Function of Circular RNAs in Fish and Their Potential Application as Biomarkers
Abstract
:1. Introduction
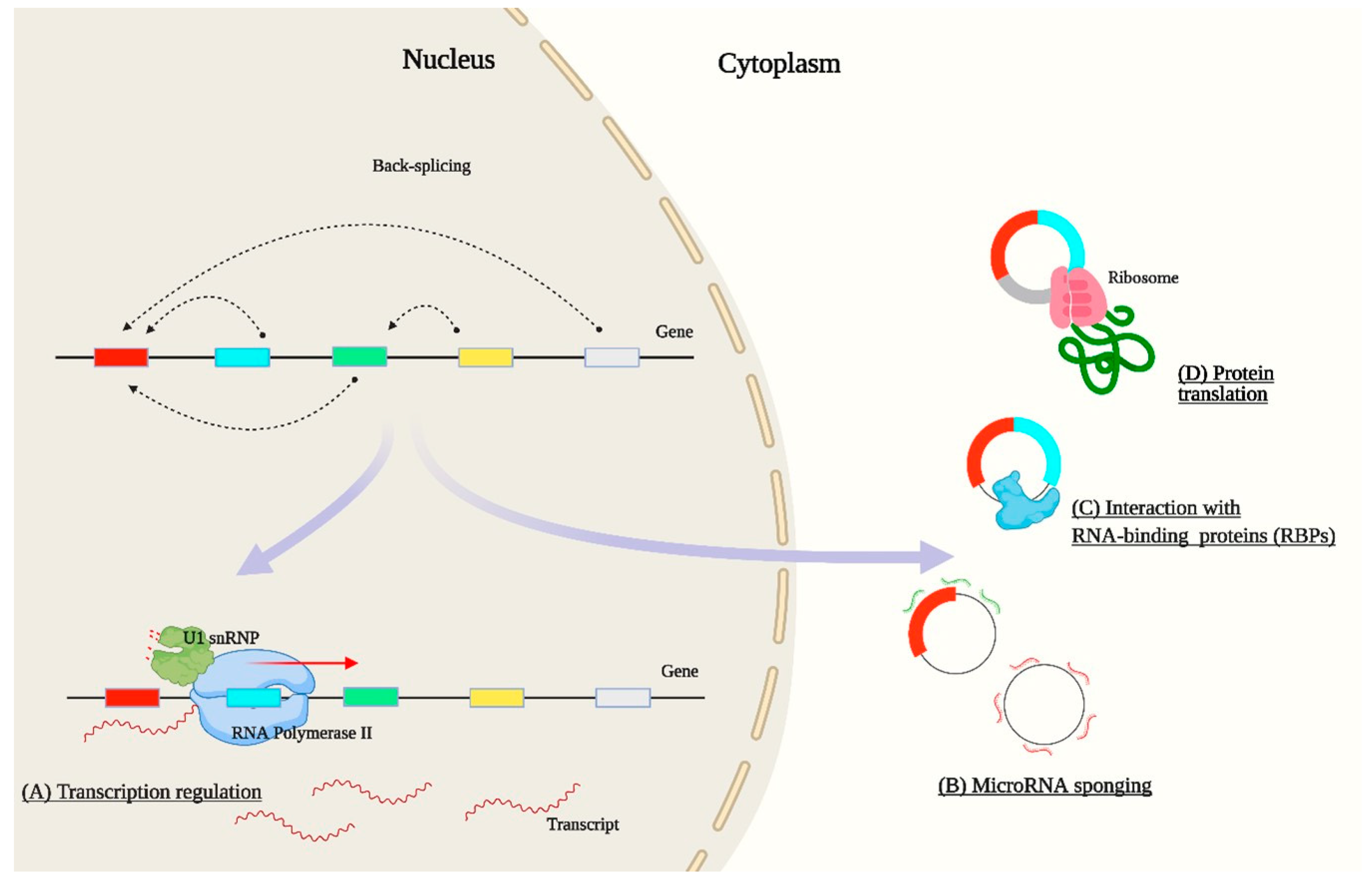
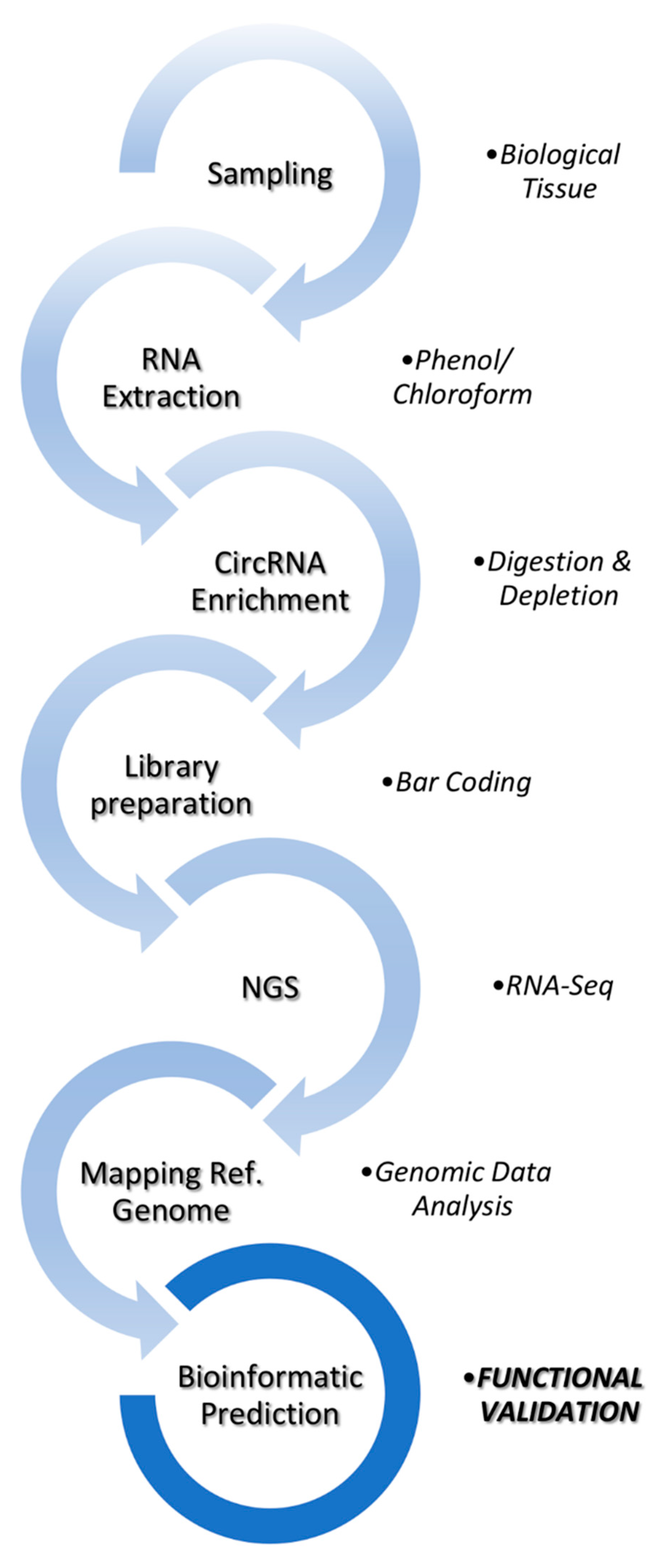
2. Bioinformatic Analysis of circRNAs
3. Role of circRNAs in Myogenesis and Growth
4. CircRNAs as Regulators of the Immune System
5. Advancements in Teleost circRNA Research
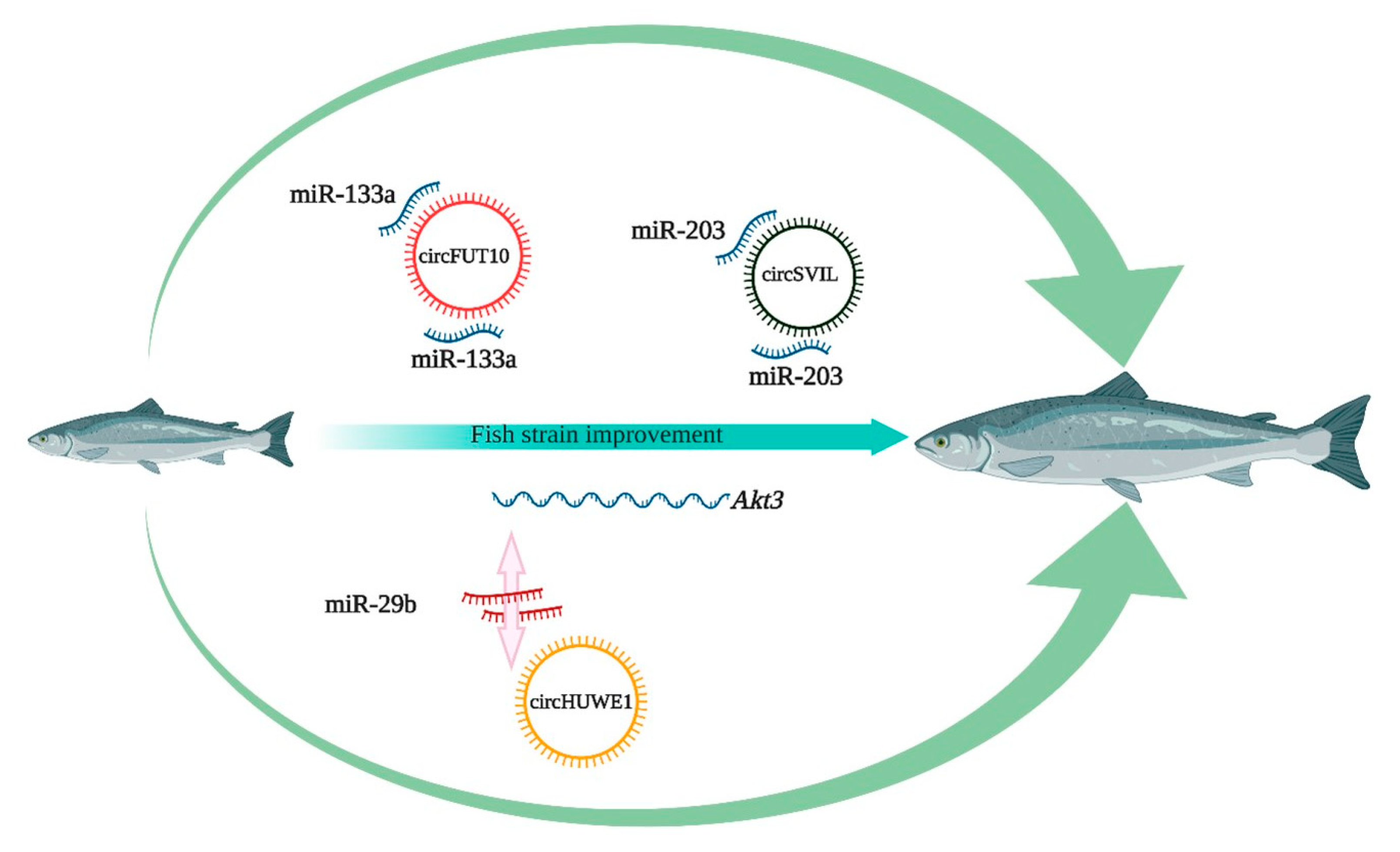
6. Potential Application of circRNAs in Aquaculture
7. Major Challenges of circRNA Research in Aquaculture
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kos, A.; Dijkema, R.; Arnberg, A.C.; van der Meide, P.H.; Schellekens, H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986, 323, 558–560. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [Green Version]
- Danan, M.; Schwartz, S.; Edelheit, S.; Sorek, R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012, 40, 3131–3142. [Google Scholar] [CrossRef]
- Hansen, T.B.; Venø, M.T.; Damgaard, C.K.; Kjems, J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016, 44, e58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.; Zhang, J.-L.; Yang, L.; Chen, L.-L. The biogenesis of nascent circular RNAs. Cell Rep. 2016, 15, 611–624. [Google Scholar] [CrossRef] [Green Version]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.-H.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Metge, F.; Dieterich, C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics 2016, 32, 1094–1096. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, S.; Feng, J.; Lei, L.; Hu, J.; Xia, L.; Wang, J.; Xiang, Y.; Liu, L.; Zhong, S.; Han, L. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Briefings Bioinf. 2017, 18, 984–992. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Yang, B.; Chen, B.; Bliim, N.; Ueberham, U.; Arendt, T.; Janitz, M. The emerging role of circular RNAs in transcriptome regulation. Genomics 2017, 109, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Zhou, D.; Dong, L.; Yang, L.; Ma, Q.; Liu, F.; Li, Y.; Xiong, S. Identification and analysis of circRNA–miRNA–mRNA regulatory network in hepatocellular carcinoma. IET Syst. Biol. 2020, 14, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Szabo, L.; Morey, R.; Palpant, N.J.; Wang, P.L.; Afari, N.; Jiang, C.; Parast, M.M.; Murry, C.E.; Laurent, L.C.; Salzman, J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015, 16, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y. Extensive translation of circular RNAs driven by N 6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Zhang, X.; Yan, M.; Li, H. Emerging role of circular RNAs in cancer. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Li, D.; Li, Z.; Yang, Y.; Zeng, X.; Li, Y.; Du, X.; Zhu, X. Circular RNAs as biomarkers and therapeutic targets in environmental chemical exposure-related diseases. Environ. Res. 2020, 180, 108825. [Google Scholar] [CrossRef]
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18, 90. [Google Scholar] [CrossRef]
- Werfel, S.; Nothjunge, S.; Schwarzmayr, T.; Strom, T.-M.; Meitinger, T.; Engelhardt, S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016, 98, 103–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.-Y.; Yang, J.-M.; Xiong, X.-D. The emerging landscape of circular RNA in cardiovascular diseases. J. Mol. Cell. Cardiol. 2018, 122, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Kun-Peng, Z.; Xiao-Long, M.; Chun-Lin, Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int. J. Biol. Sci. 2018, 14, 321. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Pan, S.; Zhou, J.; Diao, X.; Liu, S. CircRNA CDR1as knockdown inhibits progression of non-small-cell lung cancer by regulating miR-219a-5p/SOX5 axis. Thorac Cancer 2020. [Google Scholar] [CrossRef] [Green Version]
