Perturbation of the Actin Cytoskeleton in Human Hepatoma Cells Influences Interleukin-6 (IL-6) Signaling, but Not Soluble IL-6 Receptor Generation or NF-κB Activation
Abstract
:1. Introduction
2. Results
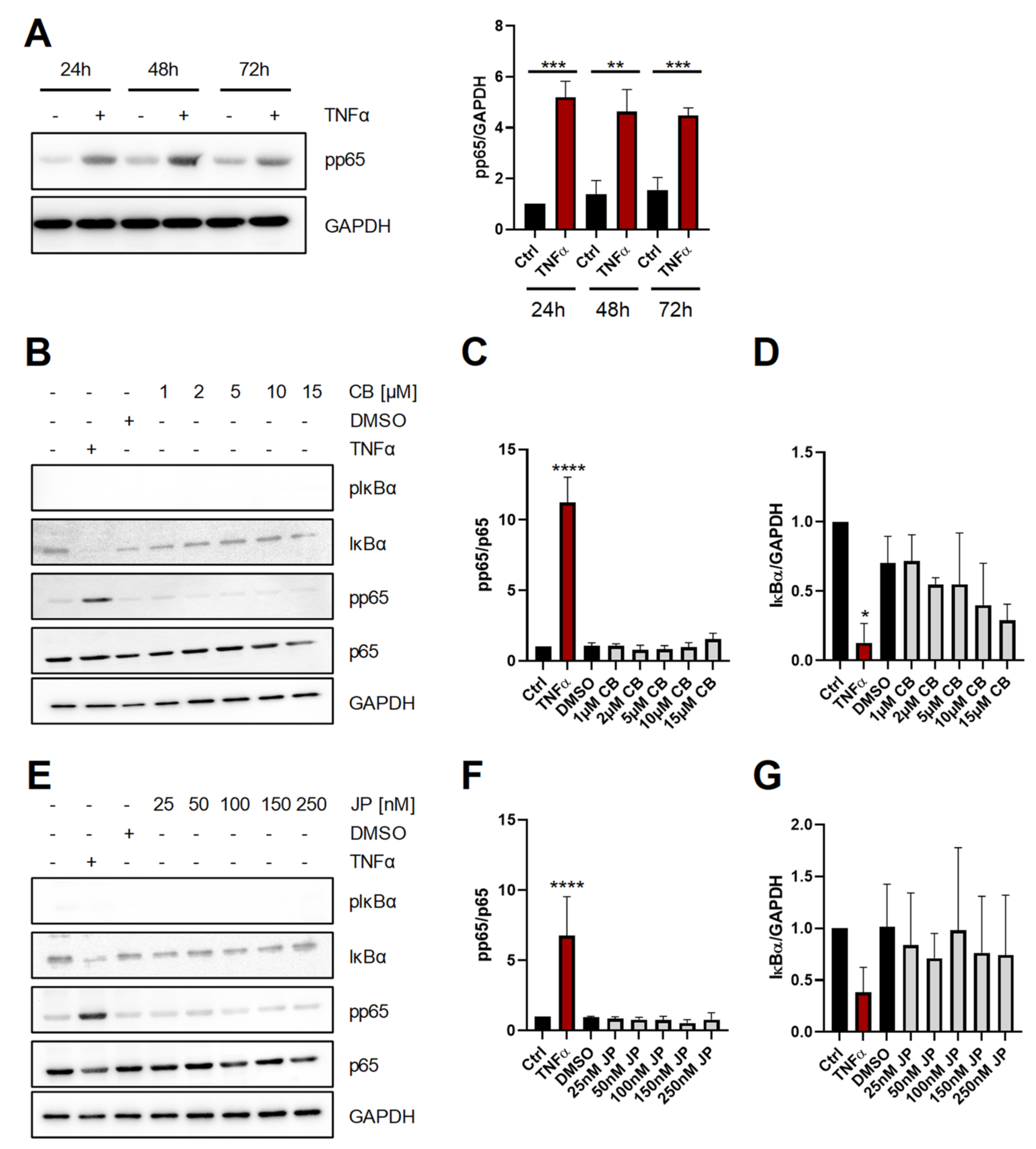
2.1. Short-Term Perturbation of the Actin Cytoskeleton Does Not Activate NF-κB
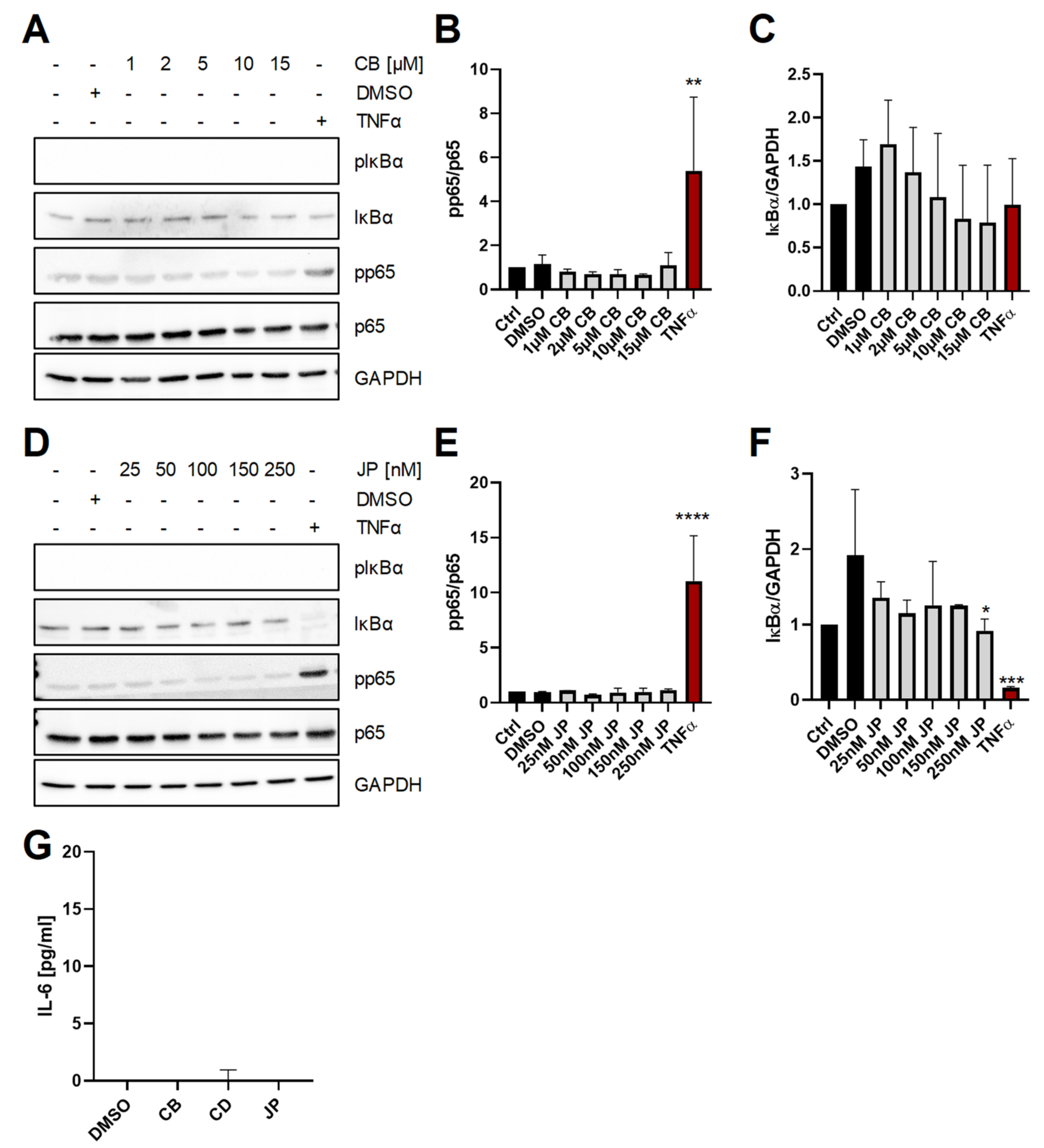
2.2. Long-Term Perturbation of the Actin Cytoskeleton Does Not Activate NF-κB or Induce IL-6 Production
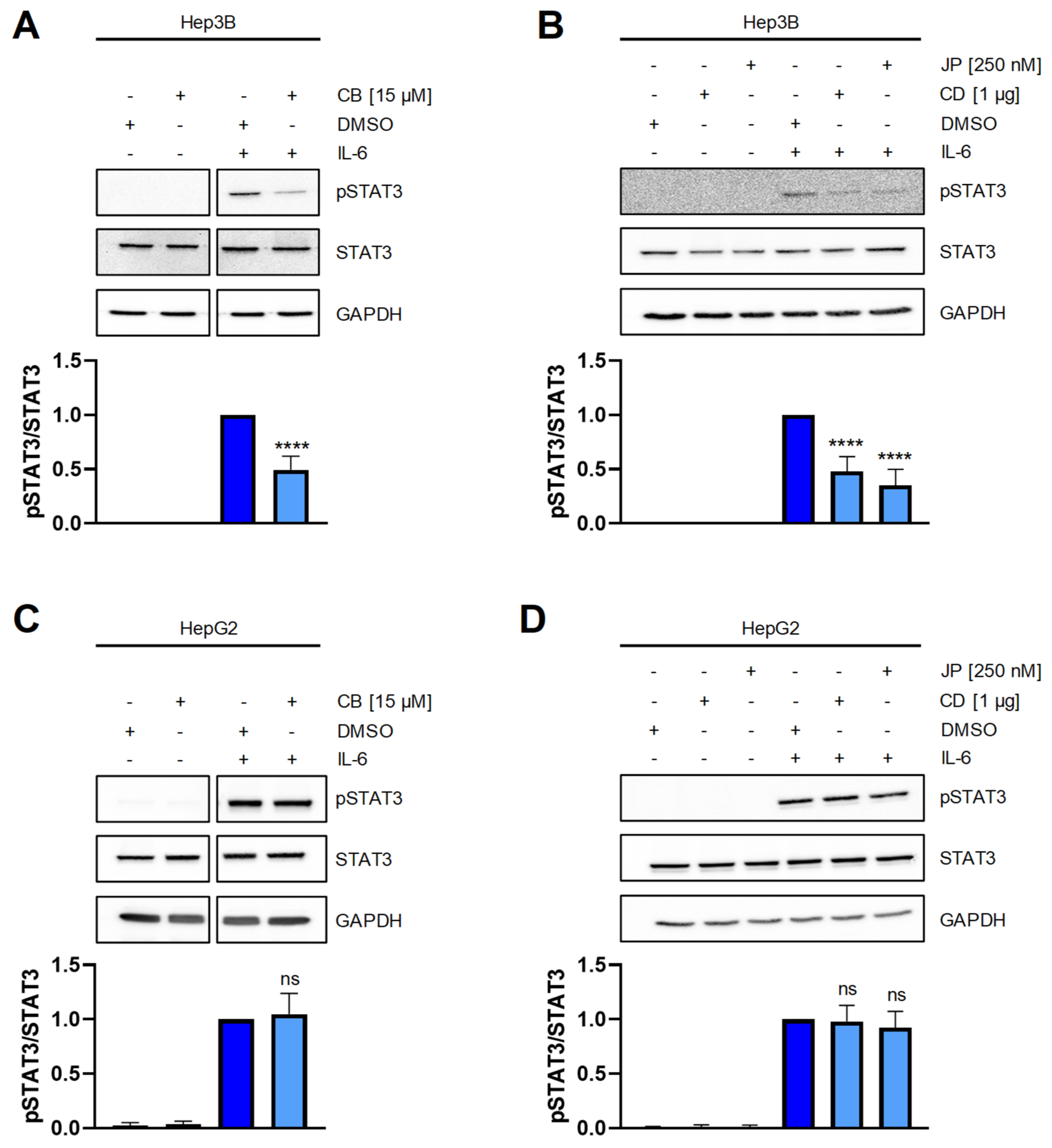
2.3. Perturbation of the Actin Cytoskeleton Interferes with STAT3 Signaling in Hep3B Cells
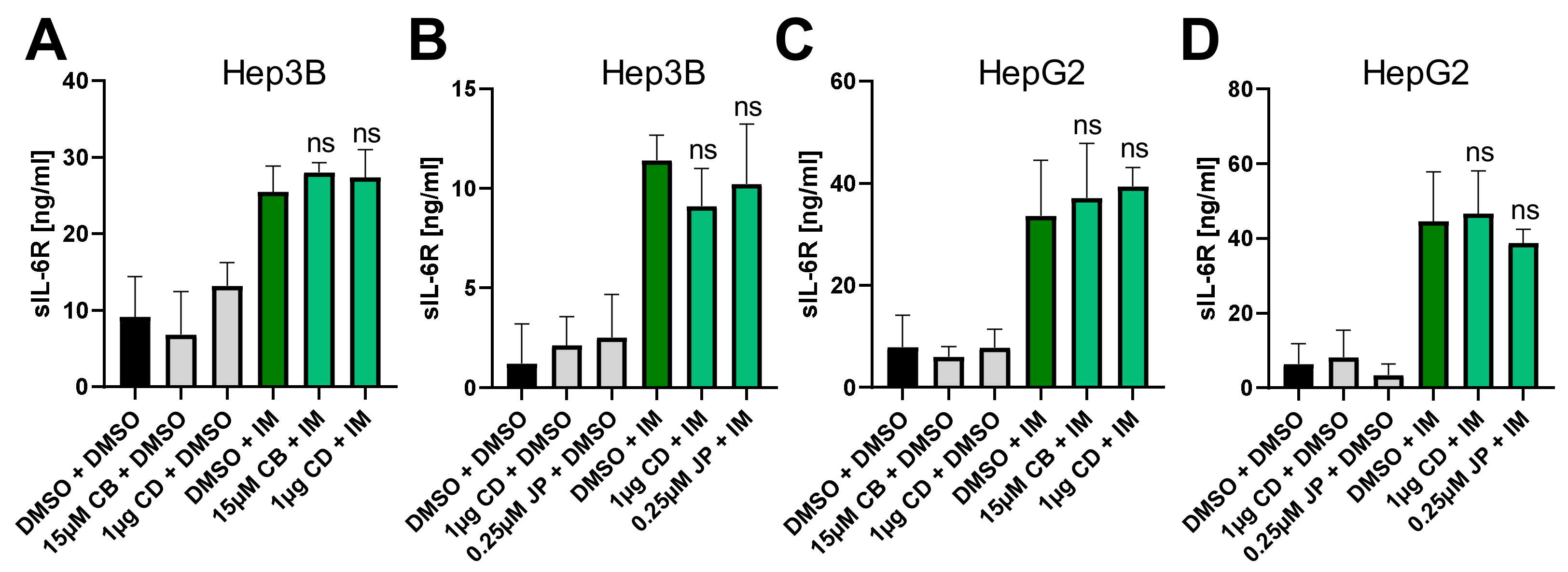
2.4. Perturbation of the Actin Cytoskeleton Does Not Influence IL-6R Proteolysis by ADAM10
3. Discussion
4. Materials and Methods
4.1. Cells Lines
4.2. Antibodies and Proteins
4.3. Stimulation and Lysis of Cells
4.4. Western Blotting and Densitometric Analysis
4.5. ELISA
4.6. Presentations of Experimental Data and Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garbers, C.; Hermanns, H.; Schaper, F.; Müller-Newen, G.; Grötzinger, J.; Rose-John, S.; Scheller, J. Plasticity and crosstalk of Interleukin 6-type cytokines. Cytokine Growth Factor Rev. 2012, 23, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Rose-John, S.; Garbers, C. Interleukin-6 and its receptors: A highly regulated and dynamic system. Cytokine 2014, 70, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Broser, M.; Rom, W.N. Activation of the interleukin 6 gene by Mycobacterium tuberculosis or lipopolysaccharide is mediated by nuclear factors NF-IL6 and NF-kappa B. Proc. Natl. Acad. Sci. USA 1994, 91, 2225–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal. Transduct Target. 