Genomics of Endometriosis: From Genome Wide Association Studies to Exome Sequencing
Abstract
1. Introduction
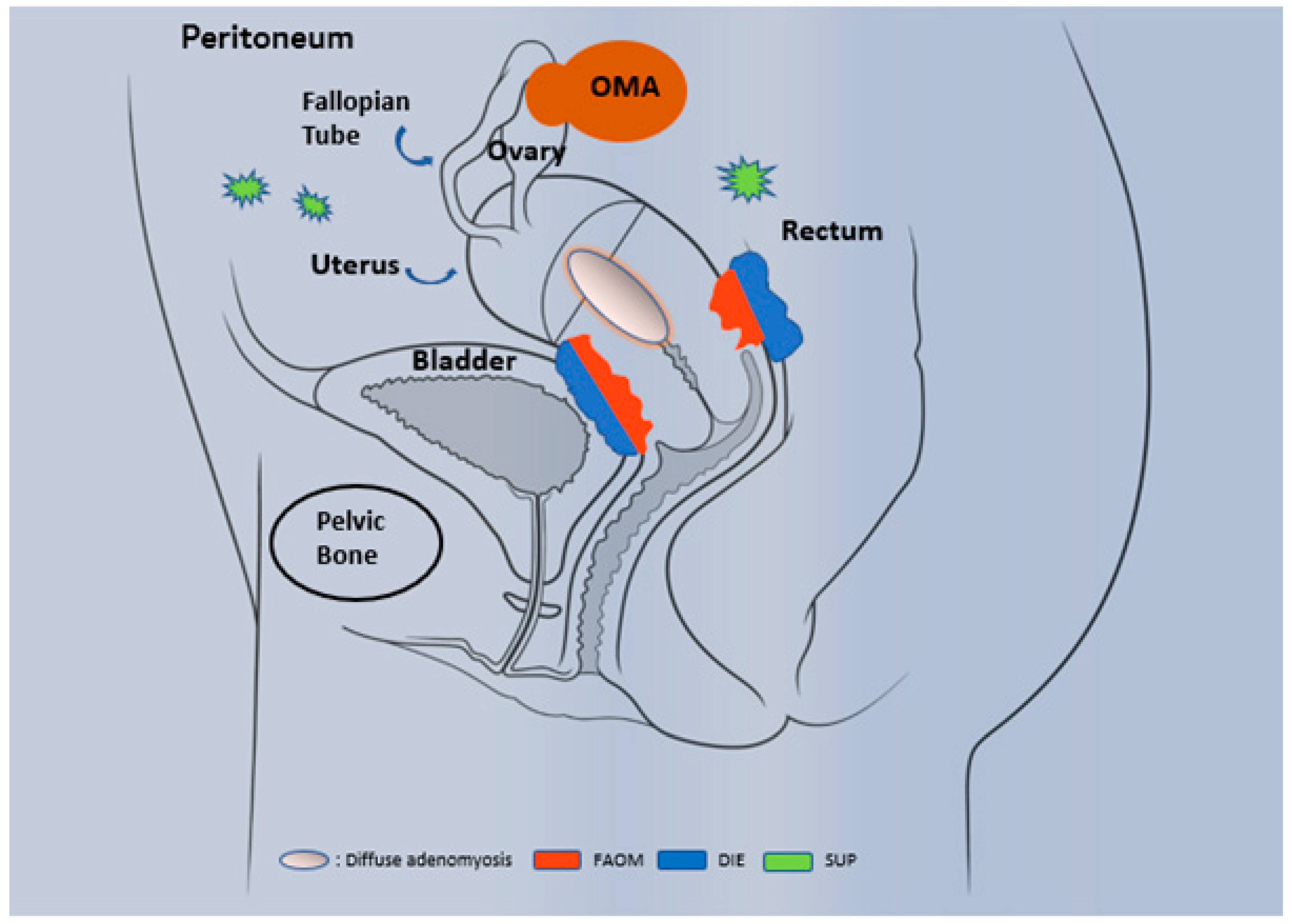
1.1. Epidemiology and Phenotypic Description
1.2. Diagnosis
1.3. Origin and Genetics
2. Procedure of This Review for the GWAS and Overview of the Results
3. Genome-wide Association Studies
3.1. Large GWAS from Heterogeneous Populations
3.2. GWAS Studies from Genetic Isolates
3.3. Small-scale GWAS and Pooling Approaches
3.4. Genetic Connections between Endometriosis and Other Diseases and Phenotypes
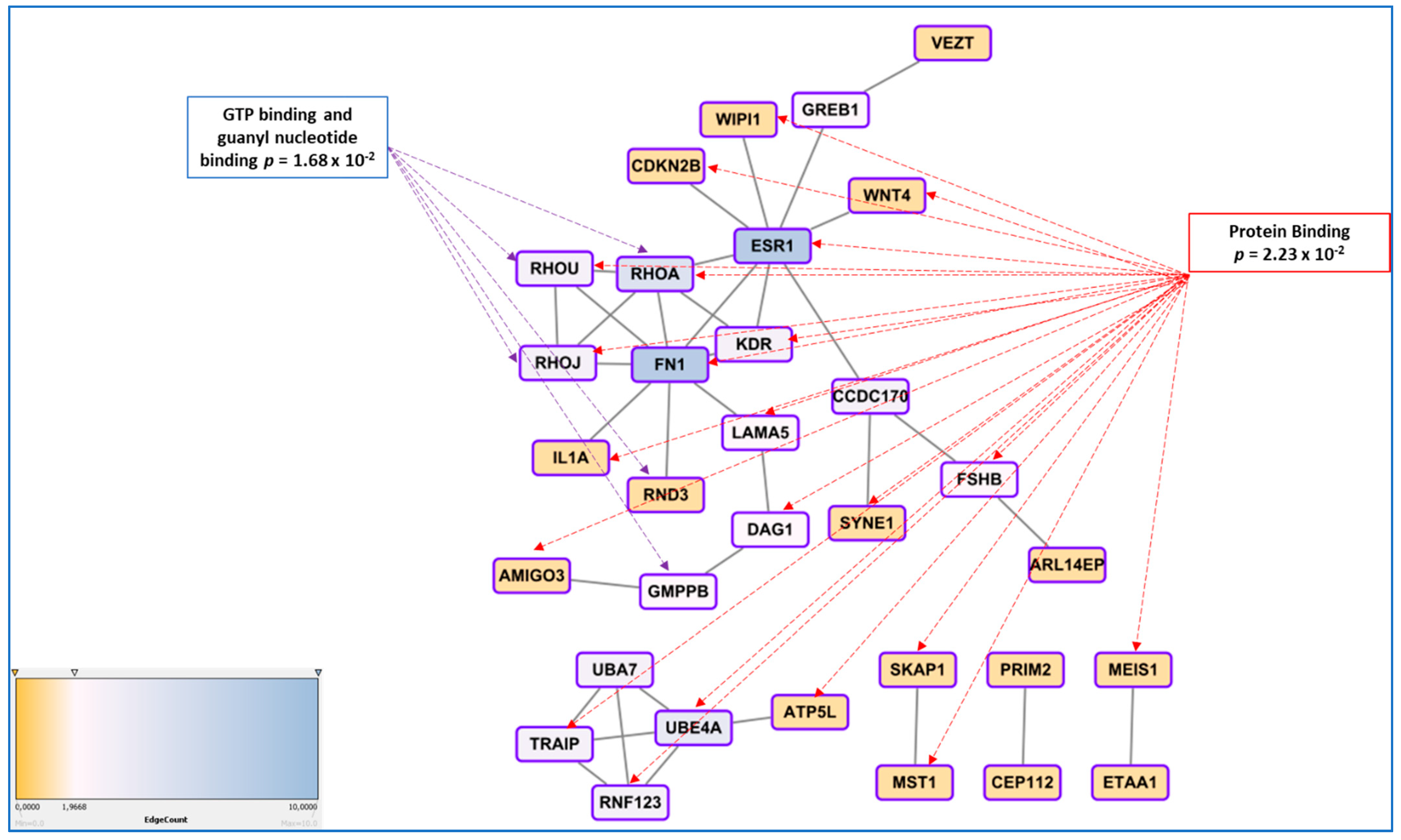
3.5. Genetic Regulation of Gene Expression and System Biology Analyses in Connection with GWAS Approaches
4. Replication Studies
5. Functional Studies
- In endometriosis, as in many complex diseases, there is a discrepancy between the calculated heritability and the sum of the SNP effects found in GWAS. This difference is explained by the concept of missing heritability [88]. More precisely, it has been estimated in 2012 that the total variation tagged by frequent SNPs as used in GWAS was 0.26, i.e., about half of the total genetic variation [55]. Missing heritability is currently explained by various hypotheses, one of the most prominent relies in the idea that GWAS are carried out using microarray platforms encompassing SNPs having a relatively high Minor Allele Frequency (MAF) and hence, will miss rare alleles that may be the one indeed associated to the disease. These rare alleles have presumably been counter-selected through the mechanisms of evolution. An indication of such possibility is provided in [41], where the authors focused on protein modifying variants analyzing 7164 cases and 21,005 controls for the discovery and 1840 cases and 129,016 controls in the replication cohort. The only locus that was replicated was GREB1 at2p25. Even in exome arrays designs, rare variants are not massively present, which could explain this relative failure. The authors logically conclude that sequencing high-risk families at the exome level is a promising way to identify novel rare variants in genes involved in endometriosis. Another major issue is the impact of inter-patient variability in the big GWAS approaches, suggesting that optimizing the clinical classification of the patients could improve the detection power of the GWAS [89,90]. Other explanations are the possible association of disease with CNVs (variations in length of large repeats), which are poorly addressed by classical arrays, epigenetics regulation that may also mask some of the gene-defined variation, as well as epistatic mechanisms that implies that for obtaining a tangible phenotype, the co-occurrence of two or more gene variants is requested [91].