- Zeng, K.; Chen, X.; Xu, M.; Liu, X.; Hu, X.; Xu, T.; Sun, H.; Pan, Y.; He, B.; Wang, S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, X.; Wei, S.; Chen, Y.; Chen, Y.; Fan, X.; Han, S.; Wu, G. hsa_circ_0013958: A circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017, 284, 2170–2182. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Zhou, Z.; Jiang, R.; Huang, J.; Chen, L.; Cao, Z.; Chu, H.; Han, B.; Cheng, Y. Silica-induced initiation of circular ZC3H4RNA/ZC3H4 pathway promotes the pulmonary macrophage activation. FASEB J. 2018, 32, 3264–3277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, Z.; Xu, S.; Guo, J. Novel potential tumor biomarkers: Circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J. Clin. Lab. Anal. 2020, e23359. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Sehgal, P.; Mathew, S.; Vellarikkal, S.K.; Singh, A.R.; Kapoor, S.; Jayarajan, R.; Scaria, V.; Sivasubbu, S. A genome-wide map of circular RNAs in adult zebrafish. Sci. Rep. 2019, 9, 3432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Guo, X.; Wang, W. Identification and characterization of circular RNAs in zebrafish. FEBS Lett. 2017, 591, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Near, T.J.; Eytan, R.I.; Dornburg, A.; Kuhn, K.L.; Moore, J.A.; Davis, M.P.; Wainwright, P.C.; Friedman, M.; Smith, W.L. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. USA 2012, 109, 13698–13703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desvignes, T.; Carey, A.; Postlethwait, J.H. Evolution of caudal fin ray development and caudal fin hypural diastema complex in spotted gar, teleosts, and other neopterygian fishes. Dev. Dyn. 2018, 247, 832–853. [Google Scholar] [CrossRef]
- Faircloth, B.C.; Sorenson, L.; Santini, F.; Alfaro, M.E. A phylogenomic perspective on the radiation of ray-finned fishes based upon targeted sequencing of ultraconserved elements (UCEs). PLoS ONE 2013, 8, e65923. [Google Scholar]
- Balon, E.K. The oldest domesticated fishes, and the consequences of an epigenetic dichotomy in fish culture. J. Ichthyol. Aquat. Biol. 2006, 11, 47–86. [Google Scholar]
- Nakajima, T.; Hudson, M.J.; Uchiyama, J.; Makibayashi, K.; Zhang, J. Common carp aquaculture in Neolithic China dates back 8000 years. Nat. Ecol. Evol. 2019, 3, 1415–1418. [Google Scholar] [CrossRef]
- Volff, J. Genome evolution and biodiversity in teleost fish. Heredity 2005, 94, 280–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennings, S.; Stentiford, G.D.; Leocadio, A.M.; Jeffery, K.R.; Metcalfe, J.D.; Katsiadaki, I.; Auchterlonie, N.A.; Mangi, S.C.; Pinnegar, J.K.; Ellis, T. Aquatic food security: Insights into challenges and solutions from an analysis of interactions between fisheries, aquaculture, food safety, human health, fish and human welfare, economy and environment. Fish Fish 2016, 17, 893–938. [Google Scholar] [CrossRef] [Green Version]
- Boudry, P.; Allal, F.; Aslam, M.L.; Bargelloni, L.; Bean, T.P.; Brard-Fudulea, S.; Brieuc, M.S.O.; Calboli, F.C.F.; Gilbey, J.; Haffray, P. Current status and potential of genomic selection to improve selective breeding in the main aquaculture species of International Council for the Exploration of the Sea (ICES) member countries. Aquacult. Rep. 2021, 20, 100700. [Google Scholar]
- De Verdal, H.; Komen, H.; Quillet, E.; Chatain, B.; Allal, F.; Benzie, J.A.; Vandeputte, M. Improving feed efficiency in fish using selective breeding: A review. Rev. Aquacult. 2018, 10, 833–851. [Google Scholar] [CrossRef]
