C.R.; Wang, Z.; Raffeld, M.; Makiya, M.A.; Palath, V.; Leung, J.; Baer, M.; et al. The eosinophil surface receptor epidermal growth factor-like module containing mucin-like hormone receptor 1 (EMR1): A novel therapeutic target for eosinophilic disorders. J. Allergy Clin. Immunol. 2014, 133, 1439–1447. [Google Scholar] [CrossRef]
- Sastre, B.; Rodrigo-Muñoz, J.M.; Garcia-Sanchez, D.A.; Cañas, J.A.; Del Pozo, V. Eosinophils: Old players in a new game. J. Investig. Allergol. Clin. Immunol. 2018, 28, 289–304. [Google Scholar] [CrossRef]
- Yoon, J.; Um, H.N.; Jang, J.; Bae, Y.A.; Park, W.J.; Kim, H.J.; Yoon, M.S.; Chung, I.Y.; Jung, Y.J. Eosinophil Activation by Toll-Like Receptor 4 Ligands Regulates Macrophage Polarization. Front. Cell Dev. Biol. 2019, 7, 329. [Google Scholar] [CrossRef]
- Zustakova, M.; Kratochvilova, L.; Slama, P. Apoptosis of eosinophil granulocytes. Biology 2020, 9, 457. [Google Scholar] [CrossRef]
- Kovalszki, A.; Weller, P.F. Eosinophilia. Prim. Care Clin. Off. Pract. 2016, 43, 607–617. [Google Scholar] [CrossRef]
- Kita, H. Eosinophils: Multifunctional and distinctive properties. Int. Arch. Allergy Immunol. 2013, 161, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Gieseck, R.L.; Wilson, M.S.; Wynn, T.A. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 2018, 18, 62–76. [Google Scholar] [CrossRef]
- Doran, E.; Cai, F.; Holweg, C.T.J.; Wong, K.; Brumm, J.; Arron, J.R. Interleukin-13 in Asthma and Other Eosinophilic Disorders. Front. Med. 2017, 4, 139. [Google Scholar] [CrossRef] [PubMed]
- Percopo, C.M.; Dyer, K.D.; Ochkur, S.I.; Luo, J.L.; Fischer, E.R.; Lee, J.J.; Lee, N.A.; Domachowske, J.B.; Rosenberg, H.F. Activated mouse eosinophils protect against lethal respiratory virus infection. Blood 2014, 123, 743–752. [Google Scholar] [CrossRef]
- Huang, L.; Gebreselassie, N.G.; Gagliardo, L.F.; Ruyechan, M.C.; Luber, K.L.; Lee, N.A.; Lee, J.J.; Appleton, J.A. Eosinophils Mediate Protective Immunity against Secondary Nematode Infection. J. Immunol. 2015, 194, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Appleton, J.A. Eosinophils in Helminth Infection: Defenders and Dupes. Trends Parasitol. 2016, 32, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Scepek, S.; Moqbel, R.; Lindau, M. Compound exocytosis and cumulative degranulation by eosinophils and their role in parasite killing. Parasitol. Today 1994, 10, 276–278. [Google Scholar] [CrossRef]
- Melo, R.C.N.; Perez, S.A.C.; Spencer, L.A.; Dvorak, A.M.; Weller, P.F. Intragranular vesiculotubular compartments are involved in piecemeal degranulation by activated human eosinophils. Traffic 2005, 6, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Carmo, L.A.S.; Bonjour, K.; Ueki, S.; Neves, J.S.; Liu, L.; Spencer, L.A.; Dvorak, A.M.; Weller, P.F.; Melo, R.C.N. CD63 is tightly associated with intracellular, secretory events chaperoning piecemeal degranulation and compound exocytosis in human eosinophils. J. Leukoc. Biol. 2016, 100, 391–401. [Google Scholar] [CrossRef]
- Dias, F.F.; Amaral, K.B.; Malta, K.K.; Silva, T.P.; Rodrigues, G.S.C.; Rosa, F.M.; Rodrigues, G.O.L.; Costa, V.V.; Chiarini-Garcia, H.; Weller, P.F.; et al. Identification of Piecemeal Degranulation and Vesicular Transport of MBP-1 in Liver-Infiltrating Mouse Eosinophils During Acute Experimental Schistosoma mansoni Infection. Front. Immunol. 2018, 9, 3019. [Google Scholar] [CrossRef] [PubMed]
- Spencer, L.A.; Melo, R.C.N.; Perez, S.A.C.; Bafford, S.P.; Dvorak, A.M.; Weller, P.F. Cytokine receptor-mediated trafficking of preformed IL-4 in eosinophils identifies an innate immune mechanism of cytokine secretion. Proc. Natl. Acad. Sci. USA 2006, 103, 3333–3338. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.S.; Radke, A.L.; Weller, P.F. Cysteinyl leukotrienes acting via granule membrane-expressed receptors elicit secretion from within cell-free human eosinophil granules. J. Allergy Clin. Immunol. 2010, 125, 477–482. [Google Scholar] [CrossRef]
- Adu, B.; Dodoo, D.; Adukpo, S.; Gyan, B.A.; Hedley, P.L.; Goka, B.; Adjei, G.O.; Larsen, S.O.; Christiansen, M.; Theisen, M. Polymorphisms in the RNASE3 gene are associated with susceptibility to cerebral malaria in Ghanaian children. PLoS ONE 2011, 6, e29465. [Google Scholar] [CrossRef]
- Diop, G.; Derbois, C.; Loucoubar, C.; Mbengue, B.; Ndao, B.N.; Thiam, F.; Thiam, A.; Ndiaye, R.; Dieye, Y.; Olaso, R.; et al. Genetic variants of RNASE3 (ECP) and susceptibility to severe malaria in Senegalese population. Malar. J. 2018, 17, 1–10. [Google Scholar] [CrossRef]
- Fabre, V.; Beiting, D.P.; Bliss, S.K.; Gebreselassie, N.G.; Gagliardo, L.F.; Lee, N.A.; Lee, J.J.; Appleton, J.A. Eosinophil Deficiency Compromises Parasite Survival in Chronic Nematode Infection. J. Immunol. 2009, 182, 1577–1583. [Google Scholar] [CrossRef]
- Huang, L.; Gebreselassie, N.G.; Gagliardo, L.F.; Ruyechan, M.C.; Lee, N.A.; Lee, J.J.; Appleton, J.A. Eosinophil-Derived IL-10 Supports Chronic Nematode Infection. J. Immunol. 2014, 193, 4178–4187. [Google Scholar] [CrossRef]
- Radonjic-Hoesli, S.; Wang, X.; de Graauw, E.; Stoeckle, C.; Styp-Rekowska, B.; Hlushchuk, R.; Simon, D.; Spaeth, P.J.; Yousefi, S.; Simon, H.U. Adhesion-induced eosinophil cytolysis requires the receptor-interacting protein kinase 3 (RIPK3)–mixed lineage kinase-like (MLKL) signaling pathway, which is counterregulated by autophagy. J. Allergy Clin. Immunol. 2017, 140, 1632–1642. [Google Scholar] [CrossRef]
- Neves, J.S.; Perez, S.A.C.; Spencer, L.A.; Melo, R.C.N.; Reynolds, L.; Ghiran, I.; Mahmudi-Azer, S.; Odemuyiwa, S.O.; Dvorak, A.M.; Moqbel, R.; et al. Eosinophil granules function extracellularly as receptor-mediated secretory organelles. Proc. Natl. Acad. Sci. USA 2008, 105, 18478–18483. [Google Scholar] [CrossRef]
- Ueki, S.; Melo, R.C.N.; Ghiran, I.; Spencer, L.A.; Dvorak, A.M.; Weller, P.F. Eosinophil extracellular DNA trap cell death mediates lytic release of free secretion-competent eosinophil granules in humans. Blood 2013, 121, 2074–2083. [Google Scholar] [CrossRef]