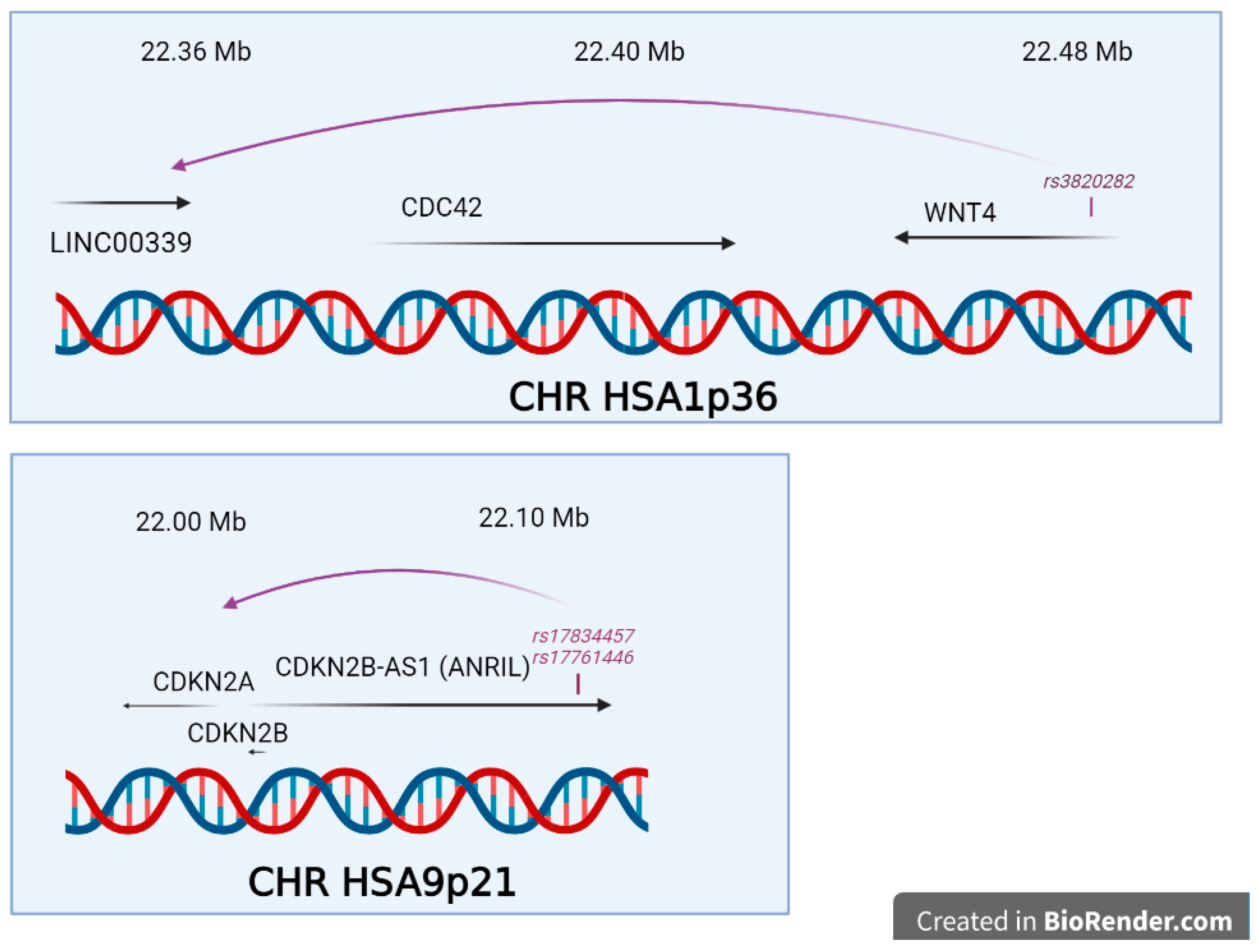
- Finding a significant SNPs does not explain how, functionally, this SNP triggers the disease risk, the situation being made even more difficult because the relative risk is generally comprised between 1.1 and 1.5, meaning that many carriers of the ‘at-risk’ variant are not stricken by the disease, while carriers of the ‘protective’ variant are not at all. This latest question was systematically addressed in a 2015 review by Fung and co-workers, and showed that once the variant is identified, a long and tedious stroll commences, from a refined mapping with additional SNPs in the region identified, a study of existing functional annotations (which is difficult when non-coding or intergenic SNPs are found, a case encountered for the chromosome 7 rs12700667 in endometriosis ), a measure of cell-type specific gene expression and protein levels, analysis of the cell-type specific local epigenetic regulation, cell models and animal models, eventually, if available, a complicated issue in the case of a human-specific disease such as endometriosis [92].
6. Exome Sequencing
6.1. Family Studies
6.2. Population Studies
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

References
- Gurates, B.; Bulun, S.E. Endometriosis: The ultimate hormonal disease. Semin. Reprod. Med. 2003, 21, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Borghese, B.; Zondervan, K.T.; Abrao, M.S.; Chapron, C.; Vaiman, D. Recent insights on the genetics and epigenetics of endometriosis. Clin. Genet. 2017, 91, 254–264. [Google Scholar] [CrossRef] [PubMed]
- de Ziegler, D.; Borghese, B.; Chapron, C. Endometriosis and infertility: Pathophysiology and management. Lancet 2010, 376, 730–738. [Google Scholar] [CrossRef]
- Pocate-Cheriet, K.; Santulli, P.; Kateb, F.; Bourdon, M.; Maignien, C.; Batteux, F.; Chouzenoux, S.; Patrat, C.; Wolf, J.P.; Bertho, G.; et al. The follicular fluid metabolome differs according to the endometriosis phenotype. Reprod. Biomed. Online 2020, 41, 1023–1037. [Google Scholar] [CrossRef]
- Revised American Fertility Society classification of endometriosis: 1985. Fertil. Steril. 1985, 43, 351–352. [CrossRef]
- Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil. Steril. 1997, 67, 817–821. [CrossRef]
- Zhang, Q.; Duan, J.; Olson, M.; Fazleabas, A.; Guo, S.W. Cellular Changes Consistent With Epithelial-Mesenchymal Transition and Fibroblast-to-Myofibroblast Transdifferentiation in the Progression of Experimental Endometriosis in Baboons. Reprod. Sci. 2016, 23, 1409–1421. [Google Scholar] [CrossRef]
- Zhang, Q.; Duan, J.; Liu, X.; Guo, S.W. Platelets drive smooth muscle metaplasia and fibrogenesis in endometriosis through epithelial-mesenchymal transition and fibroblast-to-myofibroblast transdifferentiation. Mol. Cell Endocrinol. 2016, 428, 1–16. [Google Scholar] [CrossRef]
- Gonzalez-Foruria, I.; Santulli, P.; Chouzenoux, S.; Carmona, F.; Chapron, C.; Batteux, F. Dysregulation of the ADAM17/Notch signalling pathways in endometriosis: From oxidative stress to fibrosis. Mol. Hum. Reprod. 2017, 23, 488–499. [Google Scholar] [CrossRef]
- Borghese, B.; Mondon, F.; Noel, J.C.; Fayt, I.; Mignot, T.M.; Vaiman, D.; Chapron, C. Gene expression profile for ectopic versus eutopic endometrium provides new insights into endometriosis oncogenic potential. Mol. Endocrinol. 2008, 22, 2557–2562. [Google Scholar] [CrossRef]
- Chapron, C.; Marcellin, L.; Borghese, B.; Santulli, P. Rethinking mechanisms, diagnosis and management of endometriosis. Nat. Rev. Endocrinol. 2019, 15, 666–682. [Google Scholar] [CrossRef]
- Sinaii, N.; Plumb, K.; Cotton, L.; Lambert, A.; Kennedy, S.; Zondervan, K.; Stratton, P. Differences in characteristics among 1,000 women with endometriosis based on extent of disease. Fertil. Steril. 2008, 89, 538–545. [Google Scholar] [CrossRef]
- Ballweg, M.L. Impact of endometriosis on women’s health: Comparative historical data show that the earlier the onset, the more severe the disease. Best Pract. Res. Clin. Obs. Gynaecol. 2004, 18, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Hurd, W.W. Criteria that indicate endometriosis is the cause of chronic pelvic pain. Obs. Gynecol. 1998, 92, 1029–1032. [Google Scholar] [CrossRef]
- Chapron, C.; Vercellini, P.; Barakat, H.; Vieira, M.; Dubuisson, J.B. Management of ovarian endometriomas. Hum. Reprod. Update 2002, 8, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Koninckx, P.R.; Meuleman, C.; Oosterlynck, D.; Cornillie, F.J. Diagnosis of deep endometriosis by clinical examination during menstruation and plasma CA-125 concentration. Fertil. Steril. 1996, 65, 280–287. [Google Scholar] [CrossRef]
- Guerriero, S.; Condous, G.; van den Bosch, T.; Valentin, L.; Leone, F.P.; Van Schoubroeck, D.; Exacoustos, C.; Installe, A.J.; Martins, W.P.; Abrao, M.S.; et al. Systematic approach to sonographic evaluation of the pelvis in women with suspected endometriosis, including terms, definitions and measurements: A consensus opinion from the International Deep Endometriosis Analysis (IDEA) group. Ultrasound Obs. Gynecol. 2016, 48, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, S.; Alcazar, J.L.; Pascual, M.A.; Ajossa, S.; Perniciano, M.; Piras, A.; Mais, V.; Piras, B.; Schirru, F.; Benedetto, M.G.; et al. Deep Infiltrating Endometriosis: Comparison between 2-Dimensional Ultrasonography (US), 3-Dimensional US, and Magnetic Resonance Imaging. J. Ultrasound Med. 2018, 37, 1511–1521. [Google Scholar] [CrossRef]
- Van den Bosch, T.; Van Schoubroeck, D. Ultrasound diagnosis of endometriosis and adenomyosis: State of the art. Best Pract. Res. Clin. Obs. Gynaecol. 2018, 51, 16–24. [Google Scholar] [CrossRef]
- Guerriero, S.; Saba, L.; Pascual, M.A.; Ajossa, S.; Rodriguez, I.; Mais, V.; Alcazar, J.L. Transvaginal ultrasound vs magnetic resonance imaging for diagnosing deep infiltrating endometriosis: Systematic review and meta-analysis. Ultrasound Obs. Gynecol. 2018, 51, 586–595. [Google Scholar] [CrossRef]
- Piketty, M.; Chopin, N.; Dousset, B.; Millischer-Bellaische, A.E.; Roseau, G.; Leconte, M.; Borghese, B.; Chapron, C. Preoperative work-up for patients with deeply infiltrating endometriosis: Transvaginal ultrasonography must definitely be the first-line imaging examination. Hum. Reprod. 2009, 24, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, M.O.; Podgaec, S.; Dias, J.A., Jr.; Gonzalez, M.; Abrao, M.S. Transvaginal ultrasonography with bowel preparation is able to predict the number of lesions and rectosigmoid layers affected in cases of deep endometriosis, defining surgical strategy. Hum. Reprod. 2010, 25, 665–671. [Google Scholar] [CrossRef]