- Gjedrem, T.; Rye, M. Selection response in fish and shellfish: A review. Rev. Aquacult. 2018, 10, 168–179. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, J.; Zhao, F. Circular RNA identification based on multiple seed matching. Briefings Bioinf. 2018, 19, 803–810. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Metge, F.; Czaja-Hasse, L.F.; Reinhardt, R.; Dieterich, C. FUCHS—Towards full circular RNA characterization using RNAseq. PeerJ 2017, 5, e2934. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Zhang, N.; Han, P.; Moon, B.-S.; Lai, R.K.; Wang, K.; Lu, W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016, 44, e87. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Li, G. CircCode: A powerful tool for identifying circRNA coding ability. Front. Genet. 2019, 10, 981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.-O.; Wang, H.-B.; Zhang, Y.; Lu, X.; Chen, L.-L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [Green Version]
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679. [Google Scholar] [CrossRef] [Green Version]
- Chuang, T.-J.; Wu, C.-S.; Chen, C.-Y.; Hung, L.-Y.; Chiang, T.-W.; Yang, M.-Y. NCLscan: Accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 2016, 44, e29. [Google Scholar] [CrossRef] [Green Version]
- You, X.; Conrad, T.O. Acfs: Accurate circRNA identification and quantification from RNA-Seq data. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B. Improved circRNA identification by combining prediction algorithms. Front. Cell Dev. Biol. 2018, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Lin, W.; Guo, M.; Zou, Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Nedoluzhko, A.; Sharko, F.; Rbbani, M.G.; Teslyuk, A.; Konstantinidis, I.; Fernandes, J.M. CircParser: A novel streamlined pipeline for circular RNA structure and host gene prediction in non-model organisms. PeerJ 2020, 8, e8757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Xiang, Y.; Xia, S.; Liu, H.; Wang, J.; Ozguc, F.M.; Lei, L.; Kong, R.; Diao, L.; He, C. CircView: A visualization and exploration tool for circular RNAs. Briefings Bioinf. 2019, 20, 745–751. [Google Scholar] [CrossRef]
- Emmert-Streib, F.; Yang, Z.; Feng, H.; Tripathi, S.; Dehmer, M. An Introductory Review of Deep Learning for Prediction Models With Big Data. Front. Artif. Intell. 2020, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Jackson, S.A. Machine Learning and Complex Biological Data. Genome Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Chaabane, M.; Williams, R.M.; Stephens, A.T.; Park, J.W. circDeep: Deep learning approach for circular RNA classification from other long non-coding RNA. Bioinformatics 2020, 36, 73–80. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Deng, Y.; Liu, Q.; Ye, B.; Dai, Z.; Chen, Y.; Dai, X. Identifying circular RNA and predicting its regulatory interactions by machine learning. Front. Genet. 2020, 11, 655. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.; Yang, J.; Zhao, F. Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Goody, M.F.; Henry, C.A. A need for NAD+ in muscle development, homeostasis, and aging. Skelet Muscle 2018, 8, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, I.A.; Bower, N.I.; Macqueen, D.J. Growth and the regulation of myotomal muscle mass in teleost fish. J. Exp. Biol. 2011, 214, 1617–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Collier, J.M.; Hlaing, M.; Zhang, L.; Delshad, E.H.; Bristow, J.; Bernstein, H.S. Genome-wide examination of myoblast cell cycle withdrawal during differentiation. Dev. Dyn. 2003, 226, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Gang, E.J.; Bosnakovski, D.; Simsek, T.; To, K.; Perlingeiro, R.C. Pax3 activation promotes the differentiation of mesenchymal stem cells toward the myogenic lineage. Exp. Cell. Res. 2008, 314, 1721–1733. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, Y.; Du, M. Farm animals for studying muscle development and metabolism: Dual purposes for animal production and human health. Anim. Front. 2019, 9, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Rossi, G.; Messina, G. Comparative myogenesis in teleosts and mammals. Cell. Mol. Life Sci. 2014, 71, 3081–3099. [Google Scholar] [CrossRef] [Green Version]
