Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis
Abstract
:1. Introduction
2. Functions of bHLH Factors in Plant Growth and Development in Arabidopsis
2.1. Roles in Seed Germination
2.2. Functions in the Flowering Time Control
2.3. Functions on Cell Fate Determination
3. Functions in Environmental Response
3.1. Functions in Plant Mineral Nutrition and Abiotic Stress
3.2. Functions in Stress Responses
4. Functions in the Response to Light and Phytohormones
4.1. Response to Light Signaling
4.2. Functions in JA Signaling Pathway
4.3. Functions in IAA Signaling Pathway
4.4. Roles in ABA Signaling Pathway
4.5. The Cross-Talk between Light and Phytohormones
5. Functions in Other Aspects of Plant Biology
6. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murre, C.; McCaw, P.S.; Baltimore, D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 1989, 56, 777–783. [Google Scholar] [CrossRef]
- Massari, M.E.; Murre, C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000, 20, 429–440. [Google Scholar] [CrossRef] [Green Version]
- Duek, P.D.; Fankhauser, C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005, 10, 51–54. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef] [Green Version]
- Henriksson, M.; Lüscher, B. Proteins of the Myc network: Essential regulators of cell growth and differentiation. Adv. Cancer Res. 1996, 68, 109–182. [Google Scholar] [CrossRef]
- Goding, C.R. Motif from neural crest to melanoma: Signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000, 14, 1712–1728. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.H.; Copeland, N.G.; Jenkins, N.A.; Baltimore, D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol. Cell. Biol. 1991, 11, 5603–5611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledent, V.; Vervoort, M. The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, A.; Caudy, M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays 1998, 20, 298–306. [Google Scholar] [CrossRef]
- Crozatier, M.; Valle, D.; Dubois, L.; Ibnsouda, S.; Vincent, A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr. Biol. 1996, 6, 707–718. [Google Scholar] [CrossRef] [Green Version]
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Jakoby, M.; Werber, M.; Weisshaar, B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 2003, 15, 2497–2501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heim, M.A.; Jakoby, M.; Werber, M.; Marti, C.; Weisshaar, B.; Bailey, P.C. The basic Helix-Loop-Helix transcription factor family in plants: A genome-wide study of Protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buck, M.J.; Atchley, W.R. Phylogenetic Analysis of Plant Basic Helix-Loop-Helix Proteins. J. Mol. Evol. 2003, 56, 742–750. [Google Scholar] [CrossRef]
- Castelain, M.; Hir, R.L.; Bellini, C. The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiologia Plantarum 2012, 145, 450–460. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Friedrichsen, D.M.; Nemhauser, J.; Muramitsu, T.; Maloof, J.N.; Alonso, J.; Ecker, J.R.; Furuya, M.; Chory, J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 2002, 162, 1445–1456. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Li, K.; Liu, H.; Lin, C. Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in Arabidopsis. PLoS Genet. 2013, 9, e1003861. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; Cai, Y.; Yu, D.; Liang, G. bHLH104 confers tolerance to cadmium stress in Arabidopsis thaliana. J. Integr. Plant Biol. 2018, 60, 691–702. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and diversification of Basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef] [Green Version]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, H.; Ding, L.; Soppe, W.J.J.; Xiang, Y. REVERSAL OF RDO5 1, a homolog of rice seed dormancy 4, interacts with bHLH57 and controls ABA biosynthesis and seed dormancy in Arabidopsis. Plant Cell 2020, 32, 1933–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, M.; Tepperman, J.M.; Quail, P.H. PIF3, a Phytochrome-interacting factor necessary for normal photoinduced signal transduction, as a novel basic helix-loop-helix protein. Cell 1998, 95, 657–667. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Zhu, J.; Wang, Z. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Yamaguchi, S.; Kamiya, Y.; Bae, G.; Chung, W.I.; Choi, G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006, 47, 124–139. [Google Scholar] [CrossRef]