- Nisenblat, V.; Bossuyt, P.M.; Farquhar, C.; Johnson, N.; Hull, M.L. Imaging modalities for the non-invasive diagnosis of endometriosis. Cochrane Database Syst. Rev. 2016, 2, CD009591. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.A. Metastatic or Embolic Endometriosis, due to the Menstrual Dissemination of Endometrial Tissue into the Venous Circulation. Am. J. Pathol. 1927, 3, 93–110, 143. [Google Scholar] [PubMed]
- Sarma, D.; Iyengar, P.; Marotta, T.R.; terBrugge, K.G.; Gentili, F.; Halliday, W. Cerebellar endometriosis. Ajr. Am. J. Roentgenol. 2004, 182, 1543–1546. [Google Scholar] [CrossRef]
- Treloar, S.A.; O’Connor, D.T.; O’Connor, V.M.; Martin, N.G. Genetic influences on endometriosis in an Australian twin sample. Fertil. Steril. 1999, 71, 701–710. [Google Scholar] [CrossRef]
- Saha, R.; Pettersson, H.J.; Svedberg, P.; Olovsson, M.; Bergqvist, A.; Marions, L.; Tornvall, P.; Kuja-Halkola, R. Heritability of endometriosis. Fertil. Steril. 2015, 104, 947–952. [Google Scholar] [CrossRef]
- Chettier, R.; Ward, K.; Albertsen, H.M. Endometriosis is associated with rare copy number variants. PLoS ONE 2014, 9, e103968. [Google Scholar] [CrossRef]
- Mafra, F.; Mazzotti, D.; Pellegrino, R.; Bianco, B.; Barbosa, C.P.; Hakonarson, H.; Christofolini, D. Copy number variation analysis reveals additional variants contributing to endometriosis development. J. Assist. Reprod. Genet. 2017, 34, 117–124. [Google Scholar] [CrossRef]
- Uno, S.; Zembutsu, H.; Hirasawa, A.; Takahashi, A.; Kubo, M.; Akahane, T.; Aoki, D.; Kamatani, N.; Hirata, K.; Nakamura, Y. A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat. Genet. 2010, 42, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Adachi, S.; Tajima, A.; Quan, J.; Haino, K.; Yoshihara, K.; Masuzaki, H.; Katabuchi, H.; Ikuma, K.; Suginami, H.; Nishida, N.; et al. Meta-analysis of genome-wide association scans for genetic susceptibility to endometriosis in Japanese population. J. Hum. Genet. 2010, 55, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Painter, J.N.; Anderson, C.A.; Nyholt, D.R.; Macgregor, S.; Lin, J.; Lee, S.H.; Lambert, A.; Zhao, Z.Z.; Roseman, F.; Guo, Q.; et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat. Genet. 2011, 43, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Nyholt, D.R.; Low, S.K.; Anderson, C.A.; Painter, J.N.; Uno, S.; Morris, A.P.; MacGregor, S.; Gordon, S.D.; Henders, A.K.; Martin, N.G.; et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat. Genet. 2012, 44, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Pagliardini, L.; Gentilini, D.; Vigano, P.; Panina-Bordignon, P.; Busacca, M.; Candiani, M.; Di Blasio, A.M. An Italian association study and meta-analysis with previous GWAS confirm WNT4, CDKN2BAS and FN1 as the first identified susceptibility loci for endometriosis. J. Med. Genet. 2013, 50, 43–46. [Google Scholar] [CrossRef]
- Albertsen, H.M.; Chettier, R.; Farrington, P.; Ward, K. Genome-wide association study link novel loci to endometriosis. PLoS ONE 2013, 8, e58257. [Google Scholar] [CrossRef]
- Rahmioglu, N.; Macgregor, S.; Drong, A.W.; Hedman, A.K.; Harris, H.R.; Randall, J.C.; Prokopenko, I.; The International Endogene Consortium (IEC); The GIANT Consortium; Nyholt, D.R.; et al. Genome-wide enrichment analysis between endometriosis and obesity-related traits reveals novel susceptibility loci. Hum. Mol. Genet. 2015, 24, 1185–1199. [Google Scholar] [CrossRef]
- Borghese, B.; Tost, J.; de Surville, M.; Busato, F.; Letourneur, F.; Mondon, F.; Vaiman, D.; Chapron, C. Identification of susceptibility genes for peritoneal, ovarian, and deep infiltrating endometriosis using a pooled sample-based genome-wide association study. Biomed. Res. Int. 2015, 2015, 461024. [Google Scholar] [CrossRef]
- Sapkota, Y.; Low, S.K.; Attia, J.; Gordon, S.D.; Henders, A.K.; Holliday, E.G.; MacGregor, S.; Martin, N.G.; McEvoy, M.; Morris, A.P.; et al. Association between endometriosis and the interleukin 1A (IL1A) locus. Hum. Reprod. 2015, 30, 239–248. [Google Scholar] [CrossRef]
- Wang, W.; Li, Y.; Li, S.; Wu, Z.; Yuan, M.; Wang, T.; Wang, S. Pooling-Based Genome-Wide Association Study Identifies Risk Loci in the Pathogenesis of Ovarian Endometrioma in Chinese Han Women. Reprod. Sci. 2017, 24, 400–406. [Google Scholar] [CrossRef]
- Uimari, O.; Rahmioglu, N.; Nyholt, D.R.; Vincent, K.; Missmer, S.A.; Becker, C.; Morris, A.P.; Montgomery, G.W.; Zondervan, K.T. Genome-wide genetic analyses highlight mitogen-activated protein kinase (MAPK) signaling in the pathogenesis of endometriosis. Hum. Reprod. 2017, 32, 780–793. [Google Scholar] [CrossRef]
- Sapkota, Y.; Steinthorsdottir, V.; Morris, A.P.; Fassbender, A.; Rahmioglu, N.; De Vivo, I.; Buring, J.E.; Zhang, F.; Edwards, T.L.; Jones, S.; et al. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat. Commun. 2017, 8, 15539. [Google Scholar] [CrossRef]
- Sobalska-Kwapis, M.; Smolarz, B.; Slomka, M.; Szaflik, T.; Kepka, E.; Kulig, B.; Siewierska-Gorska, A.; Polak, G.; Romanowicz, H.; Strapagiel, D.; et al. New variants near RHOJ and C2, HLA-DRA region and susceptibility to endometriosis in the Polish population-The genome-wide association study. Eur. J. Obs. Gynecol. Reprod. Biol. 2017, 217, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, Y.; Vivo, I.; Steinthorsdottir, V.; Fassbender, A.; Bowdler, L.; Buring, J.E.; Edwards, T.L.; Jones, S.; Dorien, O.; Peterse, D.; et al. Analysis of potential protein-modifying variants in 9000 endometriosis patients and 150000 controls of European ancestry. Sci. Rep. 2017, 7, 11380. [Google Scholar] [CrossRef] [PubMed]
- Painter, J.N.; O’Mara, T.A.; Morris, A.P.; Cheng, T.H.T.; Gorman, M.; Martin, L.; Hodson, S.; Jones, A.; Martin, N.G.; Gordon, S.; et al. Genetic overlap between endometriosis and endometrial cancer: Evidence from cross-disease genetic correlation and GWAS meta-analyses. Cancer Med. 2018, 7, 1978–1987. [Google Scholar] [CrossRef] [PubMed]
- Christofolini, D.M.; Mafra, F.A.; Catto, M.C.; Bianco, B.; Barbosa, C.P. New candidate genes associated to endometriosis. Gynecol. Endocrinol. 2019, 35, 62–65. [Google Scholar] [CrossRef]
- Adewuyi, E.O.; Sapkota, Y.; International Endogene Consortium (IEC); 23andMe Research Team; International Headache Genetics Consortium (IHGC); Auta, A.; Yoshihara, K.; Nyegaard, M.; Griffiths, L.R.; Montgomery, G.W.; et al. Shared Molecular Genetic Mechanisms Underlie Endometriosis and Migraine Comorbidity. Genes 2020, 11, 268. [Google Scholar] [CrossRef]
- Osinski, M.; Mostowska, A.; Wirstlein, P.; Wender-Ozegowska, E.; Jagodzinski, P.P.; Szczepanska, M. The assessment of GWAS - identified polymorphisms associated with infertility risk in Polish women with endometriosis. Ginekol. Pol. 2018, 89, 304–310. [Google Scholar] [CrossRef]