- Asaduzzaman, M.; Akolkar, D.B.; Kinoshita, S.; Watabe, S. The expression of multiple myosin heavy chain genes during skeletal muscle development of torafugu Takifugu rubripes embryos and larvae. Gene 2013, 515, 144–154. [Google Scholar] [CrossRef]
- Bassel-Duby, R.; Olson, E.N. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 2006, 75, 19–37. [Google Scholar] [CrossRef] [Green Version]
- Miska, E.A.; Karlsson, C.; Langley, E.; Nielsen, S.J.; Pines, J.; Kouzarides, T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999, 18, 5099–5107. [Google Scholar] [CrossRef] [PubMed]
- Nie, M.; Deng, Z.-L.; Liu, J.; Wang, D.-Z. Noncoding RNAs, emerging regulators of skeletal muscle development and diseases. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Li, H.; Yang, J.; Hao, D.; Dong, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Lin, F. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017, 8, e3153. [Google Scholar] [CrossRef]
- Ling, Y.; Zheng, Q.; Zhu, L.; Xu, L.; Sui, M.; Zhang, Y.; Liu, Y.; Fang, F.; Chu, M.; Ma, Y. Trend analysis of the role of circular RNA in goat skeletal muscle development. BMC Genom. 2020, 21, 1–12. [Google Scholar] [CrossRef]
- Shen, X.; Liu, Z.; Cao, X.; He, H.; Han, S.; Chen, Y.; Cui, C.; Zhao, J.; Li, D.; Wang, Y. Circular RNA profiling identified an abundant circular RNA circTMTC1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging miR-128-3p. Int. J. Biol. Sci. 2019, 15, 2265. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmohsen, K.; Panda, A.C.; De, S.; Grammatikakis, I.; Kim, J.; Ding, J.; Noh, J.H.; Kim, K.M.; Mattison, J.A.; de Cabo, R. Circular RNAs in monkey muscle: Age-dependent changes. Aging 2015, 7, 903. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Xu, H.; Li, R.; Wu, W.; Chao, Z.; Li, C.; Xia, W.; Wang, L.; Yang, J.; Xu, Y. Assessment of myoblast circular RNA dynamics and its correlation with miRNA during myogenic differentiation. Int. J. Biochem. Cell Biol. 2018, 99, 211–218. [Google Scholar] [CrossRef]
- Li, C.; Li, X.; Yao, Y.; Ma, Q.; Ni, W.; Zhang, X.; Cao, Y.; Hazi, W.; Wang, D.; Quan, R. Genome-wide analysis of circular RNAs in prenatal and postnatal muscle of sheep. Oncotarget 2017, 8, 97165. [Google Scholar] [CrossRef]
- Chen, J.; Zou, Q.; Lv, D.; Wei, Y.; Raza, M.A.; Chen, Y.; Li, P.; Xi, X.; Xu, H.; Wen, A. Comprehensive transcriptional landscape of porcine cardiac and skeletal muscles reveals differences of aging. Oncotarget 2018, 9, 1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, L.; Gu, T.; He, Y.; Zhou, C.; Hu, Q.; Xingwang, W.; Zheng, E.; Huang, S.; Xu, Z.; Yang, J. Genome-wide analysis of circular RNAs mediated ceRNA regulation in porcine embryonic muscle development. Front. Cell Dev. Biol. 2019, 7, 289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, I.F.; Climent, M.; Quintavalle, M.; Farina, F.M.; Schorn, T.; Zani, S.; Carullo, P.; Kunderfranco, P.; Civilini, E.; Condorelli, G. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ. Res. 2019, 124, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, M.; Wang, Y.; Liu, J.; Zhang, M.; Fang, X.; Chen, H.; Zhang, C. A Zfp609 circular RNA regulates myoblast differentiation by sponging miR-194-5p. Boil. Macromol. 2019, 121, 1308–1313. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, S. Amphioxus IGF-like peptide induces mouse muscle cell development via binding to IGF receptors and activating MAPK and PI3K/Akt signaling pathways. Mol. Cell. Endocrinol. 2011, 343, 45–54. [Google Scholar] [CrossRef]