- Yousefi, S.; Gold, J.A.; Andina, N.; Lee, J.J.; Kelly, A.M.; Kozlowski, E.; Schmid, I.; Straumann, A.; Reichenbach, J.; Gleich, G.J.; et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 2008, 14, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Muniz, V.S.; Silva, J.C.; Braga, Y.A.V.; Melo, R.C.N.; Ueki, S.; Takeda, M.; Hebisawa, A.; Asano, K.; Figueiredo, R.T.; Neves, J.S. Eosinophils release extracellular DNA traps in response to Aspergillus fumigatus. J. Allergy Clin. Immunol. 2018, 141, 571–585. [Google Scholar] [CrossRef]
- Gevaert, E.; Zhang, N.; Krysko, O.; Lan, F.; Holtappels, G.; De Ruyck, N.; Nauwynck, H.; Yousefi, S.; Simon, H.U.; Bachert, C. Extracellular eosinophilic traps in association with Staphylococcus aureus at the site of epithelial barrier defects in patients with severe airway inflammation. J. Allergy Clin. Immunol. 2017, 139, 1849–1860. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Masterson, J.C.; Furuta, G.T. Eosinophils, probiotics, and the microbiome. J. Leukoc. Biol. 2016, 100, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Svensson, L.; Wennerås, C. Human eosinophils selectively recognize and become activated by bacteria belonging to different taxonomic groups. Microbes Infect. 2005, 7, 720–728. [Google Scholar] [CrossRef]
- Linch, S.N.; Kelly, A.M.; Danielson, E.T.; Pero, R.; Lee, J.J.; Gold, J.A. Mouse eosinophils possess potent antibacterial properties in vivo. Infect. Immun. 2009, 77, 4976–4982. [Google Scholar] [CrossRef]
- Torrent, M.; de la Torre, B.G.; Nogués, V.M.; Andreu, D.; Boix, E. Bactericidal and membrane disruption activities of the eosinophil cationic protein are largely retained in an N-terminal fragment. Biochem. J. 2009, 421, 425–434. [Google Scholar] [CrossRef]
- Dworski, R.; Simon, H.U.; Hoskins, A.; Yousefi, S. Eosinophil and neutrophil extracellular DNA traps in human allergic asthmatic airways. J. Allergy Clin. Immunol. 2011, 127, 1260–1266. [Google Scholar] [CrossRef]
- Choi, Y.; Le Pham, D.; Lee, D.H.; Lee, S.H.; Kim, S.H.; Park, H.S. Biological function of eosinophil extracellular traps in patients with severe eosinophilic asthma. Exp. Mol. Med. 2018, 50, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.; Radonjic-Hösli, S.; Straumann, A.; Yousefi, S.; Simon, H.U. Active eosinophilic esophagitis is characterized by epithelial barrier defects and eosinophil extracellular trap formation. Allergy Eur. J. Allergy Clin. Immunol. 2015, 70, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Melo, R.C.N.; Wang, H.; Silva, T.P.; Imoto, Y.; Fujieda, S.; Fukuchi, M.; Miyabe, Y.; Hirokawa, M.; Ueki, S.; Weller, P.F. Galectin-10, the protein that forms Charcot-Leyden crystals, is not stored in granules but resides in the peripheral cytoplasm of human eosinophils. J. Leukoc. Biol. 2020, 108, 139–149. [Google Scholar] [CrossRef]
- Prince, L.R.; Graham, K.J.; Connolly, J.; Anwar, S.; Ridley, R.; Sabroe, I.; Foster, S.J.; Whyte, M.K.B. Staphylococcus aureus induces eosinophil cell death mediated by α-Hemolysin. PLoS ONE 2012, 7, e31506. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Boix, E. Host Defence RNases as Antiviral Agents against Enveloped Single Stranded RNA Viruses. Virulence 2021, 12, 444–469. [Google Scholar] [CrossRef] [PubMed]
- Phipps, S.; En Lam, C.; Mahalingam, S.; Newhouse, M.; Ramirez, R.; Rosenberg, H.F.; Foster, P.S.; Matthaei, K.I. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood 2007, 110, 1578–1586. [Google Scholar] [CrossRef]
- Samarasinghe, A.E.; Melo, R.C.N.; Duan, S.; LeMessurier, K.S.; Liedmann, S.; Surman, S.L.; Lee, J.J.; Hurwitz, J.L.; Thomas, P.G.; McCullers, J.A. Eosinophils Promote Antiviral Immunity in Mice Infected with Influenza A Virus. J. Immunol. 2017, 198, 3214–3226. [Google Scholar] [CrossRef]
- Drake, M.G.; Bivins-Smith, E.R.; Proskocil, B.J.; Nie, Z.; Scott, G.D.; Lee, J.J.; Lee, N.A.; Fryer, A.D.; Jacoby, D.B. Human and mouse eosinophils have antiviral activity against parainfluenza virus. Am. J. Respir. Cell Mol. Biol. 2016, 55, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Redes, J.L.; Percopo, C.M.; Druey, K.M.; Rosenberg, H.F. Alternaria alternata challenge at the nasal mucosa results in eosinophilic inflammation and increased susceptibility to influenza virus infection. Clin. Exp. Allergy 2018, 48, 691–702. [Google Scholar] [CrossRef]
- Flores-Torres, A.S.; Salinas-Carmona, M.C.; Salinas, E.; Rosas-Taraco, A.G. Eosinophils-Respiratory Viruses. Viral Immunol. 2019, 32, 198–207. [Google Scholar] [CrossRef]
- Rodrigo-Muñoz, J.; Sastre, B.; Cañas, J.; Gil-Martínez, M.; Redondo, N.; del Pozo, V. Eosinophil Response Against Classical and Emerging Respiratory Viruses: COVID-19. J. Investig. Allergol. Clin. Immunol. 2020, 31, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.P.; Soliman, A.; Al Masalamani, M.A.; De Sanctis, V.; Nashwan, A.J.; Sasi, S.; Ali, E.A.; Hassan, O.A.; Iqbal, F.M.; Yassin, M.A. Clinical outcome of eosinophilia in patients with covid-19: A controlled study. Acta Biomed. 2020, 91, 1–10. [Google Scholar] [CrossRef]
- Ferastraoaru, D.; Hudes, G.; Jerschow, E.; Jariwala, S.; Karagic, M.; de Vos, G.; Rosenstreich, D.; Ramesh, M. Eosinophilia in Asthma Patients Is Protective Against Severe COVID-19 Illness. J. Allergy Clin. Immunol. Pract. 2021, 9, 1152–1162. [Google Scholar] [CrossRef]
- Mateos González, M.; Sierra Gonzalo, E.; Casado Lopez, I.; Arnalich Fernández, F.; Beato Pérez, J.L.; Monge Monge, D.; Vargas Núñez, J.A.; García Fenoll, R.; Suárez Fernández, C.; Freire Castro, S.J.; et al. The Prognostic Value of Eosinophil Recovery in COVID-19: A Multicentre, Retrospective Cohort Study on Patients Hospitalised in Spanish Hospitals. J. Clin. Med. 2021, 10, 305. [Google Scholar] [CrossRef]
- Liu, F.; Xu, A.; Zhang, Y.; Xuan, W.; Yan, T.; Pan, K.; Yu, W.; Zhang, J. Patients of COVID-19 may benefit from sustained Lopinavir-combined regimen and the increase of Eosinophil may predict the outcome of COVID-19 progression. Int. J. Infect. Dis. 2020, 95, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Glickman, J.W.; Pavel, A.B.; Guttman-Yassky, E.; Miller, R.L. The role of circulating eosinophils on COVID-19 mortality varies by race/ethnicity. J. Allergy Clin. Immunol. 2021, 76, 925–927. [Google Scholar] [CrossRef]
