Selenoproteome Expression Studied by Non-Radioactive Isotopic Selenium-Labeling in Human Cell Lines
Abstract
:1. Introduction
2. Results
2.1. Fractionation of Intracellular Selenocompounds from HEK293 by Size-Exclusion Chromatography Followed by Inductively Coupled Plasma Mass Spectrometry Detection (SEC-ICP MS)
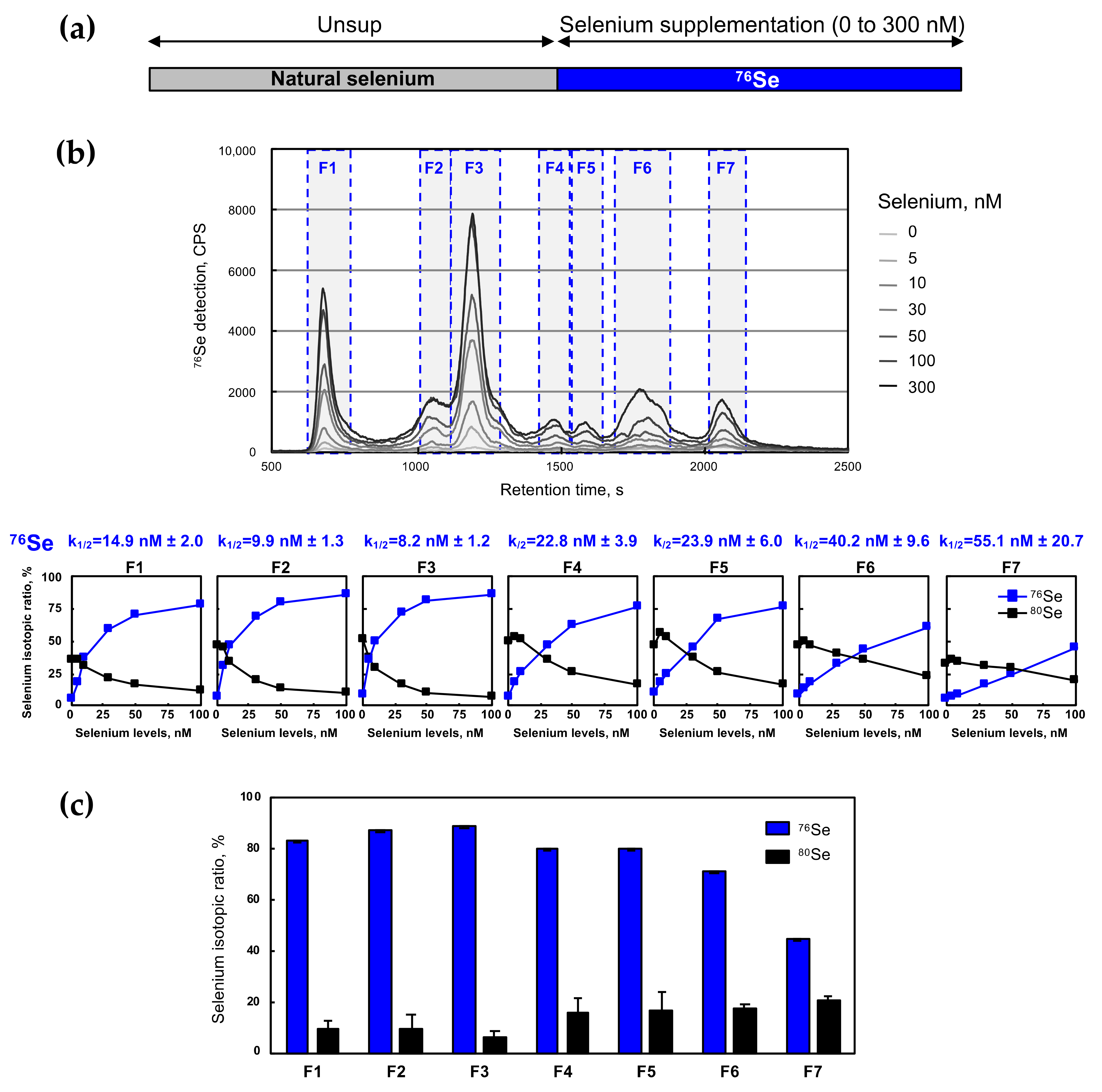
2.2. Selenium Assimilation in HEK293 Cells as a Function of Selenium Levels
2.3. The Selenoproteome Was Differentially Modulated According to the Selenium Level
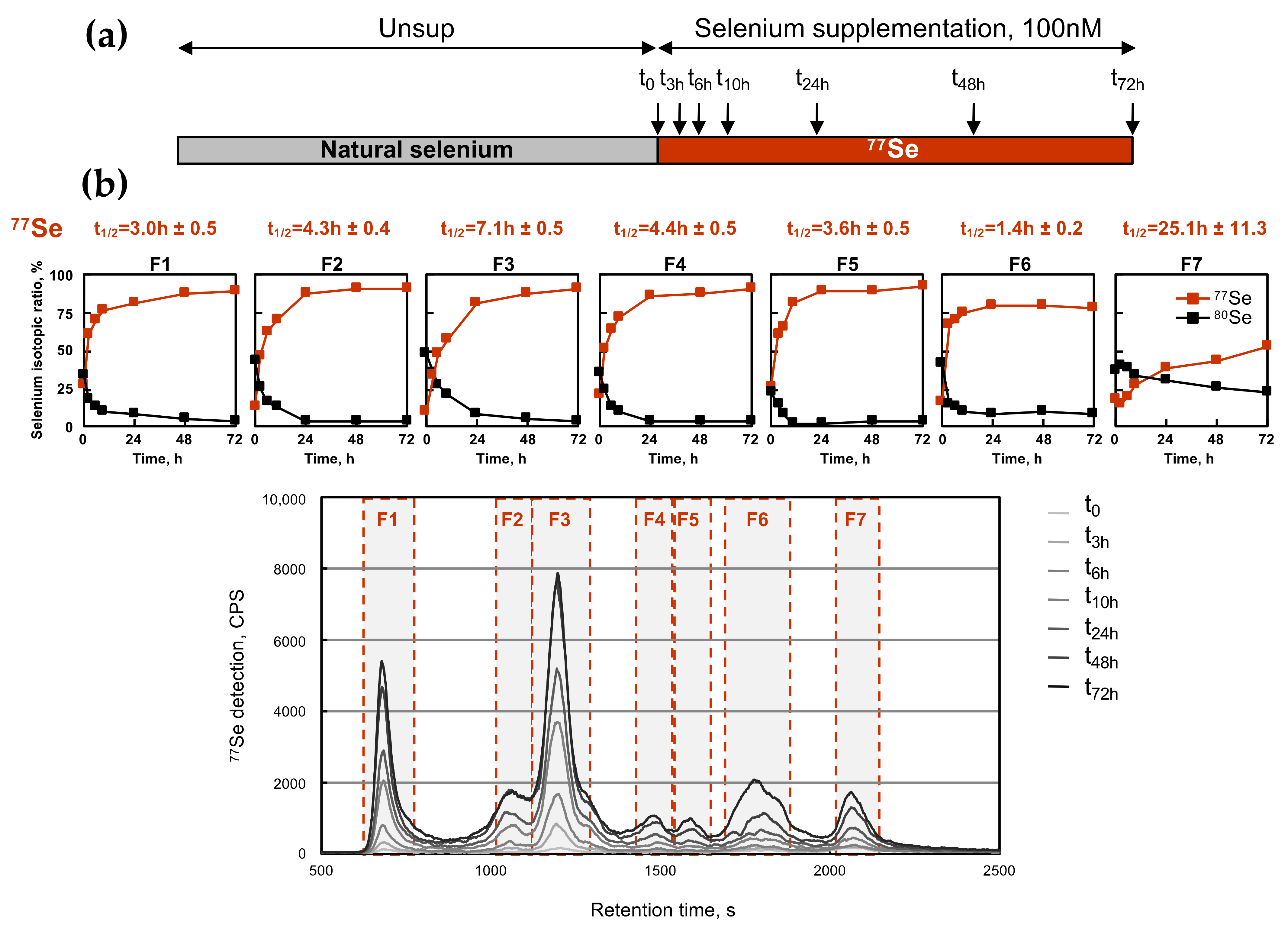
2.4. Kinetics of Selenium Assimilation by HEK293 Cells as Selenoproteins and as Low-Molecular Selenocompounds
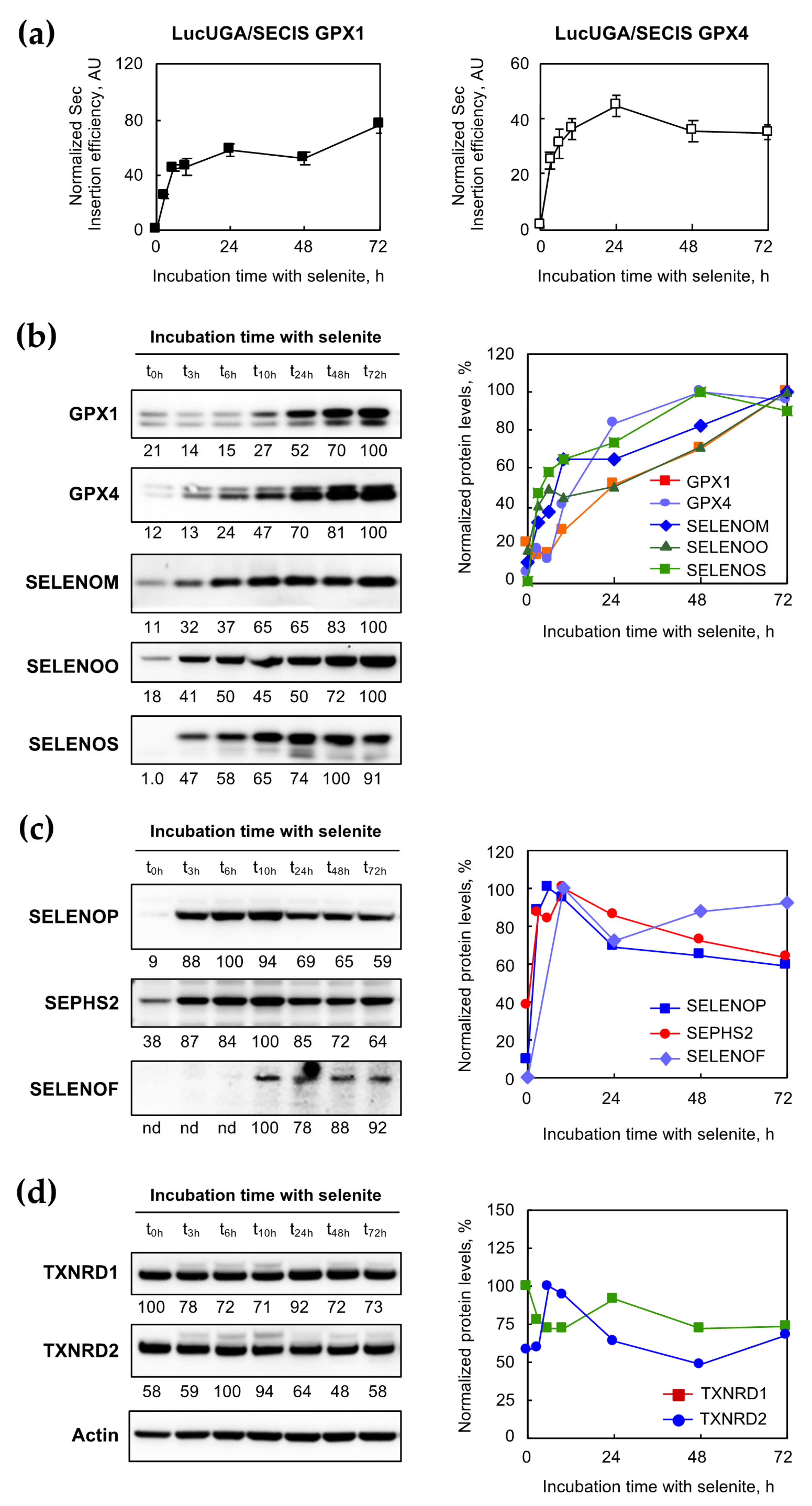
2.5. Selenocysteine Insertion in Selenoprotein Was Timely Controlled in Response to Selenium Supplementation
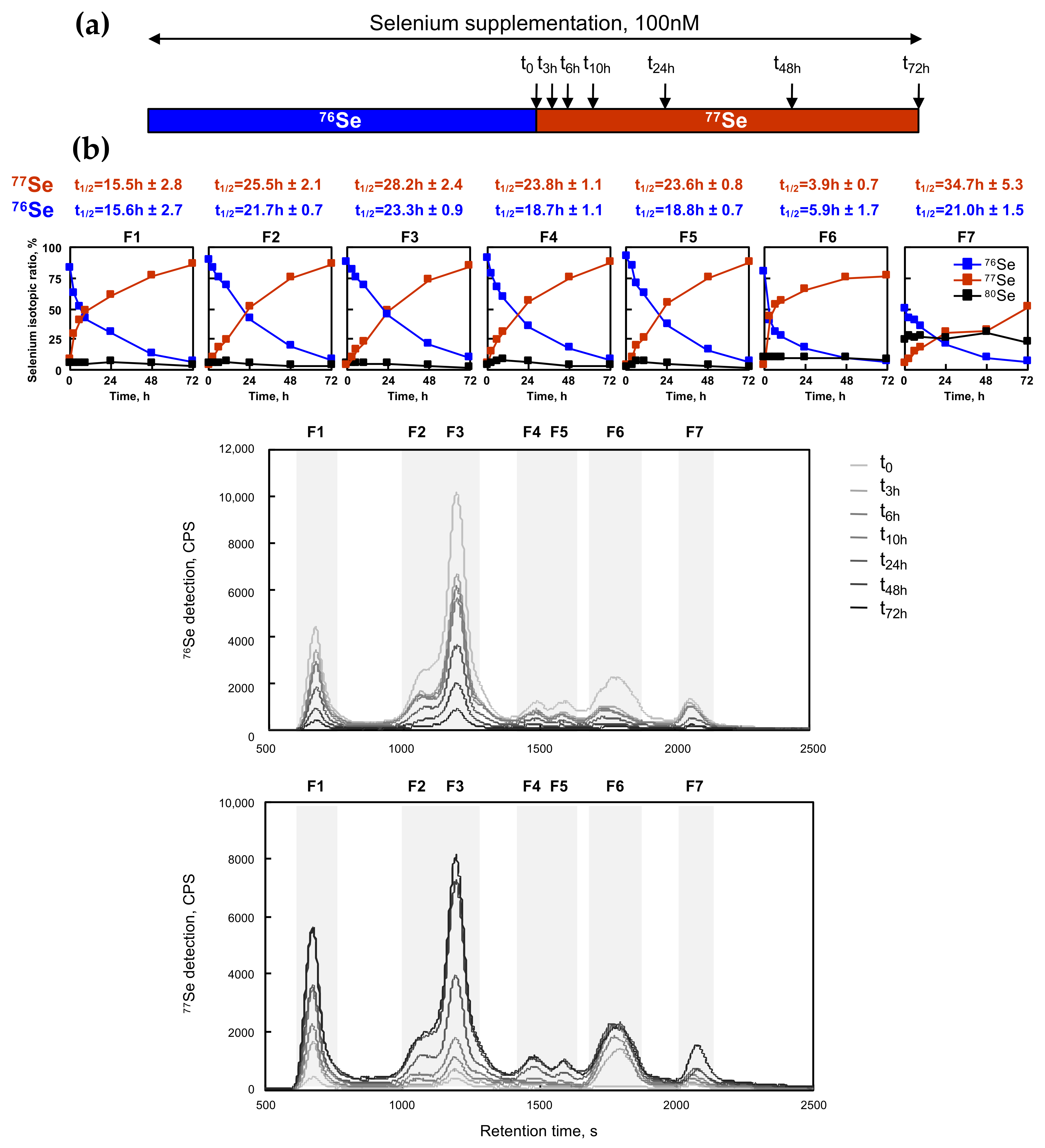
2.6. Kinetics of Selenium Exchange in HEK293 Grown in Selenium-Supplemented Conditions
3. Discussion
3.1. Hierarchy of Selenoproteins in Response to Selenium Supplementation: Kinetic and Dose–Response Analyses
3.2. Selenium-Enriched Isotope Labeling: An Alternative to Radioactive Labeling and a Novel Multiplexing Strategy for Selenoproteomic Analyses
4. Materials and Methods
4.1. Materials
4.2. Cell Culture and Incubation with Different Selenium Doses
4.3. Evaluation of Selenocysteine Insertion Efficiency
4.4. Fractionation of Selenium Containing Molecules by Size Exclusion Chromatography (SEC) with ICP MS Detection (SEC-ICP MS)
4.5. Protein Gels and Western Immunoblotting