- Gallagher, C.S.; Makinen, N.; Harris, H.R.; Rahmioglu, N.; Uimari, O.; Cook, J.P.; Shigesi, N.; Ferreira, T.; Velez-Edwards, D.R.; Edwards, T.L.; et al. Genome-wide association and epidemiological analyses reveal common genetic origins between uterine leiomyomata and endometriosis. Nat. Commun. 2019, 10, 4857. [Google Scholar] [CrossRef]
- Cardoso, J.V.; Perini, J.A.; Machado, D.E.; Pinto, R.; Medeiros, R. Systematic review of genome-wide association studies on susceptibility to endometriosis. Eur. J. Obs. Gynecol. Reprod. Biol. 2020, 255, 74–82. [Google Scholar] [CrossRef]
- Vainio, S.; Heikkila, M.; Kispert, A.; Chin, N.; McMahon, A.P. Female development in mammals is regulated by Wnt-4 signalling. Nature 1999, 397, 405–409. [Google Scholar] [CrossRef]
- Ebenesersdottir, S.S.; Sandoval-Velasco, M.; Gunnarsdottir, E.D.; Jagadeesan, A.; Guethmundsdottir, V.B.; Thordardottir, E.L.; Einarsdottir, M.S.; Moore, K.H.S.; Sigurethsson, A.; Magnusdottir, D.N.; et al. Ancient genomes from Iceland reveal the making of a human population. Science 2018, 360, 1028–1032. [Google Scholar] [CrossRef]
- Steinthorsdottir, V.; Thorleifsson, G.; Aradottir, K.; Feenstra, B.; Sigurdsson, A.; Stefansdottir, L.; Kristinsdottir, A.M.; Zink, F.; Halldorsson, G.H.; Munk Nielsen, N.; et al. Common variants upstream of KDR encoding VEGFR2 and in TTC39B associate with endometriosis. Nat. Commun. 2016, 7, 12350. [Google Scholar] [CrossRef]
- Angioni, S.; D’Alterio, M.N.; Coiana, A.; Anni, F.; Gessa, S.; Deiana, D. Genetic Characterization of Endometriosis Patients: Review of the Literature and a Prospective Cohort Study on a Mediterranean Population. Int. J. Mol. Sci. 2020, 21, 1765. [Google Scholar] [CrossRef]
- Holdsworth-Carson, S.J.; Fung, J.N.; Luong, H.T.; Sapkota, Y.; Bowdler, L.M.; Wallace, L.; Teh, W.T.; Powell, J.E.; Girling, J.E.; Healey, M.; et al. Endometrial vezatin and its association with endometriosis risk. Hum. Reprod. 2016, 31, 999–1013. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Harold, D.; Nyholt, D.R.; ANZGene Consortium; Goddard, M.E.; Zondervan, K.T.; Williams, J.; Montgomery, G.W.; Wray, N.R.; Visscher , P.W. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer’s disease, multiple sclerosis and endometriosis. Hum. Mol. Genet. 2013, 22, 832–841. [Google Scholar] [CrossRef]
- Li, Y.; Hao, N.; Wang, Y.X.; Kang, S. Association of Endometriosis-Associated Genetic Polymorphisms From Genome-Wide Association Studies With Ovarian Endometriosis in a Chinese Population. Reprod. Sci. 2017, 24, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Azimzadeh, P.; Khorram Khorshid, H.R.; Akhondi, M.M.; Shirazi, A. Association of interleukin-16 polymorphisms with disease progression and susceptibility in endometriosis. Int. J. Immunogenet. 2016, 43, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Pitonak, J.; Galova, J.; Bernasovska, J. Association of two selected polymorphisms with developed endometriosis in women from Slovakia. Bratisl. Lek. Listy 2016, 117, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, J.; Fan, Y.; Guo, Q.; Ge, L.; Yu, N.; Zheng, X.; Dou, Y.; Zheng, S. Associations between various possible promoter polymorphisms of MMPs genes and endometriosis risk: A meta-analysis. Eur. J. Obs. Gynecol. Reprod. Biol. 2016, 205, 174–188. [Google Scholar] [CrossRef]
- Zamani, M.R.; Salmaninejad, A.; Akbari Asbagh, F.; Masoud, A.; Rezaei, N. STAT4 single nucleotide gene polymorphisms and susceptibility to endometriosis-related infertility. Eur. J. Obs. Gynecol. Reprod. Biol. 2016, 203, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Devesa-Peiro, A.; Sebastian-Leon, P.; Garcia-Garcia, F.; Arnau, V.; Aleman, A.; Pellicer, A.; Diaz-Gimeno, P. Uterine disorders affecting female fertility: What are the molecular functions altered in endometrium? Fertil. Steril. 2020, 113, 1261–1274. [Google Scholar] [CrossRef]
- Hediger, M.L.; Hartnett, H.J.; Louis, G.M. Association of endometriosis with body size and figure. Fertil. Steril. 2005, 84, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Ogawa, K.; Kamatani, Y.; Murakami, Y.; Kimura, T.; Okada, Y. A Mendelian randomization study identified obesity as a causal risk factor of uterine endometrial cancer in Japanese. Cancer Sci. 2020, 111, 4646–4651. [Google Scholar] [CrossRef]
- Holdsworth-Carson, S.J.; Chung, J.; Sloggett, C.; Mortlock, S.; Fung, J.N.; Montgomery, G.W.; Dior, U.P.; Healey, M.; Rogers, P.A.; Girling, J.E. Obesity does not alter endometrial gene expression in women with endometriosis. Reprod. Biomed. Online 2020, 41, 113–118. [Google Scholar] [CrossRef]
- Garitazelaia, A.; Rueda-Martinez, A.; Arauzo, R.; de Miguel, J.; Cilleros-Portet, A.; Mari, S.; Bilbao, J.R.; Fernandez-Jimenez, N.; Garcia-Santisteban, I. A Systematic Two-Sample Mendelian Randomization Analysis Identifies Shared Genetic Origin of Endometriosis and Associated Phenotypes. Life 2021, 11, 24. [Google Scholar] [CrossRef]
- Adewuyi, E.O.; Mehta, D.; Sapkota, Y.; International Endogene Consortium; 23andMe Research Team; Auta, A.; Yoshihara, K.; Nyegaard, M.; Griffiths, L.R.; Montgomery, G.W.; et al. Genetic analysis of endometriosis and depression identifies shared loci and implicates causal links with gastric mucosa abnormality. Hum. Genet. 2021, 140, 529–552. [Google Scholar] [CrossRef] [PubMed]
- Nezhat, C.; Li, A.; Abed, S.; Balassiano, E.; Soliemannjad, R.; Nezhat, A.; Nezhat, C.H.; Nezhat, F. Strong Association Between Endometriosis and Symptomatic Leiomyomas. JSLS 2016, 20. [Google Scholar] [CrossRef] [PubMed]
- Kvaskoff, M.; Mahamat-Saleh, Y.; Farland, L.V.; Shigesi, N.; Terry, K.L.; Harris, H.R.; Roman, H.; Becker, C.M.; As-Sanie, S.; Zondervan, K.T.; et al. Endometriosis and cancer: A systematic review and meta-analysis. Hum. Reprod. Update 2021, 27, 393–420. [Google Scholar] [CrossRef]
- Worley, M.J., Jr.; Liu, S.; Hua, Y.; Kwok, J.S.; Samuel, A.; Hou, L.; Shoni, M.; Lu, S.; Sandberg, E.M.; Keryan, A.; et al. Molecular changes in endometriosis-associated ovarian clear cell carcinoma. Eur. J. Cancer 2015, 51, 1831–1842. [Google Scholar] [CrossRef]
- Govatati, S.; Kodati, V.L.; Deenadayal, M.; Chakravarty, B.; Shivaji, S.; Bhanoori, M. Mutations in the PTEN tumor gene and risk of endometriosis: A case-control study. Hum. Reprod. 2014, 29, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, K.C.; Shah, S.P.; Al-Agha, O.M.; Zhao, Y.; Tse, K.; Zeng, T.; Senz, J.; McConechy, M.K.; Anglesio, M.S.; Kalloger, S.E.; et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010, 363, 1532–1543. [Google Scholar] [CrossRef]
- Masuda, T.; Low, S.K.; Akiyama, M.; Hirata, M.; Ueda, Y.; Matsuda, K.; Kimura, T.; Murakami, Y.; Kubo, M.; Kamatani, Y.; et al. GWAS of five gynecologic diseases and cross-trait analysis in Japanese. Eur. J. Hum. Genet. 2020, 28, 95–107. [Google Scholar] [CrossRef]