- Penfield, S.; Josse, E.; Kannangara, R.; Gilday, A.D.; Halliday, K.J.; Graham, I.A. Cold and light control of seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 2005, 15, 1998–2006. [Google Scholar] [CrossRef] [Green Version]
- Groszmann, M.; Bylstra, Y.; Lampugnani, E.R.; Smyth, D.R. Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J. Exp. Bot. 2010, 61, 1495–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Yang, H.; Mockler, T.; Lin, C. Regulation of flowering time by Arabidopsis photoreceptors. Science 1998, 279, 1360–1363. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, Y.; Ma, D.; Chen, Z.; Wang, J.; Liu, H. CIB1 and CO interact to mediate CRY2-dependent regulation of flowering. EMBO Rep. 2018, 19, e45762. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Xin, R.; Kim, D.; Sung, S.; Lange, T.; Huq, E. NO FLOWERING IN SHORT DAY (NFL) is a bHLH transcription factor that promotes flowering specifically under short-day conditions in Arabidopsis. Development 2016, 143, 682–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Li, Y.; Pan, J.; Lou, D.; Hu, Y.; Yu, D. The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in Arabidopsis. Mol. Plant 2017, 10, 1461–1464. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Yang, Z.; Li, F. Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol. J. 2019, 17, 1706–1722. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alós, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Heisler, M.G.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 2001, 128, 1089–1098. [Google Scholar] [CrossRef]
- Zhao, H.; Li, X.; Ma, L. Basic helix-loop-helix transcription factors and epidermal cell fate determination in Arabidopsis. Plant Signal. Behav. 2012, 7, 1556–1560. [Google Scholar] [CrossRef] [Green Version]
- Menand, B.; Yi, K.; Jouannic, S.; Hoffmann, L.; Ryan, E.; Linstead, P.; Schaefer, D.G.; Dolan, L. An ancient mechanism controls the development of cells with a rooting function on land plants. Science 2007, 316, 1477–1480. [Google Scholar] [CrossRef] [Green Version]
- Yi, K.; Menand, B.; Bell, E.; Dolan, L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 2010, 42, 264–267. [Google Scholar] [CrossRef]
- Bruex, A.; Kainkaryam, R.M.; Wieckowski, Y.; Kang, Y.H.; Bernhardt, C.; Xia, Y.; Zheng, X.; Wang, J.Y.; Lee, M.M.; Benfey, P.; et al. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 2012, 8, e1002446. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Zhang, M.; Yang, M.; Hu, Y. Arabidopsis JAZ proteins interact with and suppress RHD6 transcription factor to regulate jasmonate-stimulated root hair development. Plant Cell 2020, 32, 1049–1062. [Google Scholar] [CrossRef]
- Lin, Q.; Ohashi, Y.; Kato, M.; Tsuge, T.; Gu, H.; Qu, L.J.; Aoyama, T. GLABRA2 directly suppresses basic helix-loop-helix transcription factor genes with diverse functions in root hair development. Plant Cell 2015, 27, 2894–2906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masucci, J.D.; Rerie, W.G.; Foreman, D.R.; Zhang, M.; Galway, M.E.; Marks, M.D.; Schiefelbein, J.W. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 1996, 122, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, C.; Lee, M.M.; Gonzalez, A.; Zhang, F.; Lloyd, A.; Schiefelbein, J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef] [Green Version]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Chen, C.L.; Cui, M.; Zhou, W.J.; Wu, H.L.; Ling, H.Q. Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Mol. Plant 2018, 11, 1166–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Wu, H.; Wang, N.; Li, J.; Zhao, W.; Du, J.; Wang, D.; Ling, H.Q. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008, 18, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Klatte, M.; Jakoby, M.; Bäumlein, H.; Weisshaar, B.; Bauer, P. Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 2007, 226, 897–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colangelo, E.P.; Guerinot, M.L. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 2004, 16, 3400–3412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakoby, M.; Wang, H.Y.; Reidt, W.; Weisshaar, B.; Bauer, P. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 2004, 577, 528–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.X.; Zhang, J.; Wang, D.W.; Ling, H.Q. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 2005, 15, 613–621. [Google Scholar] [CrossRef] [Green Version]