- Xie, G.; Ding, F.; Han, L.; Yin, D.; Lu, H.; Zhang, M. The role of peripheral blood eosinophil counts in COVID-19 patients. Allergy Eur. J. Allergy Clin. Immunol. 2021, 76, 471–482. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, X.; Liu, X.; Sun, W.; Zhou, L.; Wang, Y.; Sui, H. Clinical Characteristics and Eosinophils in Young SARS-CoV-2-Positive Chinese Travelers Returning to Shanghai. Front. Public Health 2020, 8, 368. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, Z.; Liu, S.; Gong, C.; Chen, L.; Ai, G.; Zhu, X.; Zhang, C.; Li, D. Absolute Eosinophil Count Predicts Intensive Care Unit Transfer Among Elderly COVID-19 Patients From General Isolation Wards. Front. Med. 2020, 7, 585222. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulou, V.E.; Garmpis, N.; Damaskos, C.; Valsami, S.; Dimitroulis, D.; Diamantis, E.; Farmaki, P.; Papageorgiou, C.V.; Makrodimitri, S.; Gravvanis, N.; et al. The impact of peripheral eosinophil counts and eosinophil to lymphocyte ratio (ELR) in the clinical course of covid-19 patients: A retrospective study. In Vivo 2021, 35, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhou, J.; Zhou, Q.; Hu, L.; Long, Y. Role of eosinophils in the diagnosis and prognostic evaluation of COVID-19. J. Med. Virol. 2021, 93, 1105–1110. [Google Scholar] [CrossRef]
- Mu, T.; Yi, Z.; Wang, M.; Wang, J.; Zhang, C.; Chen, H.; Bai, M.; Jiang, L.; Zhang, Y. Expression of eosinophil in peripheral blood of patients with COVID-19 and its clinical significance. J. Clin. Lab. Anal. 2021, 35, e23620. [Google Scholar] [CrossRef]
- Yan, B.; Yang, J.; Xie, Y.; Tang, X. Relationship between blood eosinophil levels and COVID-19 mortality. World Allergy Organ. J. 2021, 14, 100521. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Roncati, L.; Nasillo, V.; Lusenti, B.; Riva, G. Signals of Th2 immune response from COVID-19 patients requiring intensive care. Ann. Hematol. 2020, 99, 1419–1420. [Google Scholar] [CrossRef]
- Rodriguez, L.; Pekkarinen, P.T.; Lakshmikanth, T.; Tan, Z.; Consiglio, C.R.; Pou, C.; Chen, Y.; Mugabo, C.H.; Nguyen, N.A.; Nowlan, K.; et al. Systems-Level Immunomonitoring from Acute to Recovery Phase of Severe COVID-19. Cell Rep. Med. 2020, 1, 100078. [Google Scholar] [CrossRef]
- Pala, D.; Pistis, M. Anti-IL5 Drugs in COVID-19 Patients: Role of Eosinophils in SARS-CoV-2-Induced Immunopathology. Front. Pharmacol. 2021, 12, 622554. [Google Scholar] [CrossRef]
- Gan, J.; Li, J.; Li, S.; Yang, C. Leucocyte Subsets Effectively Predict the Clinical Outcome of Patients With COVID-19 Pneumonia: A Retrospective Case-Control Study. Front. Public Health 2020, 8, 299. [Google Scholar] [CrossRef]
- Soni, M. Evaluation of eosinopenia as a diagnostic and prognostic indicator in COVID-19 infection. Int. J. Lab. Hematol. 2020. [Google Scholar] [CrossRef]
- Djangang, N.N.; Peluso, L.; Talamonti, M.; Izzi, A.; Gevenois, P.A.; Garufi, A.; Goffard, J.C.; Henrard, S.; Severgnini, P.; Vincent, J.L.; et al. Eosinopenia in COVID-19 patients: A retrospective analysis. Microorganisms 2020, 8, 1929. [Google Scholar] [CrossRef]
- Vial, R.; Gully, M.; Bobot, M.; Scarfoglière, V.; Brunet, P.; Bouchouareb, D.; Duval, A.; Zino, H.-O.; Faraut, J.; Jehel, O.; et al. Triage of Patients Suspected of COVID-19 in Chronic Hemodialysis: Eosinophil Count Differentiates Low and High Suspicion of COVID-19. J. Clin. Med. 2020, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Outh, R.; Boutin, C.; Gueudet, P.; Suzuki, M.; Saada, M.; Aumaître, H. Eosinopenia <100/μL as a marker of active COVID-19: An observational prospective study. J. Microbiol. Immunol. Infect. 2021, 54, 61–68. [Google Scholar] [CrossRef]
- Lippi, G.; Sanchis-Gomar, F.; Henry, B.M. Response to: Eosinophil count in coronavirus disease 2019: More doubts than answers. QJM 2021, 114, 70–71. [Google Scholar] [CrossRef] [PubMed]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Barroso, B.; Valverde-Monge, M.; Cañas Jose, A.; Rodrigo-Muñoz, J.M.; Gonzalez-Cano, B.; Villalobos-Violan, V.; Betancor, D.; Gomez-Cardeñosa, A.; Vallejo-Chamorro, G.; Baptista-Serna, L.; et al. Prevalence, characteristics, and outcome of asthmatic patients with type 2 diseases in hospitalized patients with COVID-19 in Madrid, Spain. J. Investig. Allergol. Clin. Immunol. 2020, 30, 382–384. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.J.; Busse, W.W.; Bacharier, L.B.; Kattan, M.; O’Connor, G.T.; Wood, R.A.; Visness, C.M.; Durham, S.R.; Larson, D.; Esnault, S.; et al. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J. Allergy Clin. Immunol. 2020, 146, 203–206. [Google Scholar] [CrossRef]
- Camiolo, M.; Gauthier, M.; Kaminski, N.; Ray, A.; Wenzel, S.E. Expression of SARS-CoV-2 receptor ACE2 and coincident host response signature varies by asthma inflammatory phenotype. J. Allergy Clin. Immunol. 2020, 146, 315–324. [Google Scholar] [CrossRef]
- Hanon, S.; Brusselle, G.; Deschampheleire, M.; Louis, R.; Michils, A.; Peché, R.; Pilette, C.; Rummens, P.; Schuermans, D.; Simonis, H.; et al. COVID-19 and biologics in severe asthma: Data from the Belgian Severe Asthma Registry. Eur. Respir. J. 2020, 56, 2002857. [Google Scholar] [CrossRef] [PubMed]
- Rial, M.J.; Valverde, M.; del Pozo, V.; González-Barcala, F.J.; Martínez-Rivera, C.; Muñoz, X.; Olaguibel, J.M.; Plaza, V.; Curto, E.; Quirce, S.; et al. Clinical characteristics in 545 patients with severe asthma on biological treatment during the COVID-19 outbreak. J. Allergy Clin. Immunol. Pract. 2021, 9, 487–489. [Google Scholar] [CrossRef]
- Heffler, E.; Detoraki, A.; Contoli, M.; Papi, A.; Paoletti, G.; Malipiero, G.; Brussino, L.; Crimi, C.; Morrone, D.; Padovani, M.; et al. COVID-19 in Severe Asthma Network in Italy (SANI) patients: Clinical features, impact of comorbidities and treatments. Allergy 2021, 76, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Eger, K.; Hashimoto, S.; Braunstahl, G.J.; Ten Brinke, A.; Patberg, K.W.; Beukert, A.; Smeenk, F.; van der Sar–van der Brugge, S.; Weersink, E.J.M.; Bel, E.H. Poor outcome of SARS-CoV-2 infection in patients with severe asthma on biologic therapy. Respir. Med. 2021, 177, 106287. [Google Scholar] [CrossRef]