4.6. Large Scale Fraction Collection for Western Immunoblotting Analyses
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, Z.H.; Song, G.L. Roles of selenoproteins in brain function and the potential mechanism of selenium in Alzheimer’s disease. Front. Neurosci. 2021, 15, 646518. [Google Scholar] [CrossRef]
- Guillin, O.M.; Vindry, C.; Ohlmann, T.; Chavatte, L. Selenium, selenoproteins and viral infection. Nutrients 2019, 11, 2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocca, C.; Pasqua, T.; Boukhzar, L.; Anouar, Y.; Angelone, T. Progress in the emerging role of selenoproteins in cardiovascular disease: Focus on endoplasmic reticulum-resident selenoproteins. Cell Mol. Life Sci. 2019, 76, 3969–3985. [Google Scholar] [CrossRef]
- Touat-Hamici, Z.; Legrain, Y.; Sonet, J.; Bulteau, A.-L.; Chavatte, L. Alteration of selenoprotein expression during stress and in aging. In Selenium: Its Molecular Biology and Role in Human Health, 4th ed.; Hatfield, D.L., Ulrich, S., Tsuji, P.A., Gladyshev, V.N., Eds.; Springer Science & Business Media LLC: New York, NY, USA, 2016; pp. 539–551. [Google Scholar]
- Sonet, J.; Bulteau, A.-L.; Chavatte, L. Selenium and selenoproteins in human health and diseases. In Metallomics: Analytical Techniques and Speciation Methods; Michalke, B., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Hoboken, NJ, USA, 2016; pp. 364–381. [Google Scholar]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [Green Version]
- Latrèche, L.; Chavatte, L. Selenium incorporation into selenoproteins, implications in human health. Metal. Ions Biol. Med. 2008, 10, 731–737. [Google Scholar]