- Pitts, R.J.; Cernac, A.; Estelle, M. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 1999, 16, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, C.; Du, J.; Liu, H.; Cui, Y.; Zhang, Y.; He, Y.; Wang, Y.; Chu, C.; Feng, Z.; et al. Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 2012, 158, 790–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Liu, B.; Li, M.; Jin, H.; Wang, P.; Liu, J.; Xiong, F.; Wang, J.; Wang, H. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 2015, 27, 787–805. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; Zhang, H.; Li, X.; Ai, Q.; Yu, D. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 1743–1755. [Google Scholar] [CrossRef]
- Tissot, N.; Robe, K.; Gao, F.; Grant-Grant, S.; Boucherez, J.; Bellegrade, F.; Maghiaoui, A.; Marcelin, R.; Izquierdo, E.; Benhamed, M.; et al. Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. N. Phytol. 2019, 223, 1433–1446. [Google Scholar] [CrossRef]
- Long, T.A.; Tsukagoshi, H.; Busch, W.; Lahner, B.; Salt, D.E.; Benfey, P.N. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 2010, 22, 2219–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selote, D.; Samira, R.; Matthiadis, A.; Gillikin, J.W.; Long, T.A. Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol. 2015, 167, 273–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; Dubos, C. Transcriptional integration of plant responses to iron availability. J. Exp. Bot. 2021, 72, 2056–2070. [Google Scholar] [CrossRef] [PubMed]
- Martín-Barranco, A.; Spielmann, J.; Dubeaux, G.; Vert, G.; Enric Zelazny, E. Dynamic control of high-affinity iron uptake complex in root epidermal cells. Plant Physiol. 2020, 184, 1236–1250. [Google Scholar] [CrossRef]
- Tanabe, N.; Noshi, M.; Mori, D.; Nozawa, K.; Tamoi, M.; Shigeoka, S. The basic helix-loop-helix transcription factor, bHLH11 functions in the iron-uptake system in Arabidopsis thaliana. J. Plant Res. 2018, 132, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Kurbidaeva, A.; Ezhova, T.; Novokreshchenova, M. Arabidopsis thaliana ICE2 gene: Phylogeny, structural evolution and functional diversification from ICE1. Plant Sci. 2014, 229, 10–22. [Google Scholar] [CrossRef]
- Guan, Q.; Wu, J.; Yue, X.; Zhang, Y.; Zhu, J. A Nuclear calcium-sensing pathway is critical for gene regulation and salt stress tolerance in Arabidopsis. PLoS Genet. 2013, 9, e1003755. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. N. Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.; Köhler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.-I.; Asensi-Fabado, M.A.; Munne’-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuraba, Y.; Bülbül, S.; Piao, W.; Choi, G.; Paek, N. Arabidopsis EARLY FLOWERING3 increases salt tolerance by suppressing salt stress response pathways. Plant J. 2017, 92, 1106–1120. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Yi, H.; Choi, G.; Shin, B.; Song, P.S.; Choi, G. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 2003, 15, 2399–2407. [Google Scholar] [CrossRef] [Green Version]
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paik, I.; Kathare, P.K.; Kim, J.; Huq, E. Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 2017, 10, 1035–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huq, E.; Quail, P.H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002, 21, 2441–2450. [Google Scholar] [CrossRef] [Green Version]