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Garbers, C.; Aparicio-Siegmund, S.; Rose-John, S. The IL-6/gp130/STAT3 signaling axis: Recent advances towards specific inhibition. Curr. Opin. Immunol. 2015, 34, 75–82. [Google Scholar] [CrossRef]
- Garbers, C.; Heink, S.; Korn, T.; Rose-John, S. Interleukin-6: Designing specific therapeutics for a complex cytokine. Nat. Rev. Drug Discov. 2018, 17, 395–412. [Google Scholar] [CrossRef]
- Lokau, J.; Garbers, C. Biological functions and therapeutic opportunities of soluble cytokine receptors. Cytokine Growth Factor Rev. 2020, 55, 94–108. [Google Scholar] [CrossRef]
- Riethmueller, S.; Somasundaram, P.; Ehlers, J.C.; Hung, C.-W.W.; Flynn, C.M.; Lokau, J.; Agthe, M.; Düsterhöft, S.; Zhu, Y.; Grötzinger, J.; et al. Proteolytic Origin of the Soluble Human IL-6R In Vivo and a Decisive Role of N-Glycosylation. PLoS Biol. 2017, 15, e2000080. [Google Scholar] [CrossRef]
- Riethmueller, S.; Ehlers, J.C.; Lokau, J.; Düsterhöft, S.; Knittler, K.; Dombrowsky, G.; Grötzinger, J.; Rabe, B.; Rose-John, S.; Garbers, C. Cleavage Site Localization Differentially Controls Interleukin-6 Receptor Proteolysis by ADAM10 and ADAM17. Sci. Rep. 2016, 6, 25550. [Google Scholar] [CrossRef]
- Matthews, V.; Schuster, B.; Schutze, S.; Bussmeyer, I.; Ludwig, A.; Hundhausen, C.; Sadowski, T.; Saftig, P.; Hartmann, D.; Kallen, K.J.; et al. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE). J. Biol. Chem. 2003, 278, 38829–38839. [Google Scholar] [CrossRef] [Green Version]