- García-Moguel, I.; Díaz Campos, R.; Alonso Charterina, S.; Fernández Rodríguez, C.; Fernández Crespo, J. COVID-19, severe asthma, and biologics. Ann. Allergy, Asthma Immunol. 2020, 125, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Renner, A.; Marth, K.; Patocka, K.; Idzko, M.; Pohl, W. COVID-19 in two severe asthmatics receiving benralizumab: Busting the eosinophilia myth. ERJ Open Res. 2020, 6, 00457–02020. [Google Scholar] [CrossRef] [PubMed]
- Renner, A.; Marth, K.; Patocka, K.; Pohl, W. COVID-19 in a severe eosinophilic asthmatic receiving benralizumab–a case study. J. Asthma 2020, 1–3. [Google Scholar] [CrossRef]
- Roca, E.; Ventura, L.; Zattra, C.M.; Lombardi, C. EOSINOPENIA: An early, effective and relevant COVID-19 biomarker? QJM 2021, 114, 68–69. [Google Scholar] [CrossRef] [PubMed]
- Bolles, M.; Deming, D.; Long, K.; Agnihothram, S.; Whitmore, A.; Ferris, M.; Funkhouser, W.; Gralinski, L.; Totura, A.; Heise, M.; et al. A Double-Inactivated Severe Acute Respiratory Syndrome Coronavirus Vaccine Provides Incomplete Protection in Mice and Induces Increased Eosinophilic Proinflammatory Pulmonary Response upon Challenge. J. Virol. 2011, 85, 12201–12215. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.T.; Sbrana, E.; Iwata-Yoshikawa, N.; Newman, P.C.; Garron, T.; Atmar, R.L.; Peters, C.J.; Couch, R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS ONE 2012, 7, e35421. [Google Scholar] [CrossRef]
- Honda-Okubo, Y.; Barnard, D.; Ong, C.H.; Peng, B.-H.; Tseng, C.-T.K.; Petrovsky, N. Severe Acute Respiratory Syndrome-Associated Coronavirus Vaccines Formulated with Delta Inulin Adjuvants Provide Enhanced Protection while Ameliorating Lung Eosinophilic Immunopathology. J. Virol. 2015, 89, 2995–3007. [Google Scholar] [CrossRef]
- Iwata-Yoshikawa, N.; Uda, A.; Suzuki, T.; Tsunetsugu-Yokota, Y.; Sato, Y.; Morikawa, S.; Tashiro, M.; Sata, T.; Hasegawa, H.; Nagata, N. Effects of Toll-Like Receptor Stimulation on Eosinophilic Infiltration in Lungs of BALB/c Mice Immunized with UV-Inactivated Severe Acute Respiratory Syndrome-Related Coronavirus Vaccine. J. Virol. 2014, 88, 8597–8614. [Google Scholar] [CrossRef]
- Rosenberg, H.F.; Foster, P.S. Eosinophils and COVID-19: Diagnosis, prognosis, and vaccination strategies. Semin. Immunopathol. 2021, 1–10. [Google Scholar] [CrossRef]
- Kubo, H. Extracellular Vesicles in Lung Disease. Chest 2018, 153, 210–216. [Google Scholar] [CrossRef]
- Akuthota, P.; Carmo, L.A.S.; Bonjour, K.; Murphy, R.O.; Silva, T.P.; Gamalier, J.P.; Capron, K.L.; Tigges, J.; Toxavidis, V.; Camacho, V.; et al. Extracellular Microvesicle Production by Human Eosinophils Activated by “Inflammatory” Stimuli. Front. Cell Dev. Biol. 2016, 4, 117. [Google Scholar] [CrossRef] [PubMed]
- Cañas, J.A.; Sastre, B.; Mazzeo, C.; Fernández-Nieto, M.; Rodrigo-Muñoz, J.M.; González-Guerra, A.; Izquierdo, M.; Barranco, P.; Quirce, S.; Sastre, J.; et al. Exosomes from eosinophils autoregulate and promote eosinophil functions. J. Leukoc. Biol. 2017, 101, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Cañas, J.A.; Sastre, B.; Rodrigo-Muñoz, J.M.; Fernández-Nieto, M.; Barranco, P.; Quirce, S.; Sastre, J.; del Pozo, V. Eosinophil-derived exosomes contribute to asthma remodelling by activating structural lung cells. Clin. Exp. Allergy 2018, 48, 1173–1185. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo-Muñoz, J.M.; Cañas, J.A.; Sastre, B.; Rego, N.; Greif, G.; Rial, M.; Mínguez, P.; Mahíllo-Fernández, I.; Fernández-Nieto, M.; Mora, I.; et al. Asthma diagnosis using integrated analysis of eosinophil microRNAs. Allergy Eur. J. Allergy Clin. Immunol. 2019, 74, 507–517. [Google Scholar] [CrossRef]
- Rodrigo-Muñoz, J.M.; Rial, M.J.; Sastre, B.; Cañas, J.A.; Mahíllo-Fernández, I.; Quirce, S.; Sastre, J.; Cosío, B.G.; Del Pozo, V. Circulating miRNAs as diagnostic tool for discrimination of respiratory disease: Asthma, asthma-chronic obstructive pulmonary disease (COPD) overlap and COPD. Allergy 2019, 74, 2491–2494. [Google Scholar] [CrossRef]
- Jacobsen, E.A.; Jackson, D.J.; Heffler, E.; Mathur, S.K.; Bredenoord, A.J.; Pavord, I.D.; Akuthota, P.; Roufosse, F.; Rothenberg, M.E. Eosinophil Knockout Humans: Uncovering the Role of Eosinophils Through Eosinophil-Directed Biological Therapies. Annu. Rev. Immunol. 2021, 39, 719–757. [Google Scholar] [CrossRef] [PubMed]
- Nussbaum, J.C.; Van Dyken, S.J.; Von Moltke, J.; Cheng, L.E.; Mohapatra, A.; Molofsky, A.B.; Thornton, E.E.; Krummel, M.F.; Chawla, A.; Liang, H.E.; et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 2013, 502, 245–248. [Google Scholar] [CrossRef]
- Jung, Y.; Wen, T.; Mingler, M.K.; Caldwell, J.M.; Wang, Y.H.; Chaplin, D.D.; Lee, E.H.; Jang, M.H.; Woo, S.Y.; Seoh, J.Y.; et al. IL-1β in eosinophil-mediated small intestinal homeostasis and IgA production. Mucosal Immunol. 2015, 8, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, R.; Lee, E.J.; Jang, M.S.; Jeun, E.J.; Hong, C.P.; Kim, J.H.; Park, A.; Yun, C.H.; Hong, S.W.; Kim, Y.M.; et al. Small intestinal eosinophils regulate Th17 cells by producing IL-1 receptor antagonist. J. Exp. Med. 2016, 213, 555–567. [Google Scholar] [CrossRef]
- Chu, V.T.; Beller, A.; Rausch, S.; Strandmark, J.; Zänker, M.; Arbach, O.; Kruglov, A.; Berek, C. Eosinophils promote generation and maintenance of immunoglobulin-A-expressing plasma cells and contribute to gut immune homeostasis. Immunity 2014, 40, 582–593. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Sun, A.H.; Ojcius, D.M.; Hu, W.L.; Ge, Y.M.; Lin, X.; Li, L.J.; Pan, J.P.; Yan, J. Eosinophils from murine lamina propria induce differentiation of naïve T cells into regulatory T cells via TGF-β1 and retinoic acid. PLoS ONE 2015, 10, e0142881. [Google Scholar] [CrossRef]
- Gomez Torrijos, E.; Gonzalez-Mendiola, R.; Alvarado, M.; Avila, R.; Prieto-Garcia, A.; Valbuena, T.; Borja, J.; Infante, S.; Lopez, M.P.; Marchan, E.; et al. Eosinophilic Esophagitis: Review and Update. Front. Med. 2018, 5, 247. [Google Scholar] [CrossRef]