- Fairchild, C.D.; Schumaker, M.A.; Quail, P.H. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000, 14, 2377–2391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roig-Villanova, I.; Bou, J.; Sorin, C.; Devlin, P.F.; Martínez-García, J.F. Identification of primary target genes of phytochrome signaling: Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 2006, 141, 85–96. [Google Scholar] [CrossRef] [Green Version]
- Roig-Villanova, I.; Bou-Torrent, J.; Galstyan, A.; Carretero-paulet, L.; Portolés, S.; Rodríguez-Concepción, M.; Martínez-García, J. Interaction of shade avoidance and auxin response: A role for two novel atypical bHLH proteins. EMBO J. 2007, 26, 4756–4767. [Google Scholar] [CrossRef] [Green Version]
- Hao, Y.; Oh, E.; Choi, G.; Liang, Z.; Wang, Z.Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 2012, 5, 688–697. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Kim, J.; Park, E.; Kim, J.; Kang, C.; Choi, G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004, 16, 3045–3058. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.; Sun, T.; Kamiya, Y.; Choi, G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis Seeds. Plant Cell 2007, 19, 1192–1208. [Google Scholar] [CrossRef] [Green Version]
- Huq, E.; Al-Sady, B.; Hudson, M.; Kim, C.; Apel, K.; Quail, P.H. PHYTOCHROME-INTERACTING FACTOR 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 2004, 305, 1937–1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, J.; Zhu, L.; Shen, H.; Huq, E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci USA 2008, 105, 9433–9438. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Khanna, R.; Carle, C.M.; Quail, P.H. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007, 145, 1043–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khanna, R.; Huq, E.; Kikis, E.A.; Al-Sady, B.; Lanzatella, C.; Quail, P.H. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 2004, 16, 3033–3044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leivar, P.; Monte, E.; Al-Sady, B.; Carle, C.; Storer, A.; Alonso, J.M.; Ecker, J.R.; Quail, P.H. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 2008, 20, 337–352. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-lbanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; José Manuel Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [Green Version]
- Schweizer, F.; Fernández-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Zhao, M.; Morohashi, K.; Hatlestad, G.; Grotewold, E.; Lloyd, A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 2008, 135, 1991–1999. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-Domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Huang, H.; Wu, D.; Yan, J.; Qi, Y.; Song, S.; Xie, D. Arabidopsis DELLA and JAZ proteins bind the WD-Repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 2014, 26, 1118–1133. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, H.; Ai, Q.; Liang, G.; Yu, D. Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2478–2493. [Google Scholar] [CrossRef] [Green Version]
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Varaud, E.; Brioudes, F.; Szécsi, J.; Leroux, J.; Brown, S.; Perrot-Rechenmann, C.; Bendahmane, M. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 2011, 23, 973–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, K.; Wang, Y.; Zhang, N.; Jia, Q.; Wang, X.; Hou, C.; Chen, J.; Wang, S. Involvement of PACLOBUTRAZOL RESISTANCE6/KIDARI, an atypical bHLH transcription factor, in auxin responses in Arabidopsis. Front. Plant Sci. 2017, 8, 1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Ji, X.; Nie, X.; Qu, M.; Zheng, L.; Tan, Z.; Zhao, H.; Huo, L.; Liu, S.; Zhang, B.; et al. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. N. Phytol. 2015, 207, 692–709. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhu, J.; Lu, Y. Overexpression of AtbHLH112 suppresses lateral root emergence in Arabidopsis. Funct. Plant Biol. 2014, 41, 342–352. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kim, H.Y. Molecular characterization of a bHLH transcription factor involved in Arabidopsis abscisic acid-mediated response. BBA Gene Struct. Expr. 2006, 1759, 191–194. [Google Scholar] [CrossRef]