- Shen, X.; Zhang, X.; Ru, W.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. CircINSR promotes proliferation and reduces apoptosis of embryonic myoblasts by sponging miR-34a. Mol. Ther. Nucleic. Acids 2020. [Google Scholar] [CrossRef]
- Li, H.; Yang, J.; Wei, X.; Song, C.; Dong, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Ma, Y. CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J. Cell. Physiol. 2018, 233, 4643–4651. [Google Scholar] [CrossRef]
- Ouyang, H.; Chen, X.; Wang, Z.; Yu, J.; Jia, X.; Li, Z.; Luo, W.; Abdalla, B.A.; Jebessa, E.; Nie, Q. Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Res. 2018, 25, 71–86. [Google Scholar] [CrossRef]
- Ouyang, H.; Chen, X.; Li, W.; Li, Z.; Nie, Q.; Zhang, X. Circular RNA circSVIL promotes myoblast proliferation and differentiation by sponging miR-203 in chicken. Front. Genet. 2018, 9, 172. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wei, X.; Yang, J.; Dong, D.; Hao, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Ma, Y. circFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a. Mol. Ther. Nucleic Acids 2018, 11, 272–283. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Cao, X.; Dong, D.; Shen, X.; Cheng, J.; Jiang, R.; Yang, Z.; Peng, S.; Huang, Y.; Lan, X. Circular RNA TTN acts as a miR-432 sponge to facilitate proliferation and differentiation of myoblasts via the IGF2/PI3K/AKT signaling pathway. Mol. Ther. Nucleic Acids 2019, 18, 966–980. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ouyang, H.; Wang, Z.; Chen, B.; Nie, Q. A novel circular RNA generated by FGFR2 gene promotes myoblast proliferation and differentiation by sponging miR-133a-5p and miR-29b-1-5p. Cells 2018, 7, 199. [Google Scholar] [CrossRef] [Green Version]
- Peng, S.; Song, C.; Li, H.; Cao, X.; Ma, Y.; Wang, X.; Huang, Y.; Lan, X.; Lei, C.; Chaogetu, B. Circular RNA SNX29 Sponges miR-744 to Regulate Proliferation and Differentiation of Myoblasts by Activating the Wnt5a/Ca2+ Signaling Pathway. Mol. Ther. Nucleic Acids 2019, 16, 481–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, B.; Wang, J.; Ru, W.; Wu, J.; Cao, X.; Yang, H.; Huang, Y.; Lan, X.; Lei, C.; Huang, B. The circular RNA circHUWE1 sponges the miR-29b-AKT3 axis to regulate myoblast development. Mol. Ther. Nucleic Acids 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Yu, J.; Guo, L.; Byers, M.S.; Wang, Z.; Chen, X.; Xu, H.; Nie, Q. Circular RNA circHIPK3 promotes the proliferation and differentiation of chicken myoblast cells by sponging miR-30a-3p. Cells 2019, 8, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galindo-Villegas, J.; García-Moreno, D.; de Oliveira, S.; Meseguer, J.; Mulero, V. Regulation of immunity and disease resistance by commensal microbes and chromatin modifications during zebrafish development. PNAS 2012, 109, E2605–E2614. [Google Scholar] [CrossRef] [Green Version]
- Galindo-Villegas, J. Recent findings on vertebrate developmental immunity using the zebrafish model. Mol. Immunol. 2016, 69, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Li, X.; Zhang, M.; Lv, K. Microarray analysis of circular RNA expression patterns in polarized macrophages. Int. J. Mol. Med. 2017, 39, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell 2019, 76, 96–109.e9. [Google Scholar] [CrossRef]
- Wesselhoeft, R.A.; Kowalski, P.S.; Parker-Hale, F.C.; Huang, Y.; Bisaria, N.; Anderson, D.G. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 2019, 74, 508–520.e4. [Google Scholar] [CrossRef]