- Horikoshi, M.; Beaumont, R.N.; Day, F.R.; Warrington, N.M.; Kooijman, M.N.; Fernandez-Tajes, J.; Feenstra, B.; van Zuydam, N.R.; Gaulton, K.J.; Grarup, N.; et al. Genome-wide associations for birth weight and correlations with adult disease. Nature 2016, 538, 248–252. [Google Scholar] [CrossRef]
- Barban, N.; Jansen, R.; de Vlaming, R.; Vaez, A.; Mandemakers, J.J.; Tropf, F.C.; Shen, X.; Wilson, J.F.; Chasman, D.I.; Nolte, I.M.; et al. Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat. Genet. 2016, 48, 1462–1472. [Google Scholar] [CrossRef]
- Marla, S.; Mortlock, S.; Houshdaran, S.; Fung, J.; McKinnon, B.; Holdsworth-Carson, S.J.; Girling, J.E.; Rogers, P.A.W.; Giudice, L.C.; Montgomery, G.W. Genetic risk factors for endometriosis near estrogen receptor 1 and coexpression of genes in this region in endometrium. Mol. Hum. Reprod. 2021, 27. [Google Scholar] [CrossRef]
- Hirata, T.; Koga, K.; Johnson, T.A.; Morino, R.; Nakazono, K.; Kamitsuji, S.; Akita, M.; Kawajiri, M.; Kami, A.; Hoshi, Y.; et al. Japanese GWAS identifies variants for bust-size, dysmenorrhea, and menstrual fever that are eQTLs for relevant protein-coding or long non-coding RNAs. Sci. Rep. 2018, 8, 8502. [Google Scholar] [CrossRef] [PubMed]
- Olafsdottir, T.; Thorleifsson, G.; Sulem, P.; Stefansson, O.A.; Medek, H.; Olafsson, K.; Ingthorsson, O.; Gudmundsson, V.; Jonsdottir, I.; Halldorsson, G.H.; et al. Genome-wide association identifies seven loci for pelvic organ prolapse in Iceland and the UK Biobank. Commun. Biol. 2020, 3, 129. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.C.; Chen, M.J.; Chen, P.H.; Chang, C.W.; Yu, M.H.; Chen, Y.J.; Tsai, E.M.; Tsai, S.F.; Kuo, W.S.; Tzeng, C.R. Integration of genome-wide association study and expression quantitative trait locus mapping for identification of endometriosis-associated genes. Sci. Rep. 2021, 11, 478. [Google Scholar] [CrossRef]
- Fung, J.N.; Mortlock, S.; Girling, J.E.; Holdsworth-Carson, S.J.; Teh, W.T.; Zhu, Z.; Lukowski, S.W.; McKinnon, B.D.; McRae, A.; Yang, J.; et al. Genetic regulation of disease risk and endometrial gene expression highlights potential target genes for endometriosis and polycystic ovarian syndrome. Sci. Rep. 2018, 8, 11424. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.Q.; Ritchie, S.C.; Brozynska, M.; Inouye, M. Power, false discovery rate and Winner’s Curse in eQTL studies. Nucleic Acids Res. 2018, 46, e133. [Google Scholar] [CrossRef]
- Palmer, C.; Pe’er, I. Statistical correction of the Winner’s Curse explains replication variability in quantitative trait genome-wide association studies. PLoS Genet. 2017, 13, e1006916. [Google Scholar] [CrossRef]
- Sapkota, Y.; Fassbender, A.; Bowdler, L.; Fung, J.N.; Peterse, D.; Dorien, O.; Montgomery, G.W.; Nyholt, D.R.; D’Hooghe, T.M. Independent Replication and Meta-Analysis for Endometriosis Risk Loci. Twin Res. Hum. Genet. 2015, 18, 518–525. [Google Scholar] [CrossRef]
- Sundqvist, J.; Xu, H.; Vodolazkaia, A.; Fassbender, A.; Kyama, C.; Bokor, A.; Gemzell-Danielsson, K.; D’Hooghe, T.M.; Falconer, H. Replication of endometriosis-associated single-nucleotide polymorphisms from genome-wide association studies in a Caucasian population. Hum. Reprod. 2013, 28, 835–839. [Google Scholar] [CrossRef] [PubMed]
- Mafra, F.; Catto, M.; Bianco, B.; Barbosa, C.P.; Christofolini, D. Association of WNT4 polymorphisms with endometriosis in infertile patients. J. Assist. Reprod. Genet. 2015, 32, 1359–1364. [Google Scholar] [CrossRef]
- Hata, Y.; Nakaoka, H.; Yoshihara, K.; Adachi, S.; Haino, K.; Yamaguchi, M.; Nishikawa, N.; Kashima, K.; Yahata, T.; Tajima, A.; et al. A nonsynonymous variant of IL1A is associated with endometriosis in Japanese population. J. Hum. Genet. 2013, 58, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Badie, A.; Saliminejad, K.; Salahshourifar, I.; Khorram Khorshid, H.R. Interleukin 1 alpha (IL1A) polymorphisms and risk of endometriosis in Iranian population: A case-control study. Gynecol. Endocrinol. 2020, 36, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Albertsen, H.M.; Ward, K. Genes Linked to Endometriosis by GWAS Are Integral to Cytoskeleton Regulation and Suggests That Mesothelial Barrier Homeostasis Is a Factor in the Pathogenesis of Endometriosis. Reprod. Sci. 2017, 24, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Zaitlen, N.A.; Reich, D.; Patterson, N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 2010, 11, 459–463. [Google Scholar] [CrossRef]
- Uricchio, L.H. Evolutionary perspectives on polygenic selection, missing heritability, and GWAS. Hum. Genet. 2020, 139, 5–21. [Google Scholar] [CrossRef]
- Slim, L.; Chatelain, C.; Azencott, C.A.; Vert, J.P. Novel methods for epistasis detection in genome-wide association studies. PLoS ONE 2020, 15, e0242927. [Google Scholar] [CrossRef] [PubMed]
- Fung, J.N.; Holdsworth-Carson, S.J.; Sapkota, Y.; Zhao, Z.Z.; Jones, L.; Girling, J.E.; Paiva, P.; Healey, M.; Nyholt, D.R.; Rogers, P.A.; et al. Functional evaluation of genetic variants associated with endometriosis near GREB1. Hum. Reprod. 2015, 30, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Nakaoka, H.; Gurumurthy, A.; Hayano, T.; Ahmadloo, S.; Omer, W.H.; Yoshihara, K.; Yamamoto, A.; Kurose, K.; Enomoto, T.; Akira, S.; et al. Allelic Imbalance in Regulation of ANRIL through Chromatin Interaction at 9p21 Endometriosis Risk Locus. PLoS Genet. 2016, 12, e1005893. [Google Scholar] [CrossRef]
- Jordan, B.K.; Mohammed, M.; Ching, S.T.; Delot, E.; Chen, X.N.; Dewing, P.; Swain, A.; Rao, P.N.; Elejalde, B.R.; Vilain, E. Up-regulation of WNT-4 signaling and dosage-sensitive sex reversal in humans. Am. J. Hum. Genet. 2001, 68, 1102–1109. [Google Scholar] [CrossRef]
- Powell, J.E.; Fung, J.N.; Shakhbazov, K.; Sapkota, Y.; Cloonan, N.; Hemani, G.; Hillman, K.M.; Kaufmann, S.; Luong, H.T.; Bowdler, L.; et al. Endometriosis risk alleles at 1p36.12 act through inverse regulation of CDC42 and LINC00339. Hum. Mol. Genet. 2016, 25, 5046–5058. [Google Scholar] [CrossRef]
- Turco, M.Y.; Gardner, L.; Hughes, J.; Cindrova-Davies, T.; Gomez, M.J.; Farrell, L.; Hollinshead, M.; Marsh, S.G.E.; Brosens, J.J.; Critchley, H.O.; et al. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 2017, 19, 568–577. [Google Scholar] [CrossRef]
- Boretto, M.; Maenhoudt, N.; Luo, X.; Hennes, A.; Boeckx, B.; Bui, B.; Heremans, R.; Perneel, L.; Kobayashi, H.; Van Zundert, I.; et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 2019, 21, 1041–1051. [Google Scholar] [CrossRef]
- Albertsen, H.M.; Matalliotaki, C.; Matalliotakis, M.; Zervou, M.I.; Matalliotakis, I.; Spandidos, D.A.; Chettier, R.; Ward, K.; Goulielmos, G.N. Whole exome sequencing identifies hemizygous deletions in the UGT2B28 and USP17L2 genes in a threegeneration family with endometriosis. Mol. Med. Rep. 2019, 19, 1716–1720. [Google Scholar] [CrossRef]