- Li, H.; Sun, J.; Xu, Y.; Jiang, H.; Wu, X.; Li, C. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol. Biol. 2007, 65, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Guo, H.; Dai, X.; Cheng, Y.; Zheng, K.; Wang, X.; Wang, S. An ABA down-regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis. Sci. Rep. 2015, 5, 17587. [Google Scholar] [CrossRef] [Green Version]
- Zheng, K.; Wang, Y.; Wang, S. The non-DNA binding bHLH transcription factor Paclobutrazol Resistances are involved in the regulation of ABA and salt responses in Arabidopsis. Plant Physiol. Biochem. 2019, 139, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.E.; Moreno-Piovano, G.; Chan, R.L. The antagonistic basic helix-loop-helix partners BEE and IBH1 contribute to control plant tolerance to abiotic stress. Plant Sci. 2018, 271, 143–150. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, B.; Deyholos, M.K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genomics. 2009, 282, 503–516. [Google Scholar] [CrossRef]
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, E.; Zhu, J.; Bai, M.Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 2014, 3, e03031. [Google Scholar] [CrossRef]
- Mara, C.D.; Huang, T.; Irish, V.F. The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 2010, 22, 690–702. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Bai, M.Y.; Wu, J.; Zhu, J.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. ATBS1 INTERACTING FACTORs negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signal. Behav. 2013, 8, e23448. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Gao, Y.; Liu, Y.; Zhang, X.; Gu, X.; Ma, D.; Zhao, Z.; Yuan, Z.; Xue, H.; Liu, H. BES1-regulated BEE1 controls photoperiodic flowering downstream of blue light signaling pathway in Arabidopsis. N. Phytol. 2019, 223, 1407–1419. [Google Scholar] [CrossRef]
- Malinovsky, F.G.; Batoux, M.; Schwessinger, B.; Youn, J.H.; Stransfeld, L.; Win, J.; Kim, S.K.; Zipfel, C. Antagonistic regulation of growth and immunity by the arabidopsis basic helix-loop-helix transcription factor HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH INCREASED LEAF INCLINATION1 BINDING bHLH1. Plant Physiol. 2014, 164, 1443–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Cifuentes-Esquivel, N.; Bou-Torrent, J.; Galstyan, A.; Gallemí, M.; Sessa, G.; Salla Martret, M.; Roig-Villanova, I.; Ruberti, I.; Martínez-García, J.F. The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 2013, 75, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Gao, L.; Wang, H.; Chen, X.; Wang, Y.; Yang, H.; Wei, C.; Wan, X.; Xia, T. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct. Integr. Genomic 2013, 13, 75–98. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Morohashi, K.; Zhao, M.; Yang, M.; Read, B.; Lloyd, A.; Lamb, R.; Grotewold, E. Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 2007, 145, 736–746. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Xu, P.; Li, B.; Li, P.; Wen, X.; An, F.; Gong, Y.; Xin, Y.; Zhu, Z.; Wang, Y.; et al. Ethylene promoters root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 13834–13839. [Google Scholar] [CrossRef] [Green Version]
- Ohashi-Ito, K.; Bergmann, D.C. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 2006, 18, 2493–2505. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Cui, Y.; Liu, Y.; Fan, H.; Du, J.; Huang, Z.; Yuan, Y.; Wu, H.; Ling, H. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 2013, 6, 503–513. [Google Scholar] [CrossRef] [Green Version]
- Lei, R.; Li, Y.; Cai, Y.; Li, C.; Pu, M.; Lu, C.; Yang, Y.; Liang, G. bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining Fe homestasis in Arabidopsis. Mol. Plant 2020, 13, 634–649. [Google Scholar] [CrossRef]
- Ivanov, R.; Brumbarova, T.; Bauer, P. Fitting into the harsh reality: Regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 2012, 5, 27–42. [Google Scholar] [CrossRef] [Green Version]
- Matthiadis, A.; Long, T.A. Further insight into BRUTUS domain composition and functionality. Plant Signal. Behav. 2016, 11, 8. [Google Scholar] [CrossRef] [Green Version]