- Garbers, C.; Jänner, N.; Chalaris, A.; Moss, M.L.; Floss, D.M.; Meyer, D.; Koch-Nolte, F.; Rose-John, S.; Scheller, J. Species specificity of ADAM10 and ADAM17 proteins in interleukin-6 (IL-6) trans-signaling and novel role of ADAM10 in inducible IL-6 receptor shedding. J. Biol. Chem. 2011, 286, 14804–14811. [Google Scholar] [CrossRef] [Green Version]
- Flynn, C.M.; Garbers, Y.; Lokau, J.; Wesch, D.; Schulte, D.M.; Laudes, M.; Lieb, W.; Aparicio-Siegmund, S.; Garbers, C. Activation of Toll-like Receptor 2 (TLR2) induces Interleukin-6 trans-signaling. Sci. Rep. 2019, 9, 7306. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, S.; Aden, K.; Bernardes, J.P.; Conrad, C.; Tran, F.; Hoper, H.; Volk, V.; Mishra, N.; Blase, J.I.; Nikolaus, S.; et al. Therapeutic IL-6 trans-signalling inhibition by olamkicept (sgp130Fc) in patients with active inflammatory bowel disease. Gastroenterology 2021. [Google Scholar] [CrossRef]
- Papakonstanti, E.A.; Stournaras, C. Tumor necrosis factor-alpha promotes survival of opossum kidney cells via Cdc42-induced phospholipase C-gamma1 activation and actin filament redistribution. Mol. Biol. Cell 2004, 15, 1273–1286. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, A.; Blaas, K. Actin-Dynamics in Plant Cells: The Function of Actin-Perturbing Substances: Jasplakinolide, Chondramides, Phalloidin, Cytochalasins, and Latrunculins. Methods Mol. Biol. 2016, 1365, 243–261. [Google Scholar] [CrossRef] [Green Version]
- Kustermans, G.; El Benna, J.; Piette, J.; Legrand-Poels, S. Perturbation of actin dynamics induces NF-kappaB activation in myelomonocytic cells through an NADPH oxidase-dependent pathway. Biochem. J. 2005, 387, 531–540. [Google Scholar] [CrossRef]