- Gouon-Evans, V.; Lin, E.Y.; Pollard, J.W. Requirement of macrophages and eosinophils and their cytokines/chemokines for mammary gland development. Breast Cancer Res. 2002, 4, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Aupperlee, M.D.; Zhao, Y.; Tan, Y.S.; Leipprandt, J.R.; Bennett, J.; Haslam, S.Z.; Schwartz, R.C. Epidermal growth factor receptor (EGFR) signaling is a key mediator of hormone-induced leukocyte infiltration in the pubertal female mammary gland. Endocrinology 2014, 155, 2301–2313. [Google Scholar] [CrossRef]
- Vicetti Miguel, R.D.; Quispe Calla, N.E.; Dixon, D.; Foster, R.A.; Gambotto, A.; Pavelko, S.D.; Hall-Stoodley, L.; Cherpes, T.L. IL-4–secreting eosinophils promote endometrial stromal cell proliferation and prevent Chlamydia-induced upper genital tract damage. Proc. Natl. Acad. Sci. USA 2017, 114, E6892–E6901. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Molofsky, A.B.; Liang, H.E.; Ricardo-Gonzalez, R.R.; Jouihan, H.A.; Bando, J.K.; Chawla, A.; Locksley, R.M. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science 2011, 332, 243–247. [Google Scholar] [CrossRef]
- Qiu, Y.; Nguyen, K.D.; Odegaard, J.I.; Cui, X.; Tian, X.; Locksley, R.M.; Palmiter, R.D.; Chawla, A. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 2014, 157, 1292–1308. [Google Scholar] [CrossRef]
- Fabbiano, S.; Suárez-Zamorano, N.; Rigo, D.; Veyrat-Durebex, C.; Stevanovic Dokic, A.; Colin, D.J.; Trajkovski, M. Caloric Restriction Leads to Browning of White Adipose Tissue through Type 2 Immune Signaling. Cell Metab. 2016, 24, 434–446. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, P.; Cui, R.; Zhang, M.; Li, H.; Qian, C.; Sheng, C.; Qu, S.; Bu, L. Eosinophils Reduce Chronic Inflammation in Adipose Tissue by Secreting Th2 Cytokines and Promoting M2 Macrophages Polarization. Int. J. Endocrinol. 2015, 2015, 565760. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhong, L.; Lee, J.T.H.; Zhang, J.; Wu, D.; Geng, L.; Wang, Y.; Wong, C.M.; Xu, A. The FGF21-CCL11 Axis Mediates Beiging of White Adipose Tissues by Coupling Sympathetic Nervous System to Type 2 Immunity. Cell Metab. 2017, 26, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Alonzo, E.S.; Dorothee, G.; Pollard, J.W.; Sant’Angelo, D.B. Selective depletion of eosinophils or neutrophils in mice impacts the efficiency of apoptotic cell clearance in the thymus. PLoS ONE 2010, 5, e11439. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-B.; Ghiran, I.; Matthaei, K.; Weller, P.F. Airway Eosinophils: Allergic Inflammation Recruited Professional Antigen-Presenting Cells. J. Immunol. 2007, 179, 7585–7592. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, E.A.; Ochkur, S.I.; Pero, R.S.; Taranova, A.G.; Protheroe, C.A.; Colbert, D.C.; Lee, N.A.; Lee, J.J. Allergic pulmonary inflammation in mice is dependent on eosinophil-induced recruitment of effector T cells. J. Exp. Med. 2008, 205, 699–710. [Google Scholar] [CrossRef]
- Spencer, L.A.; Szela, C.T.; Perez, S.A.C.; Kirchhoffer, C.L.; Neves, J.S.; Radke, A.L.; Weller, P.F. Human eosinophils constitutively express multiple Th1, Th2, and immunoregulatory cytokines that are secreted rapidly and differentially. J. Leukoc. Biol. 2008, 85, 117–123. [Google Scholar] [CrossRef]
- Marc-Malovrh, M.; Camlek, L.; Škrgat, S.; Kern, I.; Fležar, M.; Dežman, M.; Korošec, P. Elevated eosinophils, IL5 and IL8 in induced sputum in asthma patients with accelerated FEV1 decline. Respir. Med. 2020, 162, 105875. [Google Scholar] [CrossRef]
- Willebrand, R.; Voehringer, D. IL-33-Induced Cytokine Secretion and Survival of Mouse Eosinophils Is Promoted by Autocrine GM-CSF. PLoS ONE 2016, 11, e0163751. [Google Scholar] [CrossRef]
- Barretto, K.T.; Swanson, C.M.; Nguyen, C.L.; Annis, D.S.; Esnault, S.J.; Mosher, D.F.; Johansson, M.W. Control of cytokine-driven eosinophil migratory behavior by TGF-beta-induced protein (TGFBI) and periostin. PLoS ONE 2018, 13, e0201320. [Google Scholar] [CrossRef] [PubMed]
- Heredia, J.E.; Mukundan, L.; Chen, F.M.; Mueller, A.A.; Deo, R.C.; Locksley, R.M.; Rando, T.A.; Chawla, A. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell 2013, 153, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Goh, Y.P.S.; Henderson, N.C.; Heredia, J.E.; Eagle, A.R.; Odegaard, J.I.; Lehwald, N.; Nguyen, K.D.; Sheppard, D.; Mukundan, L.; Locksley, R.M.; et al. Eosinophils secrete IL-4 to facilitate liver regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 9914–9919. [Google Scholar] [CrossRef] [PubMed]
- Reichman, H.; Moshkovits, I.; Itan, M.; Pasmanik-Chor, M.; Vogl, T.; Roth, J.; Munitz, A. Transcriptome profiling of mouse colonic eosinophils reveals a key role for eosinophils in the induction of s100a8 and s100a9 in mucosal healing. Sci. Rep. 2017, 7, 7117. [Google Scholar] [CrossRef]
- Puxeddu, I.; Berkman, N.; Ribatti, D.; Bader, R.; Haitchi, H.M.; Davies, D.E.; Howarth, P.H.; Levi-Schaffer, F. Osteopontin is expressed and functional in human eosinophils. Allergy Eur. J. Allergy Clin. Immunol. 2010, 65, 168–174. [Google Scholar] [CrossRef]
- Nhu, Q.M.; Aceves, S.S. Tissue Remodeling in Chronic Eosinophilic Esophageal Inflammation: Parallels in Asthma and Therapeutic Perspectives. Front. Med. 2017, 4, 128. [Google Scholar] [CrossRef]
- Abdala-Valencia, H.; Coden, M.E.; Chiarella, S.E.; Jacobsen, E.A.; Bochner, B.S.; Lee, J.J.; Berdnikovs, S. Shaping eosinophil identity in the tissue contexts of development, homeostasis, and disease. J. Leukoc. Biol. 2018, 104, 95–108. [Google Scholar] [CrossRef]
- Fulkerson, P.C.; Rothenberg, M.E. Eosinophil Development, Disease Involvement, and Therapeutic Suppression. Adv. Immunol. 2018, 138, 1–34. [Google Scholar] [PubMed]
- Marichal, T.; Mesnil, C.; Bureau, F. Homeostatic Eosinophils: Characteristics and Functions. Front. Med. 2017, 4, 101. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Dyer, K.D.; Foster, P.S. Eosinophils: Changing perspectives in health and disease. Nat. Rev. Immunol. 2013, 13, 9–22. [Google Scholar] [CrossRef]
- Chu, D.K.; Jimenez-Saiz, R.; Verschoor, C.P.; Walker, T.D.; Goncharova, S.; Llop-Guevara, A.; Shen, P.; Gordon, M.E.; Barra, N.G.; Bassett, J.D.; et al. Indigenous enteric eosinophils control DCs to initiate a primary Th2 immune response in vivo. J. Exp. Med. 2014, 211, 1657–1672. [Google Scholar] [CrossRef] [PubMed]