- Papp, L.V.; Lu, J.; Holmgren, A.; Khanna, K.K. From selenium to selenoproteins: Synthesis, identity, and their role in human health. Antioxid. Redox. Signal. 2007, 9, 775–806. [Google Scholar] [CrossRef]
- Gladyshev, V.N.; Arner, E.S.; Berry, M.J.; Brigelius-Flohe, R.; Bruford, E.A.; Burk, R.F.; Carlson, B.A.; Castellano, S.; Chavatte, L.; Conrad, M.; et al. Selenoprotein gene nomenclature. J. Biol. Chem. 2016, 291, 24036–24040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vindry, C.; Ohlmann, T.; Chavatte, L. Translation regulation of mammalian selenoproteins. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2480–2492. [Google Scholar] [CrossRef] [PubMed]
- Bulteau, A.-L.; Chavatte, L. Update on selenoprotein biosynthesis. Antioxid. Redox. Signal. 2015, 23, 775–794. [Google Scholar] [CrossRef]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigo, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef] [Green Version]
- Driscoll, D.M.; Copeland, P.R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003, 23, 17–40. [Google Scholar] [CrossRef]
- Touat-Hamici, Z.; Bulteau, A.L.; Bianga, J.; Jean-Jacques, H.; Szpunar, J.; Lobinski, R.; Chavatte, L. Selenium-regulated hierarchy of human selenoproteome in cancerous and immortalized cells lines. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2493–2505. [Google Scholar] [CrossRef]
- Touat-Hamici, Z.; Legrain, Y.; Bulteau, A.-L.; Chavatte, L. Selective up-regulation of human selenoproteins in response to oxidative stress. J. Biol. Chem. 2014, 289, 14750–14761. [Google Scholar] [CrossRef] [Green Version]