- Nemeth, Z.H.; Deitch, E.A.; Davidson, M.T.; Szabo, C.; Vizi, E.S.; Hasko, G. Disruption of the actin cytoskeleton results in nuclear factor-kappaB activation and inflammatory mediator production in cultured human intestinal epithelial cells. J. Cell Physiol. 2004, 200, 71–81. [Google Scholar] [CrossRef]
- Aparicio-Siegmund, S.; Deseke, M.; Lickert, A.; Garbers, C. Trans-signaling of interleukin-6 (IL-6) is mediated by the soluble IL-6 receptor, but not by soluble CD5. Biochem. Biophys. Res. Commun. 2017, 484, 808–812. [Google Scholar] [CrossRef]
- Scheller, J.; Chalaris, A.; Garbers, C.; Rose-John, S. ADAM17: A molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011, 32, 380–387. [Google Scholar] [CrossRef]
- Chalaris, A.; Garbers, C.; Rabe, B.; Rose-John, S.; Scheller, J. The soluble Interleukin 6 receptor: Generation and role in inflammation and cancer. Eur. J. Cell Biol. 2011, 90, 484–494. [Google Scholar] [CrossRef]
- Kalliolias, G.D.; Ivashkiv, L.B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 2016, 12, 49–62. [Google Scholar] [CrossRef]
- Rhee, S.; Lee, K.H.; Kwon, H.; Kang, M.-S. NF-ΚB Activation by Disruption of Microtubule Array during Myogenesis of L6 Cells. Korean J. Biol. Sci. 1997, 1, 63–69. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.J.; Isaacs, J.S.; Lee, S.; Trepel, J.; Neckers, L. Microtubule disruption utilizes an NFkappa B-dependent pathway to stabilize HIF-1alpha protein. J. Biol. Chem. 2003, 278, 7445–7452. [Google Scholar] [CrossRef] [Green Version]
- Peschon, J.; Slack, J.; Reddy, P.; Stocking, K.; Sunnarborg, S.; Lee, D.; Russell, W.; Castner, B.; Johnson, R.; Fitzner, J.; et al. An essential role for ectodomain shedding in mammalian development. Science 1998, 282, 1281–1284. [Google Scholar] [CrossRef]