- Matalliotakis, M.; Zervou, M.I.; Matalliotaki, C.; Rahmioglu, N.; Koumantakis, G.; Kalogiannidis, I.; Prapas, I.; Zondervan, K.; Spandidos, D.A.; Matalliotakis, I.; et al. The role of gene polymorphisms in endometriosis. Mol. Med. Rep. 2017, 16, 5881–5886. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Zhao, L.; Wang, L.; Wu, Z.; Mei, Q.; Nie, J.; Li, X.; Li, Y.; Fu, X.; et al. Whole-exome sequencing of endometriosis identifies frequent alterations in genes involved in cell adhesion and chromatin-remodeling complexes. Hum. Mol. Genet. 2014, 23, 6008–6021. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.A.; Kemp, J.P.; Youlten, S.E.; Laurent, L.; Logan, J.G.; Chai, R.C.; Vulpescu, N.A.; Forgetta, V.; Kleinman, A.; Mohanty, S.T.; et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019, 51, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Rumph, J.T.; Stephens, V.R.; Archibong, A.E.; Osteen, K.G.; Bruner-Tran, K.L. Environmental Endocrine Disruptors and Endometriosis. Adv. Anat. Embryol. Cell Biol. 2020, 232, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Sirohi, D.; Al Ramadhani, R.; Knibbs, L.D. Environmental exposures to endocrine disrupting chemicals (EDCs) and their role in endometriosis: A systematic literature review. Rev. Environ. Health 2021, 36, 101–115. [Google Scholar] [CrossRef] [PubMed]
| REFERENCE | PAPER PMID | ETHNICITY | STUDY TYPE | CASES (N) | CONTROLS (N) | ENDOME TRIOSIS DIAGNOSIS | CONTROLS SELECTION | STAGES OF THE STUDY | ASSOCIATED SNPs | SNP NAMES |
|---|---|---|---|---|---|---|---|---|---|---|
| (a) Discovery Study | ||||||||||
| Uno et al., 2010 [30] | 20601957 | Asian | Case-control of hospital based | 1907 | 5292 | Laparoscopy | Laparoscopy | Discovery | 1 | rs10965235 (CDKN2BAS) |
| Adachi et al., 2010 [31] | 20844546 | Asian | Meta-analysis of population and hospital based | 696 | 825 | Medical history, MRI and laparoscopy | Medical history, MRI and laparoscopy | Discovery | 5 | rs6542095, rs11677416, rs3783550 and rs3783525 (near IL1 A gene) and rs801112 (RHOU) |
| Painter et al., 2011 [32] | 3019124 | Oceania and European | Meta-analysis of population and hospital based | 3194 | 7060 | Laparoscopy | Men, medical history and laparoscopy | Discovery | 1 | rs12700667 (NFE2L3-HOXA10) |
| Nyholt et al., 2012 [33] | 23104006 | Oceania and European | Meta-analysis of population and hospital based | 4604 | 9393 | Laparoscopy | Men, medical history and laparoscopy | Discovery | 7 | rs12700667 (NFE2L3-HOXA10 ), rs75211902 (WNT4), rs1537377 (CDKN2B-AS1), rs10859871 (VEZT), rs4141819 (ETAA1), rs77399264 (ID4), rs13394619 (GREB1) |
| Pagliardini et al., 2013 [34] | 23142796 | Asian and European | Meta-analysis of population and hospital based | 305 | 2710 | Laparoscopy | Laparoscopy blood donors | Discovery | 3 | rs1333049 (CDKN2BAS), rs7521902 (WNT4), rs1250248 (FN1) |
| Albertsen et al., 2013 [35] | 23472165 | European | Case-control of hospital base | 2019 | 14,471 | Laparoscopy | Laparoscopy | Discovery | 3 | rs2235529 (LINC00339-WNT4),rs1519761 and rs6757804 (RND3) |
| Rahmioglu et al., 2014 [36] | 24676469 | American, European and Oceania | Meta-analysis of population and hospital based | 11,506 | 32,678 | Laparoscopy | Laparoscopy | Discovery | 6 | rs12700667 (NFE2L3-HOXA10),rs7521902 (WNT4), rs10859871 (VEZT) rs1537377 (CDKN2B-AS1), rs7739264 (ID4), rs13394619 (GREB1) |
| Borghese et al., 2015 [37] | 25722978 | European | Meta-analysis of population and hospital based | 60 | 20 | Laparoscopy | Laparoscopy | Discovery | 4 | rs4703908 (ZNF366),rs227849 (RUNX2/ SUPT3H), rs2479037 (VTI1A) and rs966674 (NA) |
| Sapkota et al., 2015 [38] | 26337243 | Asian, African, European and Oceania | Meta-analysis of population and hospital-based | 998 | 783 | Laparoscopy | Laparoscopy | Discovery | 6 | rs7521902 (WNT4), rs13394619 in GREB1, rs12700667 (NFE2L3-HOXA10), rs6542095 (IL1A, rs7739264 (ID4) and rs1537377 (CDKN2B-AS1) |
| Wang et al., 2016 [39] | 27506219 | Asian | Case-control of hospital based | 1448 | 1540 | Laparoscopy | Laparoscopy | Discovery | 3 | rs11692361 (MEIS1), rs10256972 (C7orf50), rs4966038 (IGF-1R) |
| Uimari et al., 2017 [40] | 28333195 | Oceania and European | Case-control of hospital-based | 3194 | 7060 | Laparoscopy | Men, medical history and laparoscopy | Discovery | 1 | rs144240142 (MAP3K4) |
| Sapkota et al., 2017 [41] | 28537267 | Asian, Oceania European and American | Meta-analysis of population and hospital-based | 17,045 | 191,596 | Laparoscopy | Medical history and laparoscopy | Discovery | 19 | rs12037376 (WNT4), rs11674184 and rs77294520 (GREB1), rs6546324 ( ETAA1), rs4762326 (VEZT), rs10167914 (IL1A), rs1903068 (KDR), rs760794 (ID4),rs74485684 (FSHB), rs12700667 (NFE2L3-HOXA10), rs1537377, rs1075727 and rs1448792 (CDKN2B-AS1), rs1250241 (FN1), rs1971256 (CCDC170), rs71575922 and rs17803970 (SYNE1) rs2206949 (ESR1), rs74491657 (7p12.3) |
| Sobalska-kwapis et al., 2017 [42] | 28881265 | European | Case-control of hospital based | 171 | 2934 | Laparoscopy | Medical History | Discovery | 19 | 18 SNPs (near of C2)and rs10129516 (PARP1P2-RHOJ) |
| Sapkota et al., 2017 [43] | 28900119 | Oceania, European and American | Meta-analysis of population and hospital based | 9000 | 150,001 | Medical History Laparoscopy | Medical History and Laparoscopy | Discovery | 3 | rs13394619 (GREB1 at 2p25.1), rs1801262 (CUBN), gene-level (CIITA et PARP4) |
| Painter et al., 2018 [44] | 29608257 | European | Case-control of hospital based | 3194 | 2057 | Laparoscopy | Laparoscopy | Discovery | 13 | rs2475335 (PTRD), rs9865110 (PDZRN3-CNTN3), rs2278868 (SKAP1), rs12303900 (KITLG-DUSP6) rs9349553(TFAP2D), rs10008492 (KLF3-TLR10), rs9530566 (LMO7-KCTD12), rs17693745 (CEP112), rs1755833 (PRIM2), rs7515106 (WNT4-ZBTB40), rs10459129 (PARP11-CCND2) rs2198894 (ZNF536-TSHZ3), rs7042500 (THEM215-APTX) |
| Christofolini et al., 2019 [45] | 30044155 | European | Case-control of hospital based | 394 | 650 | Laparoscopy | Laparoscopy | Discovery | 2 | rs10928050 (KAZN) and rs2427284 (LAM5) |
| Adewuyi et al., 2020 [46] | 32121467 | European, Asian and American | Meta-analysis of population and hospital based | 76,728 | 507,936 | Medical history and laparoscopy | Medical History | Discovery | 3 | SNPs nears ARL14EP , TRIM32, and SLC35G6 |
| (b) Replication Study | ||||||||||
| Painter et al., 2011 [32] | 3019124 | Oceanie | Meta-analysis of population and hospital based | 2392 | 2271 | Laparoscopy | Men, medical history and laparoscopy | Replication | 1 | rs12700667 (NFE2L3-HOXA10) |
| Nyholt et al., 2012 [33] | 23104006 | Asian Oceanie | Meta-analysis of population and hospital based | 1044 | 4017 | Laparoscopy | Men, medical history and laparoscopy | Replication | 7 | rs12700667 (NFE2L3-HOXA10 ), rs75211902 (WNT4), rs1537377 (CDKN2B-AS1), rs10859871 (VEZT),rs4141819 (ETAA1), rs77399264 (ID4), rs13394619 (GREB1) |