- Hindt, M.N.; Akmakjian, G.Z.; Pivarski, K.L.; Punshon, T.; Baxter, I.; Salt, D.E.; Guerinot, M.L. BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 2017, 9, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Eide, D.; Broderius, M.; Fett, J.; Guerinot, M.L. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA. 1996, 93, 5624–5628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briata, J.F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillon, A.; Shen, H.; Huq, E. Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007, 12, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brumbarova, T.; Ivanov, R. The nutrient response transcriptional regulome of Arabidopsis. Iscience 2019, 19, 358–368. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhu, Z.; An, F.; Hao, D.; Li, P.; Song, J.; Yi, C.; Guo, H. Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 2014, 26, 1105–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, J.; Giraudat, J. Abscisic acid signal transduction. Annu. Rev. Plant Phys. 1998, 49, 199–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCourt, P. Genetic analysis of hormone signaling. Annu Rev. Plant Phys. 1999, 50, 219–243. [Google Scholar] [CrossRef] [Green Version]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzo, O.; Chico, J.M.; Saénchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef] [Green Version]
- Yadav, V.; Mallappa, C.; Gangappa, S.N.; Bhatia, S.; Chattopadhyay, S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 2005, 17, 1953–1966. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Ebisu, Y.; Kinoshita, T.; Doi, M.; Okuma, E.; Murata, Y.; Shimazaki, K. bHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal. 2013, 6, ra48. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Ebisu, Y.; Shimazaki, K. Reconstitution of abscisic acid signaling from the receptor to DNA via bHLH transcription factors. Plant Physiol. 2017, 174, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Ruzinova, M.B.; Benezra, R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003, 13, 410–418. [Google Scholar] [CrossRef]
- Hyun, Y.; Lee, I. KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 2006, 61, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Fujiwara, S.; Mitsuda, N.; Ohme-Takagi, M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 2012, 24, 4483–4497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruit. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.; Kim, K.; Kim, M.E.; Kim, H.G.; Heo, G.S.; Park, O.K.; Park, Y.-I.I.; Choi, G.; Oh, E. Phytochrome and ethylene signaling integration in Arabidopsis occurs via the transcriptional regulation of genes co-targeted by PIFs and EIN3. Front. Plant Sci. 2016, 7, 1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
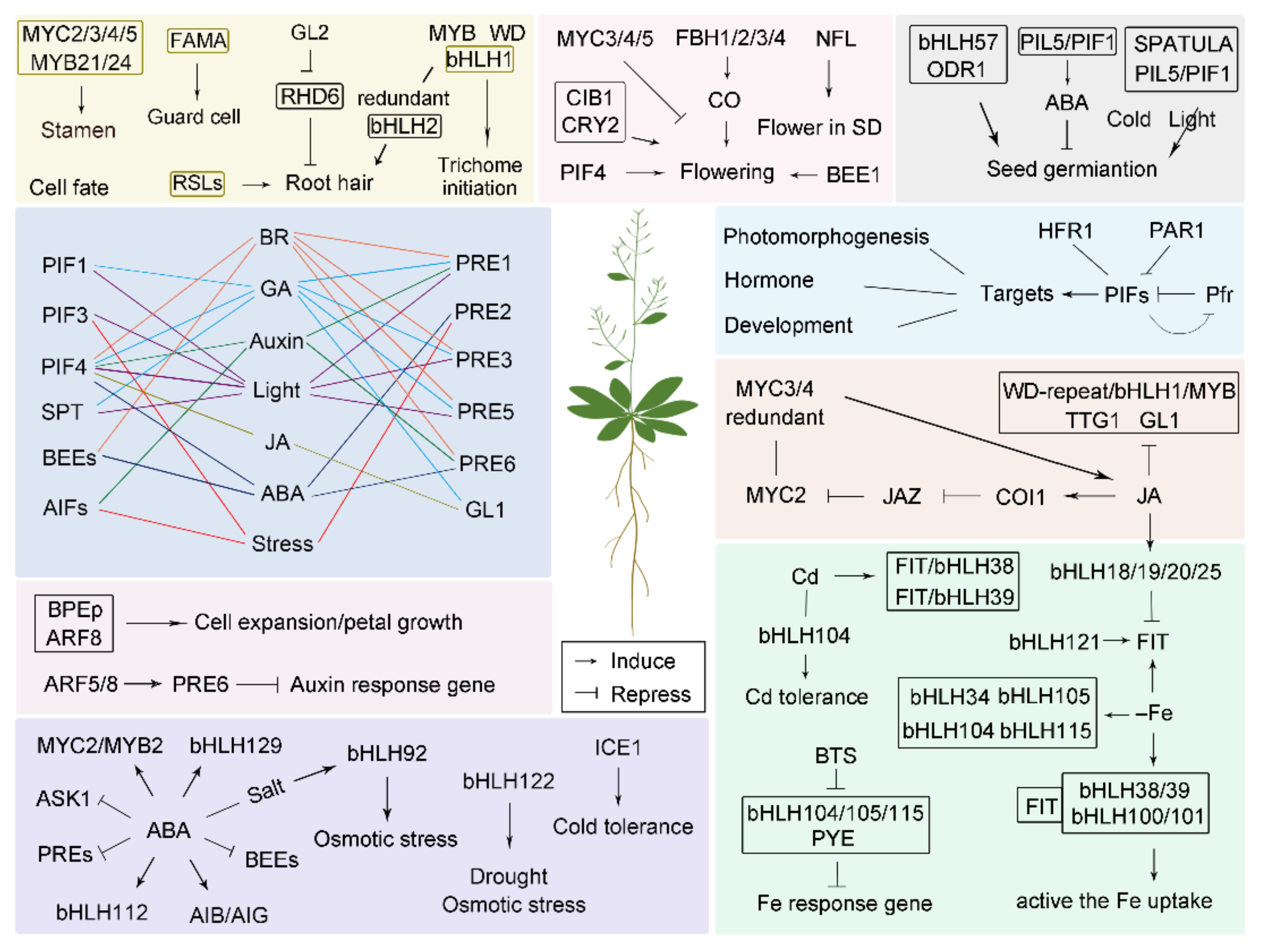
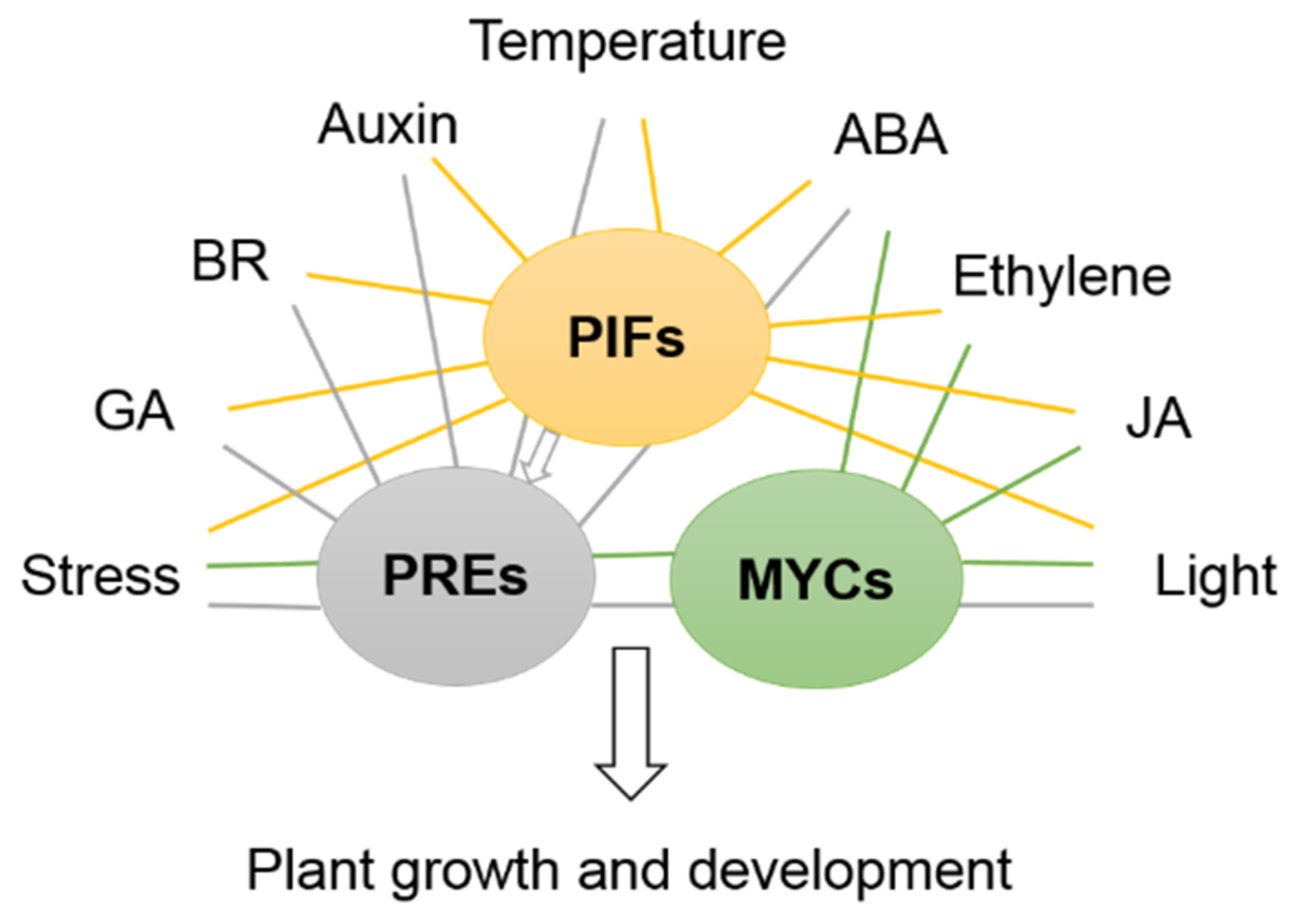
| Pathway | AGI Gene Code | Generic Name | Synonym | Functions Characterized | Group | Reference |
|---|---|---|---|---|---|---|
| Seed germination | At4g01460 | bHLH57 | Involved in seed dormancy process | Ia | [22,23] | |
| At2g20180 | bHLH15 | PIF1/PIL5 | Negative regulator of phytochrome-mediated seed germination | VIIa | [24,25,26] | |
| At4g36930 | bHLH24 | SPT | Reduced seed dormancy | VIIb | [27,28] | |
| Flowering | At1g35460 | bHLH80 | FBH1 | Regulate the effect of CO flowering time | IX | [29] |
| At4g34530 | bHLH63 | CIB1 | Promote CRY2-dependent floral initiation | XII | [30,31,32] | |
| At5g65640 | bHLH93 | NFL | Involved in GA mediated control of flowering time | IIIb | [33] | |
| At1g32640 | bHLH6 | MYC2 | Required in the JA pathway for regulating flowering time | IIIe | [34] | |
| At5g46760 | bHLH5 | MYC3 | Required in the JA pathway for regulating flowering time | IIIe | [34] | |
| At4g17880 | bHLH4 | MYC4 | Required in the JA pathway for regulating flowering time | IIIe | [34] | |
| At1g18400 | bHLH44 | BEE1 | Regulate photoperiodic flowering | XII | [35] | |
| At2g43010 | bHLH9 | PIF4 | Accelerate the flowering by activating FT at high temperature | XII | [36] | |
| At4g36930 | bHLH24 | SPT | Play a role in floral morphogenesis processes | VIIa | [37] | |
| Cell fate | At1g66470 | bHLH83 | RHD6 | ROOT HAIR DEFECTIVE6 | VIIb | [13,38,39] |
| At5g37800 | bHLH86 | RSL1 | Partially redundant and involved in root hair development | VIIIc | [40] | |