- Le Gall, S.; Maretzky, T.; Issuree, P.; Niu, X.-D.; Reiss, K.; Saftig, P.; Khokha, R.; Lundell, D.; Blobel, C. ADAM17 is regulated by a rapid and reversible mechanism that controls access to its catalytic site. J. Cell Sci. 2010, 123, 3913–3922. [Google Scholar] [CrossRef] [Green Version]
- Brenner, B.; Gulbins, E.; Busch, G.L.; Koppenhoefer, U.; Lang, F.; Linderkamp, O. L-selectin regulates actin polymerisation via activation of the small G-protein Rac2. Biochem. Biophys. Res. Commun. 1997, 231, 802–807. [Google Scholar] [CrossRef]
- Ivetic, A.; Florey, O.; Deka, J.; Haskard, D.; Ager, A.; Ridley, A. Mutagenesis of the ezrin-radixin-moesin binding domain of L-selectin tail affects shedding, microvillar positioning, and leukocyte tethering. J. Biol. Chem. 2004, 279, 33263–33272. [Google Scholar] [CrossRef] [Green Version]
- Möller-Hackbarth, K.; Dewitz, C.; Schweigert, O.; Trad, A.; Garbers, C.; Rose-John, S.; Scheller, J. A disintegrin and metalloprotease (ADAM) 10 and ADAM17 are major sheddases of T cell immunoglobulin and mucin domain 3 (Tim-3). J. Biol. Chem. 2013, 288, 34529–34544. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Kuang, S.; Song, Q.L.; Liu, M.; Sun, X.X.; Yu, Q. Cucurbitacin I inhibits STAT3, but enhances STAT1 signaling in human cancer cells in vitro through disrupting actin filaments. Acta Pharm. Sin. 2018, 39, 425–437. [Google Scholar] [CrossRef] [Green Version]