- Wu, T.-H.; Shi, L.; Lowe, A.W.; Nicolls, M.R.; Kao, P.N. Inducible expression of immediate early genes is regulated through dynamic chromatin association by NF45/ILF2 and NF90/NF110/ILF3. PLoS ONE 2019, 14, e0216042. [Google Scholar] [CrossRef] [Green Version]
- Aktaş, T.; Ilık, İ.A.; Maticzka, D.; Bhardwaj, V.; Rodrigues, C.P.; Mittler, G.; Manke, T.; Backofen, R.; Akhtar, A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 2017, 544, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.-K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 2019, 177, 865–880.e21. [Google Scholar] [CrossRef]
- Zheng, Z.-M. Circular RNAs and RNase L in PKR activation and virus infection. Cell Biosci. 2019, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Wang, S.; Ye, B.; Du, Y.; Li, C.; Xiong, Z.; Qu, Y.; Fan, Z. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 2018, 48, 688–701.e7. [Google Scholar] [CrossRef] [Green Version]
- Haque, S.; Ames, R.M.; Moore, K.; Lee, B.P.; Jeffery, N.; Harries, L.W. Islet-expressed circular RNAs are associated with type 2 diabetes status in human primary islets and in peripheral blood. BMC Med. Genom. 2020, 13, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhu, X.; Niu, X.; Chen, L.; Ge, C. Elevated hsa_circRNA_101015, hsa_circRNA_101211, and hsa_circRNA_103470 in the Human Blood: Novel Biomarkers to Early Diagnose Acute Pancreatitis. Biomed Res. Int. 2020, 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Li, K.; Lai, W.; Li, X.; Wang, H.; Yang, J.; Chu, S.; Wang, H.; Kang, C.; Qiu, Y. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin. Chim. Acta 2018, 480, 17–25. [Google Scholar] [CrossRef]
- Yao, R.; Zou, H.; Liao, W. Prospect of circular RNA in hepatocellular carcinoma: A novel potential biomarker and therapeutic target. Front. Oncol. 2018, 8, 332. [Google Scholar] [CrossRef]
- Yang, C.M.; Qiao, G.L.; Song, L.N.; Bao, S.; Ma, L.J. Circular RNAs in gastric cancer: Biomarkers for early diagnosis. Oncol. Lett. 2020, 20, 465–473. [Google Scholar] [CrossRef]
- Zuo, L.; Zhang, L.; Zu, J.; Wang, Z.; Han, B.; Chen, B.; Cheng, M.; Ju, M.; Li, M.; Shu, G. Circulating circular RNAs as biomarkers for the diagnosis and prediction of outcomes in acute ischemic stroke. Stroke 2020, 51, 319–323. [Google Scholar] [CrossRef]
- Su, H.; Chu, Q.; Zheng, W.; Chang, R.; Gao, W.; Zhang, L.; Xu, T. Circular RNA circPIKfyve acts as a sponge of miR-21-3p to enhance antiviral immunity through regulating MAVS in teleost fish. J. Virol. 2021. [Google Scholar] [CrossRef]
- He, L.; Zhang, A.; Xiong, L.; Li, Y.; Huang, R.; Liao, L.; Zhu, Z. Deep circular RNA sequencing provides insights into the mechanism underlying grass carp reovirus infection. Int. J. Mol. Sci. 2017, 18, 1977. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Yuan, R.; Liang, Z.; Zhang, T.; Zhu, M.; Zhang, X.; Geng, W.; Fang, P.; Jiang, M.; Wang, Z. Comprehensive analysis of circRNA expression pattern and circRNA–mRNA–miRNA network in Ctenopharyngodon idellus kidney (CIK) cells after grass carp reovirus (GCRV) infection. Aquaculture 2019, 512, 734349. [Google Scholar] [CrossRef]
- Pan, J.; Zhang, X.; Zhang, Y.; Yan, B.; Dai, K.; Zhu, M.; Liang, Z.; Dai, Y.; Zhang, M.; Zhang, Z. Grass carp reovirus encoding circular RNAs with antiviral activity. Aquaculture 2021, 533, 736135. [Google Scholar] [CrossRef]
- Fan, B.; Chen, F.; Li, Y.; Wang, Z.; Wang, Z.; Lu, Y.; Wu, Z.; Jian, J.; Wang, B. A comprehensive profile of the tilapia (Oreochromis niloticus) circular RNA and circRNA–miRNA network in the pathogenesis of meningoencephalitis of teleosts. Mol. Omics 2019, 15, 233–246. [Google Scholar] [CrossRef]