- Abdala Valencia, H.; Loffredo, L.F.; Misharin, A.V.; Berdnikovs, S. Phenotypic plasticity and targeting of Siglec-FhighCD11clow eosinophils to the airway in a murine model of asthma. Allergy Eur. J. Allergy Clin. Immunol. 2016, 71, 267–271. [Google Scholar] [CrossRef]
- Aoki, A.; Hirahara, K.; Kiuchi, M.; Nakayama, T. Eosinophils: Cells known for over 140 years with broad and new functions. Allergol. Int. 2021, 70, 3–8. [Google Scholar] [CrossRef]
- Mesnil, C.; Raulier, S.; Paulissen, G.; Xiao, X.; Birrell, M.A.; Pirottin, D.; Janss, T.; Starkl, P.; Ramery, E.; Henket, M.; et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J. Clin. Investig. 2016, 126, 3279–3295. [Google Scholar] [CrossRef]
- Ogawa, M.; Ishihara, T.; Isobe, Y.; Kato, T.; Kuba, K.; Imai, Y.; Uchino, Y.; Tsubota, K.; Arita, M. Eosinophils promote corneal wound healing via the 12/15-lipoxygenase pathway. FASEB J. 2020, 34, 12492–12501. [Google Scholar] [CrossRef]
- Brigger, D.; Riether, C.; van Brummelen, R.; Mosher, K.I.; Shiu, A.; Ding, Z.; Zbären, N.; Gasser, P.; Guntern, P.; Yousef, H.; et al. Eosinophils regulate adipose tissue inflammation and sustain physical and immunological fitness in old age. Nat. Metab. 2020, 2, 688–702. [Google Scholar] [CrossRef]
- Kuang, F.L. Approach to Patients with Eosinophilia. Med. Clin. N. Am. 2020, 104, 1–14. [Google Scholar] [CrossRef]
- Khoury, P.; Akuthota, P.; Ackerman, S.J.; Arron, J.R.; Bochner, B.S.; Collins, M.H.; Kahn, J.E.; Fulkerson, P.C.; Gleich, G.J.; Gopal-Srivastava, R.; et al. Revisiting the NIH Taskforce on the Research needs of Eosinophil-Associated Diseases (RE-TREAD). J. Leukoc. Biol. 2018, 104, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Dellon, E.S.; Liacouras, C.A.; Molina-Infante, J.; Furuta, G.T.; Spergel, J.M.; Zevit, N.; Spechler, S.J.; Attwood, S.E.; Straumann, A.; Aceves, S.S.; et al. Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference. Gastroenterology 2018, 155, 1022–1033.e10. [Google Scholar] [CrossRef] [PubMed]
- Lwin, T.; Melton, S.D.; Genta, R.M. Eosinophilic gastritis: Histopathological characterization and quantification of the normal gastric eosinophil content. Mod. Pathol. 2011, 24, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Dellon, E.S.; Hirano, I. Epidemiology and Natural History of Eosinophilic Esophagitis. Gastroenterology 2018, 154, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Lyles, J.; Rothenberg, M. Role of genetics, environment, and their interactions in the pathogenesis of eosinophilic esophagitis. Curr. Opin. Immunol. 2019, 60, 46–53. [Google Scholar] [CrossRef]
- Cañas, J.A.; Tabares, A.; Barbero, C.; García-Sánchez, D.; Sastre, B.; Rodrigo-Muñoz, J.M.; Mahíllo-Fernández, I.; Rayo, A.; Borrell, B.; Cilleruelo, M.L.; et al. Proton-pump Inhibitor Response Prediction Using Esophageal microRNAs in Children With Eosinophilic Esophagitis. J. Pediatric Gastroenterol. Nutr. 2020, 71, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Manohar, M.; Kandikattu, H.K.; Upparahalli Venkateshaiah, S.; Yadavalli, C.S.; Mishra, A. Eosinophils in the pathogenesis of pancreatic disorders. Semin. Immunopathol. 2021, 1–12. [Google Scholar] [CrossRef]
- Manohar, M.; Verma, A.K.; Venkateshaiah, S.U.; Sanders, N.L.; Mishra, A. Pathogenic mechanisms of pancreatitis. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 10–25. [Google Scholar] [CrossRef]
- Kakodkar, S.; Omar, H.; Cabrera, J.; Chi, K. Eosinophilic Pancreatitis Diagnosed With Endoscopic Ultrasound. ACG Case Rep. J. 2015, 2, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Bastid, C.; Sahel, J.; Choux, R.; Payan, M.J.; Sarles, H. Eosinophilic pancreatitis: Report of a case. Pancreas 1990, 5, 104–107. [Google Scholar] [CrossRef]
- Grisaru-Tal, S.; Itan, M.; Klion, A.D.; Munitz, A. A new dawn for eosinophils in the tumour microenvironment. Nat. Rev. Cancer 2020, 20, 594–607. [Google Scholar] [CrossRef] [PubMed]
- Reichman, H.; Itan, M.; Rozenberg, P.; Yarmolovski, T.; Brazowski, E.; Varol, C.; Gluck, N.; Shapira, S.; Arber, N.; Qimron, U.; et al. Activated eosinophils exert antitumorigenic activities in colorectal cancer. Cancer Immunol. Res. 2019, 7, 388–400. [Google Scholar] [CrossRef]
- Carretero, R.; Sektioglu, I.M.; Garbi, N.; Salgado, O.C.; Beckhove, P.; Hämmerling, G.J. Eosinophils orchestrate cancer rejection by normalizing tumor vessels and enhancing infiltration of CD8 + T cells. Nat. Immunol. 2015, 16, 609–617. [Google Scholar] [CrossRef]
- Hollande, C.; Boussier, J.; Ziai, J.; Nozawa, T.; Bondet, V.; Phung, W.; Lu, B.; Duffy, D.; Paradis, V.; Mallet, V.; et al. Inhibition of the dipeptidyl peptidase DPP4 (CD26) reveals IL-33-dependent eosinophil-mediated control of tumor growth. Nat. Immunol. 2019, 20, 257–264. [Google Scholar] [CrossRef]
- D’Angelo, G.; Hotz, A.M.; Todeschin, P. Acute lymphoblastic leukemia with hypereosinophilia and 9p21 deletion: Case report and review of the literature. Lab. Hematol. 2008, 14, 7–9. [Google Scholar] [CrossRef]
- Sahu, K.K.; Malhotra, P.; Khadwal, A.; Sachdeva, M.S.; Sharma, P.; Varma, N.; Varma, S.C. Hypereosinophilia in Acute Lymphoblastic Leukemia: Two Cases with Review of Literature. Indian J. Hematol. Blood Transfus. 2015, 31, 460–465. [Google Scholar] [CrossRef]
- Diny, N.L.; Rose, N.R.; Čiháková, D. Eosinophils in Autoimmune Diseases. Front. Immunol. 2017, 8, 484. [Google Scholar] [CrossRef] [PubMed]
- Hallam, C.; Pritchard, D.I.; Trigg, S.; Eady, R.P. Rat eosinophil-mediated antibody-dependent cellular cytotoxicity: Investigations of the mechanisms of target cell lysis and inhibition by glucocorticoids. Clin. Exp. Immunol. 1982, 48, 641–648. [Google Scholar]
- Noguchi, H.; Kephart, G.M.; Colby, T.V.; Gleich, G.J. Tissue eosinophilia and eosinophil degranulation in syndromes associated with fibrosis. Am. J. Pathol. 1992, 140, 521–528. [Google Scholar] [PubMed]
- del Pozo, V.; de Andrés, B.; Martín, E.; Cárdaba, B.; Fernández, J.C.; Gallardo, S.; Tramón, P.; Palomino, P.; Lahoz, C.; Leyva-Cobian, F. Eosinophil as antigen-presenting cell: Activation of T cell clones and T cell hybridoma by eosinophils after antigen processing. Eur. J. Immunol. 1992, 22, 1919–1925. [Google Scholar] [CrossRef]