- Qiu, G.H.; Xie, X.; Xu, F.; Shi, X.; Wang, Y.; Deng, L. Distinctive pharmacological differences between liver cancer cell lines HepG2 and Hep3B. Cytotechnology 2015, 67, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mackiewicz, A.; Schooltink, H.; Heinrich, P.C.; Rose-John, S. Complex of soluble human IL-6-receptor/IL-6 up-regulates expression of acute-phase proteins. J. Immunol. 1992, 149, 2021–2027. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgieva, E.; Leber, S.L.; Wex, C.; Garbers, C. Perturbation of the Actin Cytoskeleton in Human Hepatoma Cells Influences Interleukin-6 (IL-6) Signaling, but Not Soluble IL-6 Receptor Generation or NF-κB Activation. Int. J. Mol. Sci. 2021, 22, 7171. https://doi.org/10.3390/ijms22137171
Georgieva E, Leber SL, Wex C, Garbers C. Perturbation of the Actin Cytoskeleton in Human Hepatoma Cells Influences Interleukin-6 (IL-6) Signaling, but Not Soluble IL-6 Receptor Generation or NF-κB Activation. International Journal of Molecular Sciences. 2021; 22(13):7171. https://doi.org/10.3390/ijms22137171
Chicago/Turabian StyleGeorgieva, Elizabeta, Stefan L. Leber, Cora Wex, and Christoph Garbers. 2021. "Perturbation of the Actin Cytoskeleton in Human Hepatoma Cells Influences Interleukin-6 (IL-6) Signaling, but Not Soluble IL-6 Receptor Generation or NF-κB Activation" International Journal of Molecular Sciences 22, no. 13: 7171. https://doi.org/10.3390/ijms22137171
APA StyleGeorgieva, E., Leber, S. L., Wex, C., & Garbers, C. (2021). Perturbation of the Actin Cytoskeleton in Human Hepatoma Cells Influences Interleukin-6 (IL-6) Signaling, but Not Soluble IL-6 Receptor Generation or NF-κB Activation. International Journal of Molecular Sciences, 22(13), 7171. https://doi.org/10.3390/ijms22137171