- Legrain, Y.; Touat-Hamici, Z.; Chavatte, L. Interplay between selenium levels, selenoprotein expression, and replicative senescence in WI-38 human fibroblasts. J. Biol. Chem. 2014, 289, 6299–6310. [Google Scholar] [CrossRef] [Green Version]
- Papp, L.V.; Lu, J.; Striebel, F.; Kennedy, D.; Holmgren, A.; Khanna, K.K. The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function. Mol. Cell Biol. 2006, 26, 4895–4910. [Google Scholar] [CrossRef] [Green Version]
- Hammad, G.; Legrain, Y.; Touat-Hamici, Z.; Duhieu, S.; Cornu, D.; Bulteau, A.-L.; Chavatte, L. Interplay between Selenium Levels and Replicative Senescence in WI-38 Human Fibroblasts: A Proteomic Approach. Antioxidants 2018, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allmang, C.; Wurth, L.; Krol, A. The selenium to selenoprotein pathway in eukaryotes: More molecular partners than anticipated. Biochim. Biophys. Acta 2009, 1790, 1415–1423. [Google Scholar] [CrossRef]
- Hatfield, D.L.; Gladyshev, V.N. How selenium has altered our understanding of the genetic code. Mol. Cell Biol. 2002, 22, 3565–3576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, M.J.; Tujebajeva, R.M.; Copeland, P.R.; Xu, X.M.; Carlson, B.A.; Martin, G.W., 3rd; Low, S.C.; Mansell, J.B.; Grundner-Culemann, E.; Harney, J.W.; et al. Selenocysteine incorporation directed from the 3’UTR: Characterization of eukaryotic EFsec and mechanistic implications. Biofactors 2001, 14, 17–24. [Google Scholar] [CrossRef]