| At4g33880 | bHLH85 | RSL2 | Partially redundant and involved in root hair development | VIIIc | [40] | |
| At2g14760 | bHLH84 | RSL3 | RHD6-LIKE 3, required for root-hair growth | VIIIc | [40,41,42] | |
| At1g27740 | bHLH54 | RSL4 | Promote postmitotic cell growth in root-hair cells | VIIIc | [40,41,42] | |
| At5g58010 | bHLH82 | LRL3 | Regulate root hair development. | XI | [40,41,42] | |
| At5g41315 | bHLH1 | GL3 | Trigger the trichome initiation pathways | IIIf | [41,43,44] | |
| At1g63650 | bHLH2 | EGL3 | Regulate trichome and root hair development | IIIf | [45,46] | |
| At5g46830 | bHLH28 | MYC5 | Calcium-binding transcription factor involved in salt stress signaling | IIIe | [47] | |
| At1g32640 | bHLH6 | MYC2 | Positive regulator of lateral root formation. | IIIe | [47] | |
| At5g46760 | bHLH5 | MYC3 | Form the bHLH-MYB complex to regulate the stamen development | IIIe | [47] | |
| At4g17880 | bHLH4 | MYC4 | form the bHLH-MYB complex to regulate the stamen development | IIIe | [47] | |
| At3g24140 | bHLH97 | FAMA | Promote differentiation of stomatal guard cells | Ia | [48] | |
| Plant mineral nutrition and abiotic stress | At3g56970 | bHLH38 | ORG2 | Regulate the Fe-deficiency response | Ib | [49] |
| At3g56980 | bHLH39 | ORG3 | Regulate the Fe-deficiency response | Ib | [49] | |
| At2g41240 | bHLH100 | A key regulator of iron-deficiency responses | Ib | [50] | ||
| At5g04150 | bHLH101 | A key regulator of iron-deficiency responses | Ib | [50] | ||
| At2g28160 | bHLH29 | FIT | Regulate iron uptake responses | IIIa | [49,50,51,52,53] | |
| At3g19860 | bHLH121 | URI | Act as an essential part of the iron deficiency signaling pathway | IVb | [54] | |
| At3g23210 | bHLH34 | IDT1 | Involved in Fe regulation. | IVc | [55,56,57,58] | |
| At4g14410 | bHLH104 | Positively regulate Fe deficiency response | IVc | [55,56,57,58] | ||
| At5g54680 | bHLH105 | ILR3 | Plays an important role in Fe homeostasis | IVc | [55,56,57,58] | |
| At1g51070 | bHLH115 | Involved in response to Fe | IVc | [55,56,57,58] | ||
| At3g47640 | bHLH47 | PYE | Regulate response to iron deficiency in Arabidopsis roots | IVb | [59,60,61,62] | |
| At4g36060 | bHLH11 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein | IVb | [63] | ||
| Stress response | At3g26744 | bHLH116 | ICE1 | Improve cold tolerance through an ABA independent pathway | IIIb | [64] |
| At3g06590 | bHLH148 | AIF2/RITF1 | Involved in the detoxification of ROS which generated by salt stress | Orphans | [65] | |
| At1g61660 | bHLH122 | Mediate multiple response to improve stress tolerance | IX | [66,67] | ||
| At2g43010 | bHLH9 | PIF4 | Accelerate the flowering by activating FT at high temperature | XII | [68,69,70] | |
| Light signaling | At2g46970 | bHLH124 | PIL1 | Associated with APRR1/TOC1 and is a member of PIF3 family | VIIa | [24,25] |
| At3g59060 | bHLH65 | PIF5/PIL6 | Involved in shade avoidance | VIIa | [24,25] | |
| At3g62090 | bHLH132 | PIF6/PIL2 | Associated with APRR1/TOC1 and is a member of PIF3 family | VIIa | [24,25] | |
| At1g09530 | bHLH8 | PIF3 | Interact with photoreceptors phyA and phyB. | VIIa | [24,71,72] | |
| At2g43010 | bHLH9 | PIF4 | Interact with active PhyB protein | VIIa | [24,25,73,74] | |
| At1g02340 | bHLH26 | HFR1 | Involved in phytochrome signaling | VIIb | [75] | |
| At2g42870 | bHLH165 | PAR1 | Control plant development and as a negative regulator of SAS | Orphans | [76,77,78] | |
| At3g58850 | bHLH166 | PAR2 | Control plant development and as a negative regulator of SAS | Orphans | [76,77,78] | |
| At2g20180 | bHLH15 | PIF1/PIL5 | A key negative regulator of phytochrome-mediated response | VIIa | [79,80,81,82] | |
| At5g61270 | bHLH72 | PIF7 | Interacts specifically with Pfr form of phyB | VIIb | [83,84,85] | |
| JA signaling | At1g32640 | bHLH6 | MYC2 | Regulates diverse JA-dependent functions | IIIe | [86,87] |
| At5g46760 | bHLH5 | MYC3 | Act together with MYC2 and MYC4 to activate JA-responses | IIIe | [86,87] | |
| At4g17880 | bHLH4 | MYC4 | Act together with MYC2 and MYC3 to activate JA-responses | IIIe | [86,87] | |
| At5g41315 | bHLH1 | GL3 | Repressed by JAZs | IIIf | [88,89,90,91] | |