- Xiu, Y.; Jiang, G.; Zhou, S.; Diao, J.; Liu, H.; Li, C. Identification of potential immune-related circRNA-miRNA-mRNA regulatory network in intestine of Paralichthys olivaceus during Edwardsiella tarda infection. Front. Genet. 2019, 10, 731. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Dai, Y.; Zhang, X.; Dai, K.; Liu, B.; Yuan, R.; Feng, Y.; Liang, Z.; Zhu, M.; Zhang, M. Identification and characterization of novel type of RNAs, circRNAs in crucian carp Carassius auratus gibelio. Fish Shellfish Immunol. 2019, 94, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hu, Y.; Yin, J.; Yan, X.Y.; Chen, W.J.; Jiang, C.Y.; Hu, X.S.; Wang, X.Y.; Zhu, J.G.; Yu, Z.B. Profiles analysis reveals circular RNAs involving zebrafish physiological development. J. Cell. Physiol. 2019, 234, 15922–15933. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Xiao, S.; Qiu, C.; Wang, Z. Transcriptome-wide identification and functional investigation of circular RNA in the teleost large yellow croaker (Larimichthys crocea). Mar. Geonom. 2017, 32, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Signaling in control of cell growth and metabolism. Cold Spring Harbor Perspect. Biol. 2012, 4, a006783. [Google Scholar] [CrossRef] [Green Version]
- Quan, J.; Kang, Y.; Luo, Z.; Zhao, G.; Li, L.; Liu, Z. Integrated analysis of the responses of a circRNA-miRNA-mRNA ceRNA network to heat stress in rainbow trout (Oncorhynchus mykiss) liver. BMC Genom. 2021, 22, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Valente, L.M.; Cabral, E.M.; Sousa, V.; Cunha, L.M.; Fernandes, J.M. Plant protein blends in diets for Senegalese sole affect skeletal muscle growth, flesh texture and the expression of related genes. Aquaculture 2016, 453, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Valente, L.M.; Moutou, K.A.; Conceicao, L.E.; Engrola, S.; Fernandes, J.M.; Johnston, I.A. What determines growth potential and juvenile quality of farmed fish species? Rev. Aquacult. 2013, 5, S168–S193. [Google Scholar] [CrossRef] [Green Version]
- Yan, B.; Guo, J.-T.; Zhao, L.-H.; Zhao, J.-L. microRNA expression signature in skeletal muscle of Nile tilapia. Aquaculture 2012, 364, 240–246. [Google Scholar] [CrossRef]
- Yan, X.; Ding, L.; Li, Y.; Zhang, X.; Liang, Y.; Sun, X.; Teng, C.-B. Identification and profiling of microRNAs from skeletal muscle of the common carp. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Zhao, Z.; Jia, J.; Yang, G.; Sun, C.; Li, W. miR-181b-5p may regulate muscle growth in tilapia by targeting myostatin b. Front. Endocrinol. 2019, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Circular RNAs as therapeutic agents and targets. Front. Physiol. 2018, 9, 1262. [Google Scholar]
- Gutierrez, A.P.; Houston, R.D. Quantitative trait locus mapping in aquaculture species: Principles and practice. In Bioinformatics in Aquaculture: Principles and Methods; John Wiley & Sons, Ltd.: Chichester, UK, 2017. [Google Scholar]
- Laghari, M.Y.; Lashari, P.; Zhang, Y.; Sun, X. Identification of quantitative trait loci (QTLs) in aquaculture species. Rev. Fish. Sci. Aquacult. 2014, 22, 221–238. [Google Scholar] [CrossRef]
- Liu, F.; Sun, F.; Xia, J.H.; Li, J.; Fu, G.H.; Lin, G.; Tu, R.J.; Wan, Z.Y.; Quek, D.; Yue, G.H. A genome scan revealed significant associations of growth traits with a major QTL and GHR2 in tilapia. Sci. Rep. 2014, 4, 7256. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Gan, M.; Tang, Q.; Tang, G.; Jiang, Y.; Li, M.; Chen, L.; Bai, L.; Shuai, S.; Wang, J. Comprehensive analysis of lncRNAs and circRNAs reveals the metabolic specialization in oxidative and glycolytic skeletal muscles. Int. J. Mol. Sci. 2019, 20, 2855. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, I.; Karedath, T.; Al-Dasim, F.M.; Malek, J.A. Identification of human genetic variants controlling circular RNA expression. RNA 2019, 25, 1765–1778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Ran, Y.; Tao, C.; Li, S.; Chen, J.; Yang, E. Detection of circular RNA expression and related quantitative trait loci in the human dorsolateral prefrontal cortex. Genome Boil. 2019, 20, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Liu, X.; Dai, S.; Xiao, H.; Wang, D. High Efficiency Targeting of Non-coding Sequences Using CRISPR/Cas9 System in Tilapia. G3 Genes Genomes Genet. 2019, 9, 287–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Species | Common Name | Tissue/Cell Type | RNA Treatment | Detected circRNAs | Validated circRNAs | Reference |
|---|---|---|---|---|---|---|
| Bos taurus | Cattle | Skeletal muscle | RNase R+ rRNA− | 12,981 | 17 | [74] |
| Capra aegagrus hircus | Wild goat | Skeletal muscle | RNase R+ rRNA− | 9090 | 4 | [75] |
| Gallus gallus | Chicken | Skeletal muscle | RNase R+ rRNA− | 13,377 | 8 | [76] |
| Homo sapiens | Human | C2C12 myoblasts | rRNA− | 2175 | 31 | [77] |
| Macaca mulatta | Rhesus monkey | Primary myoblasts | RNase R | 2100 | 29 | [78] |
| Mus musculus | House mouse | C2C12 myoblasts | RNase R+ rRNA− | 37,751 | 10 | [79] |
| C2C12 myoblasts | rRNA− | 1592 | 31 | [77] | ||
| Oreochromis niloticus | Nile tilapia | Skeletal muscle | rRNA− | 622 | - | [57] |
| Ovis aries | Sheep | Skeletal muscle | RNase R+ | 6000 | 10 | [80] |
| Sus scrofa | Pig | Skeletal muscle | rRNA− | 4402 | 2 | [81] |
| Skeletal muscle | RNase R+ rRNA− | 7968 | 6 | [82] |
| CircRNA | Species | Tissue/Cell | Biological Role | Mode of Action | Reference |
|---|---|---|---|---|---|
| Circ-ZNF609 | Homo sapiens | C2C12 | Myoblast proliferation | Protein encoding | [77] |
| CircTTN | Bos taurus | Skeletal muscle | miRNA sponge | [91] | |
| CircINSR | Bos taurus | Skeletal muscle | miRNA sponge | [86] | |
| CircFUT10 | Bos taurus | Skeletal muscle | miRNA sponge | [87] | |
| CircSVIL | Gallus gallus | Skeletal muscle | miRNA sponge | [89] | |
| CircFGFR2 | Gallus gallus | DF-1 | miRNA sponge | [92] | |
| CircRBFOX2 | Gallus gallus | Skeletal muscle | miRNA sponge | [88] | |
| CircSNX29 | Bos taurus | Skeletal muscle | Myoblast differentiation | miRNA sponge | [93] |
| CircHUWE1 | Bos taurus | Skeletal muscle | [94] | ||
| CircLMO7 | Bos taurus | Skeletal muscle | [74] | ||
| CircHIPK3 | Gallus gallus | Skeletal muscle | [95] |
| Species | Common Name | Infection Type | Tissue | DE * circRNAs | Reference |
|---|---|---|---|---|---|
| Ctenopharyngodon idellus | Grass carp | Viral | Spleen | 41 | [113] |
| Kidney | 76 | [114] | |||
| Kidney | - | [115] | |||
| Oreochromis niloticus | Nile tilapia | Bacterial | Brain | 837 | [116] |
| Paralichthys olivaceus | Japanese flounder | Bacterial | Intestine | 62 | [117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rbbani, G.; Nedoluzhko, A.; Galindo-Villegas, J.; Fernandes, J.M.O. Function of Circular RNAs in Fish and Their Potential Application as Biomarkers. Int. J. Mol. Sci. 2021, 22, 7119. https://doi.org/10.3390/ijms22137119
Rbbani G, Nedoluzhko A, Galindo-Villegas J, Fernandes JMO. Function of Circular RNAs in Fish and Their Potential Application as Biomarkers. International Journal of Molecular Sciences. 2021; 22(13):7119. https://doi.org/10.3390/ijms22137119
Chicago/Turabian StyleRbbani, Golam, Artem Nedoluzhko, Jorge Galindo-Villegas, and Jorge M. O. Fernandes. 2021. "Function of Circular RNAs in Fish and Their Potential Application as Biomarkers" International Journal of Molecular Sciences 22, no. 13: 7119. https://doi.org/10.3390/ijms22137119