- Latreche, L.; Duhieu, S.; Touat-Hamici, Z.; Jean-Jean, O.; Chavatte, L. The differential expression of glutathione peroxidase 1 and 4 depends on the nature of the SECIS element. RNA Biol. 2012, 9, 681–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vindry, C.; Guillin, O.; Mangeot, P.E.; Ohlmann, T.; Chavatte, L. A Versatile Strategy to Reduce UGA-Selenocysteine Recoding Efficiency of the Ribosome Using CRISPR-Cas9-Viral-Like-Particles Targeting Selenocysteine-tRNA[Ser]Sec Gene. Cells 2019, 8, 574. [Google Scholar] [CrossRef] [Green Version]
- Sonet, J.; Bierla, K.; Bulteau, A.L.; Lobinski, R.; Chavatte, L. Comparison of analytical methods using enzymatic activity, immunoaffinity and selenium-specific mass spectrometric detection for the quantitation of glutathione peroxidase 1. Anal. Chim. Acta 2018, 1011, 11–19. [Google Scholar] [CrossRef]
- Sonet, J.; Mounicou, S.; Chavatte, L. Detection of selenoproteins by laser ablation inductively coupled plasma mass spectrometry (LA-ICP MS) in immobilized pH gradient (IPG) strips. Methods Mol. Biol. 2018, 1661, 205–217. [Google Scholar]
- Bianga, J.; Touat-Hamici, Z.; Bierla, K.; Mounicou, S.; Szpunar, J.; Chavatte, L.; Lobinski, R. Speciation analysis for trace levels of selenoproteins in cultured human cells. J. Proteom. 2014, 108, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Bianga, J.; Ballihaut, G.; Pecheyran, C.; Touat, Z.; Preud’homme, H.; Mounicou, S.; Chavatte, L.; Lobinski, R.; Szpunar, J. Detection of selenoproteins in human cell extracts by laser ablation-ICP MS after separation by polyacrylamide gel electrophoresis and blotting. J. Anal. At. Spectrom. 2012, 27, 25–32. [Google Scholar] [CrossRef]