| Albertsen et al., 2013 [35] | 23472165 | European | Meta-analysis of population and hospital based | 505 | 1811 | Laparoscopy | Laparoscopy | Replication | 3 | rs2235529 (LINC00339-WNT4),rs1519761 and rs6757804 (RND3) |
| Sapkota et al., 2015 [38] | 26337243 | Asian, African | Meta-analysis of population and hospital based | 998 | 783 | Laparoscopy | Laparoscopy | Replication | 6 | rs7521902 (WNT4), rs13394619 in GREB1 rs12700667 (NFE2L3-HOXA10) |
| Osiński et al., 2018 [47] | 30010178 | European | Case-control of hospital base | 315 | 406 | Laparoscopy | Medical History | Replication | 2 | rs12700667 (NFE2L3-HOXA10) and rs4141819 (ETAA1) |
| (c) Connection between Endometriosis and Others Pathologies | ||||||||||
| Painter et al., 2011 [32] | 3019124 | Oceanie and European | Meta-analysis of population and hospital based | 3194 | 7060 | Laparoscopy | Men, medical history and laparoscopy | Discovery | 1 | rs12700667 (NFE2L3-HOXA10) |
| Gallagher et al., 2019 [48] | 31649266 | European | Case-control of hospital base | 35,474 | 267,505 | Medical history, MRI | Medical history, MRI | Discovery | 4 | rs58415480 (ESR1), rs11031006 (FSHB), rs35417544 (GREB1), rs7412010 (WNT4) |
| Adewuyi et al., 2020 [46] | 32959083 | European, Asian and Americanian | Meta-analysis of population and hospital based | 187,810 | 521,301 | Laparoscopy | Medical History and laparoscopy | Discovery | 26 | rs116810322 (TRIM26), rs11793648 (TRIM32), rs9891297 (SLC35G6), rs1395455 (CSF3R), rs1620977 (NEGR1), rs12121863 (CRB1), rs9586 (KLHDC8B), rs9835157 (IP6K1), rs12512642 ( SCLT1), rs13164188 (NUDT12), rs7933594 (TYR), rs6680839 (TNR), rs72740410 (BRINP3), rs13118306 (CC2D2A), rs2134025 (TACR3) rs9347896 (C6orf1), rs11784932 (GSDMC), rs9538160 (PCDH17), rs35625885 (NR2F2), rs6808036 (CCDC36), rs323509 (NUDT12), rs6788293 (LINC00620) |
| Locus | Gene(s) of Interest | SNP | Cases (N) | Controls (N) | RA | OA | Relative Risk | p-Value | Paper PMID |
|---|---|---|---|---|---|---|---|---|---|
| 1p31.1 | NEGR1 | rs1620977 | 187,810 | 521,301 | A | G | 1.027 | 10−8 (10−2) * | 32959083 |
| 1p34.3 | CSF3R | rs1395455 | 187,810 | 521,301 | A | G | 1.025 | 10−8 (10−2) * | 32959083 |
| 1p36.12 | WNT4 | rs7521902 | 305 | 2710 | A | C | 1.2 | 10−9 | 23142796 |
| rs7521902 | 11,506 | 32,678 | A | C | 1.18 | 10−15 | 24676469 | ||
| rs4654783 | 2019 | 14,471 | A | G | 1.21 | 10−9 | 23472165 ** | ||
| rs2235529 | 2019 | 14,471 | A | G | 1.29 | 10−9 | 23472165 ** | ||
| rs7521902 | 4604 | 9393 | A | C | 1.18 | 10−8 | 23104006 | ||
| rs7521902 | 1044 | 4017 | A | C | NA | 10−5 | 23104006 ** | ||
| rs7521902 | 998 | 783 | A | C | 1.18 | 10−8 | 26337243 | ||
| rs7521902 | 998 | 783 | A | C | 1.3 | 10−3 | 26337243 ** | ||
| rs7521902 | 23,671 | 68,894 | A | C | 1.29 | 10−15 | 27470151 | ||
| rs7739264 | 998 | 783 | T | C | 1.14 | 10−7 | 26337243 | ||
| rs7515106 | 3194 | 2057 | C | T | 1.09 | 10−5 | 29608257 ** | ||
| rs12037376 | 17,045 | 191,596 | A | G | 1.16 | 10−17 | 28537267 | ||
| rs7412010 | 35,474 | 267,505 | C | G | 1.13 | 10−29 | 31649266 | ||
| 1p36.21 | KAZN | rs10928050 | 394 | 650 | A | G | 1.31 | 10−2 | 30044155 ** |
| 1q25.1 | TNR | rs6680839 | 187,810 | 521,301 | T | C | 0.975 | 10−10 (10−4) * | 32959083 |
| 1q31.1 | BRINP3 | rs72740410 | 187,810 | 521,301 | T | C | 1.048 | 10−8 (10−2) * | 32959083 |
| 1q31.3 | CRB1 | rs12121863 | 187,810 | 521,301 | A | T | 0.971 | 10−8 (10−2) * | 32959083 |
| 1q42.13 | RHOU | rs801112 | 696 | 825 | A | T | 1.65 | 10−6 | 20844546 |
| 2p14 | ETAA1 | rs6546324 | 998 | 191,596 | A | C | 1.08 | 10−9 | 28537267 |
| rs4141819 | 4604 | 9393 | C | T | 1.15 | 10−8 | 23104006 | ||
| rs4141819 | 1044 | 4017 | C | T | NA | 10−2 | 23104006 ** | ||
| rs4141819 | 11,506 | 32,678 | C | T | 1.08 | 10−6 | 24676469 | ||
| rs4141819 | 315 | 406 | C | T | 1.35 | 10−2 | 30010178 ** | ||
| 2p14 | MEIS1 | rs11692361 | 1448 | 1540 | C | T | 0.70 | 10−7 | 27506219 |
| 2p25.1 | GREB1 | rs13394619 | 11,506 | 32,678 | G | A | 1.13 | 10−8 | 24676469 |
| rs13394619 | 998 | 783 | G | A | 1.15 | 10−9 | 26337243 | ||
| rs13394619 | 998 | 783 | G | A | 1.13 | 10−2 | 26337243 ** | ||
| rs13394619 | 7164 | 21,005 | A | G | 0.89 | 10−9 | 28900119 | ||
| rs13394619 | 4604 | 9393 | A | G | 1.15 | 10−8 | 23104006 | ||
| rs13394619 | 1044 | 4017 | A | G | NA | 10−3 | 23104006 ** | ||
| rs11674184 | 17,045 | 191,596 | T | G | 1.13 | 10−17 | 28537267 | ||
| rs77294520 | 17,045 | 191,596 | C | G | 1.16 | 10−13 | 28537267 | ||
| rs35417544 | 35,474 | 267,505 | T | C | 1.09 | 10−13 | 31649266 | ||
| 2q13 | IL1A | rs6542095 | 696 | 825 | C | T | 1.5 | 10−6 | 20844546 |
| rs6542095 | 998 | 783 | C | T | 1.22 | 10−10 | 26337243 ** | ||
| rs6542095 | 998 | 783 | C | T | 1.26 | 10−2 | 26337243 | ||
| rs6542095 | 7164 | 21,005 | T | C | 0.90 | 10−7 | 28900119 | ||
| rs11677416 | 696 | 825 | T | A | 2.0 | 10−6 | 20844546 | ||
| rs10167914 | 17,045 | 191,596 | G | A | 1.15 | 10−9 | 28537267 | ||
| rs3783550 | 696 | 825 | C | A | 1.51 | 10−6 | 20844546 | ||
| rs3783525 | 696 | 825 | A | T | 1.52 | 10−6 | 20844546 | ||
| 2q23.3 | RND3 | rs1519761 | 2019 | 14,471 | C | A | 1.20 | 10−7 | 23472165 ** |
| rs6734792 | 2019 | 14,471 | G | A | 1.20 | 10−8 | 23472165 ** | ||
| rs1519761 | 2019 | 14,471 | G | A | 1.20 | 10−8 | 23472165 ** | ||
| rs6757804 | 2019 | 14,471 | G | A | 1.20 | 10−8 | 23472165 ** | ||
| rs6734792 | 11,506 | 32,678 | C | T | 1.10 | 10−6 | 24676469 | ||
| 2q35 | FN1 | rs1250241 | 17,045 | 191,596 | T | A | 1.23 | 10−9 | 28537267 |
| rs1250248 | 305 | 2710 | A | G | 1.13 | 10−9 | 23142796 | ||
| rs1250248 | 1129 | 831 | A | G | 1.17 | 10−2 | 23315067 ** | ||
| rs1250248 | 11,506 | 32,678 | A | G | 1.11 | 10−4 | 24676469 | ||
| 3p13-3p12.3 | PDZRN3-CNTN3 | rs9865110 | 3194 | 2057 | C | A | 1.1 | 10−6 | 29608257 |
| 3p21.31 | KLHDC8B | rs9586 | 187,810 | 521,301 | T | C | 0.976 | 10−8 (10−4) * | 32959083 |
| 3p21.31 | IP6K1 | rs9835157 | 187,810 | 521,301 | A | G | 1.034 | 10−8 (10−4) * | 32959083 |
| 3p21.31 | CCDC36 | rs6808036 | 187,810 | 521,301 | T | G | 1.044 | 10−8 (10−5) * | 32959083 |
| 3p25.1 | LINC00620 | rs6788293 | 187,810 | 521,301 | T | C | 0.952 | 10−8 (10−2) * | 32959083 |
| 4p14 | KLF3-TLR10 | rs10008492 | 3194 | 2057 | T | C | 1.14 | 10−5 | 29608257 |
| 4p15.32 | CC2D2A | rs13118306 | 187,810 | 521,301 | C | G | 0.977 | 10−8 (10−3) * | 32959083 |
| 4q12 | KDR | rs1903068 | 17,045 | 191,596 | A | G | 1.11 | 10−15 | 28537267 |
| 4q24 | TACR3 | rs2134025 | 187,810 | 521,301 | A | G | 1.029 | 10−8 (10−4) * | 32959083 |
| 4q28.2 | SCLT1 | rs12512642 | 187,810 | 521,301 | T | C | 1.28 | 10−8 (10−2) * | 32959083 |
| 5 | NA | rs966674 | 288 | 259 | C | G | 2.95 | 10−3 | 25722978 |