| At2g22750 | bHLH18 | Induced by JA and inhibit the transcription of the FIT | IVa | [92] | ||
| At2g22760 | bHLH19 | Induced by JA and inhibit the transcription of the FIT | IVa | [92] | ||
| At2g22770 | bHLH20 | NAI1 | Induced by JA and inhibit the transcription of the FIT | IVa | [92] | |
| At4g37850 | bHLH25 | Induced by JA and inhibit the transcription of the FIT | IVa | [92] | ||
| IAA signaling | At1g59640 | bHLH31 | BPEp | Involved in the control of petal size | XII | [93,94] |
| At1g26945 | bHLH163 | PRE6 | Involved in ABA and salts responses | XV | [93,95] | |
| ABA signaling | At1g61660 | bHLH112 | Mediate multiple response to improve stress tolerance | X | [96,97] | |
| At2g46510 | bHLH17 | AIB | Involved in response to ABA, repress MYC2-activated leaf senescence | IIId | [98,99] | |
| At1g32640 | bHLH6 | MYC2 | Induced by dehydration stress, ABA and blue light | IIIe | [17] | |
| At2g43140 | bHLH129 | Regulate root elongation and ABA response. | IX | [100] | ||
| PREs | Involved in the regulation of ABA and salt responses | XV | [101] | |||
| BEEs | Repressed by ABA and responses to abiotic stress | Va | [102] | |||
| At5g43650 | bHLH92 | Involved in salt and osmotic stress tolerance | IVd | [103] | ||
| Cross-talk between light and phyto-hormones | At5g39860 | bHLH136 | PRE1/BNQ1 | Mediate brassinosteroid regulation of cell elongation | XV | [25,104,105] |
| At5g15160 | bHLH134 | PRE2/BNQ3 | PHD finger family protein | XV | [95] | |
| At3g47710 | bHLH161 | PRE4/BNQ3 | Required for appropriate regulation of flowering time and regulating light responses. | XV | [106] | |
| At3g28857 | bHLH164 | PRE5 | Involved in the regulation of the light, GA, BR signaling pathways | XV | [25,104] | |
| At1g26945 | bHLH163 | PRE6/KIDARI | Interacts with HFR1 and negatively regulates its activity. | XV | [95] | |
| At2g43060 | bHLH158 | IBH1 | ILI1 binding bHLH 1 | Orphans | [107,108] | |
| At2g18300 | bHLH64 | HBI1 | Involved in positive regulation of cell elongation and proliferation | XII | [104] | |
| At2g43010 | bHLH9 | PIF4 | Negatively regulate phyB mediated responses and involved in SAS | VIIa | [25,104] | |
| At1g18400 | bHLH44 | BEE1 | Function in the early response to BRs | XII | [18,109] | |
| At4g36540 | bHLH58 | BEE2 | Function redundant with BEE1/3 | XII | [18,110] | |
| At1g73830 | bHLH50 | BEE3 | Function redundant with BEE1/2 | XII | [18] | |
| At5g08130 | bHLH46 | BIM1 | BES1-INTERACTING MYC-LIKE 1, involved in BRs signaling | Va | [111,112] | |
| At1g69010 | bHLH102 | BIM2 | Involved in brassinosteroid signaling and modulated SAS | Va | [111,112] | |
| At5g38860 | bHLH141 | BIM3 | Involved in brassinosteroid signaling and modulated SAS | Va | [111,112] | |
| Other aspects | At4g16430 | bHLH3 | JAM3 | Repress MYC2-activated leaf senescence, negatively regulate JA response | IIId | [47] |
| At1g01260 | bHLH13 | JAM2/MYC7E | Repress MYC2-activated leaf senescence, negatively regulate JA response | IIId | [47] | |
| At4g00870 | bHLH14 | Repress MYC2-activated leaf senescence, negatively regulate JA responses | IIId | [47] | ||
| At2g46510 | bHLH17 | AIB | Involved in response to ABA, repress MYC2-activated leaf senescence | IIId | [47] | |
| PIFs | Promoted leaf senescence | VIIa | [113] | |||
| MYCs | Positive regulator of Positively regulates flavonoid biosynthesis | IIIe | [67,114,115,116,117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. https://doi.org/10.3390/ijms22137152
Hao Y, Zong X, Ren P, Qian Y, Fu A. Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. International Journal of Molecular Sciences. 2021; 22(13):7152. https://doi.org/10.3390/ijms22137152
Chicago/Turabian StyleHao, Yaqi, Xiumei Zong, Pan Ren, Yuqi Qian, and Aigen Fu. 2021. "Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis" International Journal of Molecular Sciences 22, no. 13: 7152. https://doi.org/10.3390/ijms22137152
APA StyleHao, Y., Zong, X., Ren, P., Qian, Y., & Fu, A. (2021). Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. International Journal of Molecular Sciences, 22(13), 7152. https://doi.org/10.3390/ijms22137152