- Ballihaut, G.; Mounicou, S.; Lobinski, R. Multitechnique mass-spectrometric approach for the detection of bovine glutathione peroxidase selenoprotein: Focus on the selenopeptide. Anal. Bioanal. Chem. 2007, 388, 585–591. [Google Scholar] [CrossRef]
- Yim, S.H.; Tobe, R.; Turanov, A.A.; Carlson, B.A. Radioactive 75Se labeling and detection of selenoproteins. Methods Mol. Biol. 2018, 1661, 177–192. [Google Scholar] [PubMed]
- Korotkov, K.V.; Kumaraswamy, E.; Zhou, Y.; Hatfield, D.L.; Gladyshev, V.N. Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 2001, 276, 15330–15336. [Google Scholar] [CrossRef] [Green Version]
- Vindry, C.; Ohlmann, T.; Chavatte, L. Selenium metabolism, regulation, and sex differences in mammals. In Selenium, Molecular and Integartive Toxicology; Michalke, B., Ed.; Springer International Publishing AG, Part of Springer Nature: Cham, Switzerland, 2018; pp. 89–107. [Google Scholar]
- Sonet, J.; Mosca, M.; Bierla, K.; Modzelewska, K.; Flis-Borsuk, A.; Suchocki, P.; Ksiazek, I.; Anuszewska, E.; Bulteau, A.L.; Szpunar, J.; et al. Selenized plant oil is an efficient source of selenium for selenoprotein biosynthesis in human cell lines. Nutrients 2019, 11, 1524. [Google Scholar] [CrossRef] [Green Version]
- Latreche, L.; Jean-Jean, O.; Driscoll, D.M.; Chavatte, L. Novel structural determinants in human SECIS elements modulate the translational recoding of UGA as selenocysteine. Nucleic Acids Res. 2009, 37, 5868–5880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bermano, G.; Arthur, J.R.; Hesketh, J.E. Selective control of cytosolic glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase mRNA stability by selenium supply. FEBS Lett. 1996, 387, 157–160. [Google Scholar] [CrossRef] [Green Version]