| 5q13.1 | ZNF366 | rs4703908 | 288 | 259 | C | G | 2.22 | 10−3 | 25722978 |
| 5q21.2 | NUDT12 | rs13164188 | 187,810 | 521,301 | T | C | 1.025 | 10−9 (10−2) * | 32959083 |
| 5q21.2 | NUDT12 | rs323509 | 187,810 | 521,301 | A | C | 1.042 | 10−8 (10−2) * | 32959083 |
| 6p11.2 | PRIM2 | rs1755833 | 3194 | 2057 | A | G | 0.93 | 10−5 | 29608257 |
| 6p12.3 | TFAP2D | rs9349553 | 3194 | 2057 | T | C | 1.09 | 10−6 | 29608257 |
| 6p21.1 | RUNX2/SUPT3H | rs227849 | 288 | 259 | A | G | 2.31 | 10−3 | 25722978 |
| 6p21.33 | C2 | rs644045 | 171 | 2934 | T | C | 1.90 | 10−10 | 28881265 |
| 6p22.1 | TRIM26 | rs116810322 | 187,810 | 521,301 | T | C | 1.042 | 10−9 (10−5) | 32959083 |
| 6p22.3 | ID4 | rs7739264 | 11,506 | 32,678 | T | C | 1.11 | 10−10 | 24676469 |
| rs7739264 | 4604 | 9393 | T | C | 1.17 | 10−7 | 23104006 | ||
| rs7739264 | 1044 | 4017 | T | C | NA | 10−4 | 23104006 ** | ||
| rs7739264 | 998 | 783 | T | C | 1.14 | 10−7 | 26337243 | ||
| rs7739264 | 23,671 | 68,894 | T | C | 1.10 | 10−10 | 27470151 | ||
| rs760794 | 17,045 | 191,596 | T | C | 1.17 | 10−7 | 28537267 | ||
| rs6907340 | 2019 | 14,471 | A | G | 1.12 | 10−7 | 23472165 | ||
| 6q12 | ESY | rs12206488 | 187,810 | 521,301 | A | G | 0.976 | 10−9 (10−3) * | 32959083 |
| 6q24.2 | HIVEP2 | rs2328370 | 187,810 | 521,301 | A | C | 1.023 | 10−9 (10−3) * | 32959083 |
| 6q25.1 | CCDC170 | rs1971256 | 17,045 | 191,596 | C | T | 1.09 | 10−8 | 28537267 |
| 6q25.1 | SYNE1 | rs71575922 | 17,045 | 191,596 | G | C | 1.11 | 10−8 | 28537267 |
| rs17803970 | 17,045 | 191,596 | A | T | 1.15 | 10−8 | 28537267 | ||
| 6q25.1-q25.2 | ESR1 | rs2206949 | 17,045 | 191,596 | T | C | 1.10 | 10−7 | 28537267 |
| rs58415480 | 35,474 | 267,505 | C | G | 1.19 | 10−7 | 31649266 | ||
| 6q26 | MAP3K4 | rs144240142 | 3194 | 7060 | T | C | 1.71 | 10−8 | 28333195 |
| 6q27 | C6orf1 | rs9347896 | 187,810 | 521,301 | A | G | 1.29 | 10−8 (10−2) * | 32959083 |
| 7p15.2 | NFE2L3/HOXA10 | rs12700667 | 3194 | 7060 | A | G | 1.22 | 10−7 | 3019124 |
| rs12700667 | 2392 | 2271 | A | G | 1.17 | 10−3 | 3019124 ** | ||
| rs12700667 | 4604 | 9393 | A | G | 1.18 | 10−10 | 23104006 | ||
| rs12700667 | 1044 | 4017 | A | G | NA | 10−9 | 23104006 ** | ||
| rs12700667 | 998 | 783 | A | G | 1.19 | 10−10 | 26337243 | ||
| rs12700667 | 11,506 | 32,678 | A | G | 1.13 | 10−9 | 24676469 | ||
| rs12700667 | 17,045 | 191,596 | A | G | 1.1 | 10−10 | 28537267 | ||
| rs12700667 | 315 | 406 | A | G | 1.3 | 10−2 | 30010178 ** | ||
| 7p22.3 | C7orf50 | rs10256972 | 1448 | 1540 | A | C | 1.30 | 10−6 | 27506219 |
| 7q31.1 | DNAJB9 | rs11561993 | 187,810 | 521,301 | T | C | 1.025 | 10−9 (10−3) * | 32959083 |
| 8q24.21 | GSDMC | rs11784932 | 187,810 | 521,301 | A | C | 1.026 | 10−8 (10−4) * | 32959083 |
| 9p21.1 | THEM215-APTX | rs7042500 | 3194 | 2057 | A | G | 0.9 | 10−5 | 29608257 |
| 9p21.1 | ACO1 | rs13299293 | 187,810 | 521,301 | A | T | 0.976 | 10−9 (10−3) * | 32959083 |
| 9p21.3 | CDKN2B-AS1 | rs10965235 | 1907 | 5292 | C | A | 1.44 | 10−12 | 20601957 |
| rs1537377 | 11,506 | 32,678 | C | T | 1.12 | 10−8 | 24676469 | ||
| rs1537377 | 17,045 | 191,596 | T | C | 1.21 | 10−9 | 28537267 | ||
| rs1537377 | 4604 | 9393 | C | T | 1.15 | 10−6 | 23104006 | ||
| rs1537377 | 1044 | 4017 | C | T | NA | 10−4 | 23104006 ** | ||
| rs1448792 | 17,045 | 191,596 | G | A | 1.08 | 10−8 | 28537267 | ||
| rs10757272 | 17,045 | 191,596 | C | T | 1.07 | 10−7 | 28537267 | ||
| 9p24.1-p23 | PTRD | rs2475335 | 3194 | 2057 | T | C | 1.11 | 10−8 | 29608257 |
| 9p24.1-p23 | PTPRD | rs1931391 | 187,810 | 521,301 | T | G | 1.031 | 10−8 (10−2) * | 32959083 |
| 9q33.1 | TRIM32 | rs11793648 | 46,262 | 364,789 | NA | NA | NA | 10−6 | 32121467 |
| 10p13 | CUBN | rs1801232 | 7164 | 21,005 | T | G | 0.86 | 10−7 | 28900119 |
| 10q11.21 | HNRNPA3P1 | rs10508881 | 2019 | 14,471 | A | G | 1.19 | 10−7 | 23472165 ** |
| 10q11.21 | LOC100130539 | ||||||||
| 10q25.2 | VTI1A | rs2479037 | 288 | 259 | C | T | 4.36 | 10−3 | 25722978 |
| 11p14.1 | FSHB | rs74485684 | 17,045 | 191,596 | G | A | 1.11 | 10−8 | 28537267 |
| rs11031006 | 35,474 | 267,505 | A | G | 1,10 | 10−15 | 31649266 | ||
| 11p14.1 | ARL14EP | rs4071559 | 46,262 | 364,789 | NA | NA | NA | 10−7 | 32121467 |
| 11q14.3 | TYR | rs7933594 | 187,810 | 521,301 | C | G | 1.024 | 10−9 (10−2) * | 32959083 |
| 12p13.32 | PARP11-CCND2 | rs10459129 | 3194 | 2057 | A | G | 0.9 | 10−5 | 29608257 |
| 12q21.32-12q21.33 | KITLG-DUSP6 | rs12303900 | 3194 | 2057 | G | T | 1.28 | 10−7 | 29608257 |
| 12q22 | VEZT | rs10859871 | 11,506 | 32,678 | C | A | 1.18 | 10−15 | 24676469 |
| rs10859871 | 4604 | 9393 | C | A | 1.20 | 10−9 | 23104006 | ||
| rs10859871 | 23,671 | 68,894 | C | A | 1.19 | 10−20 | 27470151 | ||
| rs10859871 | 998 | 783 | C | A | 1.16 | 10−7 | 26337243 | ||
| rs10859871 | 1044 | 4017 | C | T | NA | 10−6 | 23104006 ** | ||
| rs4762326 | 17,045 | 191,596 | T | C | 1.08 | 10−8 | 28537267 | ||
| 13q21.1 | PCDH17 | rs9538160 | 187,810 | 521,301 | A | G | 0.976 | 10−8 (10−3) * | 32959083 |
| 13q22.2-13q22.3 | LMO7-KCTD12 | rs9530566 | 3194 | 2057 | C | A | 1.08 | 10−5 | 29608257 |
| 14q23.2-14q23.2 | PARP1P2/RHOJ | rs10129516 | 171 | 2934 | T | C | 3.01 | 10−10 | 28881265 |
| 15q26.2 | NR2F2 | rs35625885 | 187,810 | 521,301 | A | G | 0.965 | 10−8 (10−2) * | 32959083 |
| 15q26.3 | IGF-1R | rs4966038 | 1448 | 1540 | C | G | 1.40 | 10−9 | 27506219 |
| 17p13.1 | SLC35G6 | rs9891297 | 46,262 | 364,789 | NA | NA | NA | 10−6 | 32121467 |
| 17q21.32 | SKAP1 | rs2278868 | 3194 | 2057 | C | T | 0.92 | 10−6 | 29608257 |
| 17q24.1 | CEP112 | rs17693745 | 3194 | 2057 | T | C | 1.08 | 10−5 | 29608257 |
| 19q12 | ZNF536-TSHZ3 | rs2198894 | 3194 | 2057 | T | C | 1.09 | 10−5 | 29608257 |
| 20q13.33 | LAMA5 | rs2427284 | 394 | 650 | A | G | 0.49 | 10−3 | 30044155 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lalami, I.; Abo, C.; Borghese, B.; Chapron, C.; Vaiman, D. Genomics of Endometriosis: From Genome Wide Association Studies to Exome Sequencing. Int. J. Mol. Sci. 2021, 22, 7297. https://doi.org/10.3390/ijms22147297
Lalami I, Abo C, Borghese B, Chapron C, Vaiman D. Genomics of Endometriosis: From Genome Wide Association Studies to Exome Sequencing. International Journal of Molecular Sciences. 2021; 22(14):7297. https://doi.org/10.3390/ijms22147297
Chicago/Turabian StyleLalami, Imane, Carole Abo, Bruno Borghese, Charles Chapron, and Daniel Vaiman. 2021. "Genomics of Endometriosis: From Genome Wide Association Studies to Exome Sequencing" International Journal of Molecular Sciences 22, no. 14: 7297. https://doi.org/10.3390/ijms22147297
APA StyleLalami, I., Abo, C., Borghese, B., Chapron, C., & Vaiman, D. (2021). Genomics of Endometriosis: From Genome Wide Association Studies to Exome Sequencing. International Journal of Molecular Sciences, 22(14), 7297. https://doi.org/10.3390/ijms22147297