- Bernard, P.; Venot, J.; Constant, F.; Bonnetblane, J.M. Blood eosinophilia as a severity marker for bullous pemphigoid. J. Am. Acad. Dermatol. 1987, 16, 879–881. [Google Scholar] [CrossRef]
- Tsukadaira, A. Eosinophil active cytokines and surface analysis of eosinophils in churg-strauss syndrome. Allergy Asthma Proc. 1999, 20, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Loktionov, A. Eosinophils in the gastrointestinal tract and their role in the pathogenesis of major colorectal disorders. World J. Gastroenterol. 2019, 25, 3503–3526. [Google Scholar] [CrossRef] [PubMed]
- Carey, E.J.; Ali, A.H.; Lindor, K.D. Primary biliary cirrhosis. Lancet 2015, 386, 1565–1575. [Google Scholar] [CrossRef]
- Kang, E.G.; Narayana, P.K.; Pouliquen, I.J.; Lopez, M.C.; Ferreira-Cornwell, M.C.; Getsy, J.A. Efficacy and safety of mepolizumab administered subcutaneously for moderate to severe atopic dermatitis. Allergy 2020, 75, 950–953. [Google Scholar] [CrossRef]
- Fokkens, W.J.; Lund, V.J.; Hopkins, C.; Hellings, P.W.; Kern, R.; Reitsma, S.; Toppila-Salmi, S.; Bernal-Sprekelsen, M.; Mullol, J.; Alobid, I.; et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2020. Rhinology 2020, 58, 1–464. [Google Scholar] [CrossRef]
- Simon, H.U.; Yousefi, S.; Schranz, C.; Schapowal, A.; Bachert, C.; Blaser, K. Direct demonstration of delayed eosinophil apoptosis as a mechanism causing tissue eosinophilia. J. Immunol. 1997, 158, 3902–3908. [Google Scholar] [PubMed]
- Bochner, B.S.; Stevens, W.W. Biology and function of eosinophils in chronic rhinosinusitis with or without nasal polyps. Allergy Asthma Immunol. Res. 2021, 13, 8–22. [Google Scholar] [CrossRef]
- Tay, T.R.; Radhakrishna, N.; Hore-Lacy, F.; Smith, C.; Hoy, R.; Dabscheck, E.; Hew, M. Comorbidities in difficult asthma are independent risk factors for frequent exacerbations, poor control and diminished quality of life. Respirology 2016, 21, 1384–1390. [Google Scholar] [CrossRef]
- Denlinger, L.C.; Phillips, B.R.; Ramratnam, S.; Ross, K.; Bhakta, N.R.; Cardet, J.C.; Castro, M.; Peters, S.P.; Phipatanakul, W.; Aujla, S.; et al. Inflammatory and comorbid features of patients with severe asthma and frequent exacerbations. Am. J. Respir. Crit. Care Med. 2017, 195, 302–313. [Google Scholar] [CrossRef]
- Ponikau, J.U.; Sherris, D.A.; Kephart, G.M.; Kern, E.B.; Gaffey, T.A.; Tarara, J.E.; Kita, H. Features of airway remodeling and eosinophilic inflammation in chronic rhinosinusitis: Is the histopathology similar to asthma? J. Allergy Clin. Immunol. 2003, 112, 877–882. [Google Scholar] [CrossRef]
- Fahy, J.V. Type 2 inflammation in asthma-present in most, absent in many. Nat. Rev. Immunol. 2015, 15, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Wardlaw, A.J.; Dunnette, S.; Gleich, G.J.; Collins, J.V.; Kay, A.B. Eosinophils and mast cells in bronchoalveolar lavage in subjects with mild asthma. Relationship to bronchial hyperreactivity. Am. Rev. Respir. Dis. 1988, 137, 62–69. [Google Scholar] [CrossRef]
- Wardlaw, A.L. Molecular basis for selective eosinophil trafficking in asthma: A multistep paradigm. J. Allergy Clin. Immunol. 1999, 104, 917–926. [Google Scholar] [CrossRef]
- Nakagome, K.; Nagata, M. Involvement and Possible Role of Eosinophils in Asthma Exacerbation. Front. Immunol. 2018, 9, 2220. [Google Scholar] [CrossRef] [PubMed]
- Walsh, G.M. Eosinophil apoptosis and clearance in asthma. J. Cell Death 2013, 6, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.R.F.; Moqbel, R.; Walsh, G.M.; Kay, A.B.; Wardlaw, A.J. Adhesion to fibronectin prolongs eosinophil survival. J. Exp. Med. 1993, 177, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Green, R.H.; Brightling, C.E.; McKenna, S.; Hargadon, B.; Parker, D.; Bradding, P.; Wardlaw, A.J.; Pavord, I.D. Asthma exacerbations and sputum eosinophil counts: A randomised controlled trial. Lancet 2002, 360, 1715–1721. [Google Scholar] [CrossRef]
- Demarche, S.F.; Schleich, F.N.; Paulus, V.A.; Henket, M.A.; Van Hees, T.J.; Louis, R.E. Asthma Control and Sputum Eosinophils: A Longitudinal Study in Daily Practice. J. Allergy Clin. Immunol. Pract. 2017, 5, 1335–1343. [Google Scholar] [CrossRef]
- Hanania, N.A.; Alpan, O.; Hamilos, D.L.; Condemi, J.J.; Reyes-Rivera, I.; Zhu, J.; Rosen, K.E.; Eisner, M.D.; Wong, D.A.; Busse, W. Omalizumab in severe allergic asthma inadequately controlled with standard therapy: A randomized trial. Ann. Intern. Med. 2011, 154, 573–582. [Google Scholar] [CrossRef]
- Szefler, S.J.; Wenzel, S.; Brown, R.; Erzurum, S.C.; Fahy, J.V.; Hamilton, R.G.; Hunt, J.F.; Kita, H.; Liu, A.H.; Panettieri, R.A.; et al. Asthma outcomes: Biomarkers. J. Allergy Clin. Immunol. 2012, 129, S9–S23. [Google Scholar] [CrossRef] [PubMed]
- Ortega, H.; Katz, L.; Gunsoy, N.; Keene, O.; Yancey, S. Blood eosinophil counts predict treatment response in patients with severe eosinophilic asthma. J. Allergy Clin. Immunol. 2015, 136, 825–826. [Google Scholar] [CrossRef]
- Zeiger, R.S.; Schatz, M.; Dalal, A.A.; Chen, W.; Sadikova, E.; Suruki, R.Y.; Kawatkar, A.A.; Qian, L. Blood Eosinophil Count and Outcomes in Severe Uncontrolled Asthma: A Prospective Study. J. Allergy Clin. Immunol. Pract. 2017, 5, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Kaner, R.J.; Guiot, J.; Maher, T.M.; Tomassetti, S.; Moiseev, S.; Kuwana, M.; Brown, K.K. Diagnostic and Prognostic Biomarkers for Chronic Fibrosing Interstitial Lung Diseases With a Progressive Phenotype. Chest 2020, 158, 646–659. [Google Scholar] [CrossRef]
- Travis, W.D.; Costabel, U.; Hansell, D.M.; King, T.E.; Lynch, D.A.; Nicholson, A.G.; Ryerson, C.J.; Ryu, J.H.; Selman, M.; Wells, A.U.; et al. An official American Thoracic Society/European Respiratory Society statement: Update of the international multidisciplinary classification of the idiopathic interstitial pneumonias. Am. J. Respir. Crit. Care Med. 2013, 188, 733–748. [Google Scholar] [CrossRef]