- Bermano, G.; Nicol, F.; Dyer, J.A.; Sunde, R.A.; Beckett, G.J.; Arthur, J.R.; Hesketh, J.E. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem. J. 1995, 311 Pt 2, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Carlson, B.A.; Lee, B.J.; Tsuji, P.A.; Tobe, R.; Park, J.M.; Schweizer, U.; Gladyshev, V.N.; Hatfield, D.L. Selenocysteine tRNA [Ser]Sec: From nonsense suppressor tRNA to the quintessential constituent in selenoprotein biosynthesis. In Selenium: Its Molecular Biology and Role in Human Health, 4th ed.; Hatfield, D.L., Ulrich, S., Tsuji, P.A., Gladyshev, V.N., Eds.; Springer Science & Business Media LLC: New York, NY, USA, 2016; pp. 3–12. [Google Scholar]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Moghaddas Gholami, A.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yang, W.; Huang, Q.; Qiang, J.; Hart, J.R.; Wang, W.; Hu, J.; Zhu, J.; Liu, N.; Zhang, Y. Selenocysteine-specific mass spectrometry reveals tissue-distinct slenoproteomes and candidate selenoproteins. Cell Chem. Biol. 2018, 25, 1380–1388.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
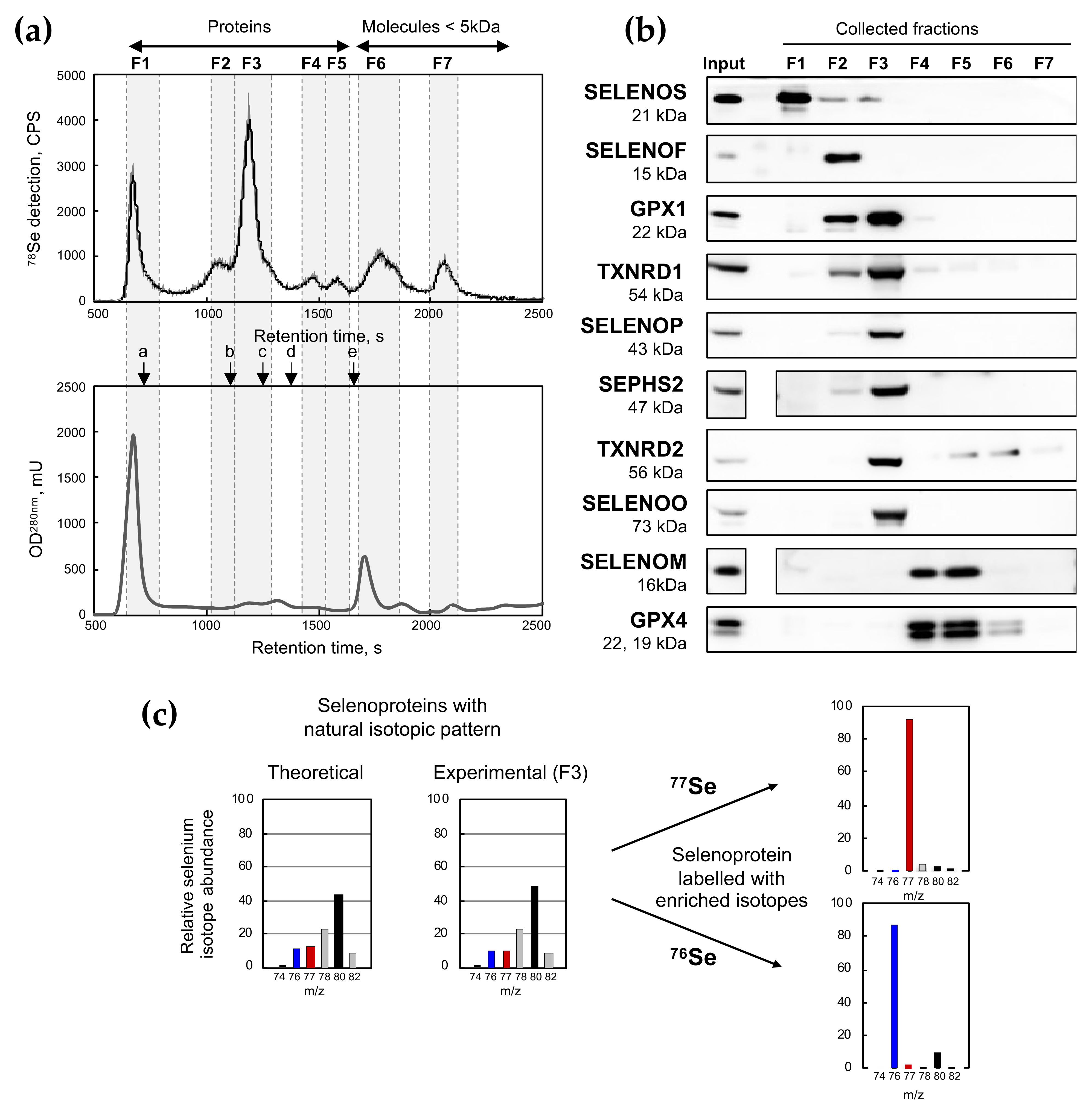

| Fraction N° | Retention Time, s | MW Range, kDa | Selenium Signal Repartition, % | Selenoproteins Detected by Westernblot (Figure 1) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dpl | Dpl + 100 nM Se | |||||||||