- Akuthota, P.; Weller, P.F. Eosinophilic pneumonias. Clin. Microbiol. Rev. 2012, 25, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Guiot, J.; Moermans, C.; Henket, M.; Corhay, J.L.; Louis, R. Blood Biomarkers in Idiopathic Pulmonary Fibrosis. Lung 2017, 195, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Kuang, F.L.; Klion, A.D. Biologic Agents for the Treatment of Hypereosinophilic Syndromes. J. Allergy Clin. Immunol. Pract. 2017, 5, 1502–1509. [Google Scholar] [CrossRef] [PubMed]
- Fulkerson, P.C.; Rothenberg, M.E. Targeting eosinophils in allergy, inflammation and beyond. Nat. Rev. Drug Discov. 2013, 12, 117–129. [Google Scholar] [CrossRef]
- Ilmarinen, P.; Kankaanranta, H. Eosinophil apoptosis as a therapeutic target in allergic asthma. Basic Clin. Pharmacol. Toxicol. 2014, 114, 109–117. [Google Scholar] [CrossRef]
- Gotlib, J. World Health Organization-defined eosinophilic disorders: 2017 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2017, 92, 1243–1259. [Google Scholar] [CrossRef]
- Kankaanranta, H.; Zhang, X.; Tumelius, R.; Ruotsalainen, M.; Haikala, H.; Nissinen, E.; Moilanen, E. Antieosinophilic activity of simendans. J. Pharmacol. Exp. Ther. 2007, 323, 31–38. [Google Scholar] [CrossRef]
- Izumi, K.; Bieber, K.; Ludwig, R.J. Current clinical trials in pemphigus and pemphigoid. Front. Immunol. 2019, 10, 978. [Google Scholar] [CrossRef] [PubMed]
- Fala, L. Nucala (Mepolizumab): First IL-5 Antagonist Monoclonal Antibody FDA Approved for Maintenance Treatment of Patients with Severe Asthma. Am. Health Drug Benefits 2016, 9, 106–110. [Google Scholar]
- Zhang, J.; Kuvelkar, R.; Murgolo, N.J.; Taremi, S.S.; Chou, C.C.; Wang, P.; Billah, M.M.; Egan, R.W. Mapping and characterization of the epitope(s) of Sch 55700, a humanized mAb, that inhibits human IL-5. Int. Immunol. 1999, 11, 1935–1943. [Google Scholar] [CrossRef][Green Version]
- Castro, M.; Zangrilli, J.; Wechsler, M.E.; Bateman, E.D.; Brusselle, G.G.; Bardin, P.; Murphy, K.; Maspero, J.F.; O’Brien, C.; Korn, S. Reslizumab for inadequately controlled asthma with elevated blood eosinophil counts: Results from two multicentre, parallel, double-blind, randomised, placebo-controlled, phase 3 trials. Lancet Respir. Med. 2015, 3, 355–366. [Google Scholar] [CrossRef]
- Flood-Page, P.T.; Menzies-Gow, A.N.; Kay, A.B.; Robinson, D.S. Eosinophil’s role remains uncertain as anti-interleukin-5 only partially depletes numbers in asthmatic airway. Am. J. Respir. Crit. Care Med. 2003, 167, 199–204. [Google Scholar] [CrossRef]
- Liu, W.; Ma, X.; Zhou, W. Adverse events of benralizumab in moderate to severe eosinophilic asthma: A meta-analysis. Medicine 2019, 98, e15868. [Google Scholar] [CrossRef] [PubMed]
- Kolbeck, R.; Kozhich, A.; Koike, M.; Peng, L.; Andersson, C.K.; Damschroder, M.M.; Reed, J.L.; Woods, R.; Dall’acqua, W.W.; Stephens, G.L.; et al. MEDI-563, a humanized anti-IL-5 receptor alpha mAb with enhanced antibody-dependent cell-mediated cytotoxicity function. J. Allergy Clin. Immunol. 2010, 125, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Maggi, L.; Rossettini, B.; Montaini, G.; Matucci, A.; Vultaggio, A.; Mazzoni, A.; Palterer, B.; Parronchi, P.; Maggi, E.; Liotta, F.; et al. Omalizumab dampens type 2 inflammation in a group of long-term treated asthma patients and detaches IgE from FcεRI. Eur. J. Immunol. 2018, 48, 2005–2014. [Google Scholar] [CrossRef] [PubMed]
- Ridolo, E.; Pucciarini, F.; Nizi, M.C.; Makri, E.; Kihlgren, P.; Panella, L.; Incorvaia, C. Mabs for treating asthma: Omalizumab, mepolizumab, reslizumab, benralizumab, dupilumab. Hum. Vaccines Immunother. 2020, 16, 2349–2356. [Google Scholar] [CrossRef]
- Delgado, J.; Dávila, I.J.; Domínguez-Ortega, J. Clinical recommendations for the management of biological treatments in severe asthma patients: A consensus statement. J. Investig. Allergol. Clin. Immunol. 2021, 31, 36–43. [Google Scholar] [CrossRef]
- Dorey-Stein, Z.L.; Shenoy, K.V. Tezepelumab as an emerging therapeutic option for the treatment of severe asthma: Evidence to date. Drug Des. Dev. Ther. 2021, 15, 331–338. [Google Scholar] [CrossRef] [PubMed]
- 2021 GINA Main Report-Global Initiative for Asthma-GINA. Available online: https://ginasthma.org/gina-reports/ (accessed on 30 April 2021).
- Siddiqui, S.H.; Guasconi, A.; Vestbo, J.; Jones, P.; Agusti, A.; Paggiaro, P.; Wedzicha, J.A.; Singh, D. Blood eosinophils: A biomarker of response to extrafine beclomethasone/formoterol in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2015, 192, 523–525. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Lee, S.H.; Lee, S.Y.; Kim, M.K.; Lee, B.J.; Werkström, V.; Barker, P.; Zangrilli, J.G. Efficacy and Safety of Benralizumab for Korean Patients With Severe, Uncontrolled Eosinophilic Asthma. Allergy Asthma Immunol. Res. 2019, 11, 508–518. [Google Scholar] [CrossRef] [PubMed]
- McGregor, M.C.; Krings, J.G.; Nair, P.; Castro, M. Role of biologics in asthma. Am. J. Respir. Crit. Care Med. 2019, 199, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.J.; Korn, S.; Mathur, S.K.; Barker, P.; Meka, V.G.; Martin, U.J.; Zangrilli, J.G. Safety of Eosinophil-Depleting Therapy for Severe, Eosinophilic Asthma: Focus on Benralizumab. Drug Saf. 2020, 43, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Gleich, G.J.; Klion, A.D.; Lee, J.J.; Weller, P.F. The consequences of not having eosinophils. Allergy 2013, 68, 829–835. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigo-Muñoz, J.M.; Gil-Martínez, M.; Sastre, B.; del Pozo, V. Emerging Evidence for Pleiotropism of Eosinophils. Int. J. Mol. Sci. 2021, 22, 7075. https://doi.org/10.3390/ijms22137075
Rodrigo-Muñoz JM, Gil-Martínez M, Sastre B, del Pozo V. Emerging Evidence for Pleiotropism of Eosinophils. International Journal of Molecular Sciences. 2021; 22(13):7075. https://doi.org/10.3390/ijms22137075
Chicago/Turabian StyleRodrigo-Muñoz, José M., Marta Gil-Martínez, Beatriz Sastre, and Victoria del Pozo. 2021. "Emerging Evidence for Pleiotropism of Eosinophils" International Journal of Molecular Sciences 22, no. 13: 7075. https://doi.org/10.3390/ijms22137075
APA StyleRodrigo-Muñoz, J. M., Gil-Martínez, M., Sastre, B., & del Pozo, V. (2021). Emerging Evidence for Pleiotropism of Eosinophils. International Journal of Molecular Sciences, 22(13), 7075. https://doi.org/10.3390/ijms22137075