| Start | End | Ave | SD | Ave | SD | |||||
| F1 | 600 | 760 | >480 | 12.0 | ± | 0.6 | 17.5 | ± | 0.9 | SelenoS |
| F2 | 1000 | 1120 | 140–75 | 4.7 | ± | 0.2 | 10.2 | ± | 0.5 | SelenoF, Gpx1, TR1 |
| F3 | 1120 | 1300 | 75–30 | 25.4 | ± | 1.3 | 37.3 | ± | 1.9 | Gpx1, Txnrd1, SelenoP, Sephs2, Txnrd2, SelenoO |
| F4 | 1410 | 1540 | 8–16 | 3.5 | ± | 0.2 | 5.8 | ± | 0,3 | SelenoM, Gpx4 |
| F5 | 1540 | 1650 | 4–8 | 2.7 | ± | 0.1 | 4.4 | ± | 0.2 | SelenoM, Gpx4, Txnrd2 |
| F6 | 1660 | 1870 | 1.5–4 | 24.7 | ± | 1.2 | 16.5 | ± | 0.8 | Gpx4, Txnrd2 |
| F7 | 1990 | 2130 | 0.38–0.80 | 27.1 | ± | 1.4 | 8.4 | ± | 0.4 | n.d. |
| Fraction N° | Dose–Response (Figure 2) | Kinetics of Selenium-Labeling (Figure 4) | Kinetics of Selenium Exchange (Figure 6) | |
|---|---|---|---|---|
| k1/2 76Se, nM | t1/2 77Se, h | t1/2 77Se, h | t1/2 76Se, h | |
| F1 | 14.9 ± 2.0 | 3.0 ± 0.5 | 15.5 ± 2.8 | 15.6 ± 2.7 |
| F2 | 9.9 ± 1.3 | 4.3 ± 0.4 | 25.5 ± 2.1 | 21.7 ± 0.7 |
| F3 | 8.2 ± 1.2 | 7.1 ± 0.5 | 28.2 ± 2.4 | 23.3 ± 0.9 |
| F4 | 22.8 ± 3.9 | 4.4 ± 0.5 | 23.8 ± 1.1 | 18.7 ± 1.1 |
| F5 | 23.9 ± 6.0 | 3.6 ± 0.5 | 23.6 ± 0.8 | 18.8 ± 0.7 |
| F6 | 40.2 ± 9.6 | 1.4 ± 0.2 | 3.9 ± 0.7 | 5.9 ± 1.7 |
| F7 | 55.1 ± 20.7 | 25.1 ± 11.3 | 34.7 ± 5.3 | 21.0 ± 1.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sonet, J.; Bulteau, A.-L.; Touat-Hamici, Z.; Mosca, M.; Bierla, K.; Mounicou, S.; Lobinski, R.; Chavatte, L. Selenoproteome Expression Studied by Non-Radioactive Isotopic Selenium-Labeling in Human Cell Lines. Int. J. Mol. Sci. 2021, 22, 7308. https://doi.org/10.3390/ijms22147308
Sonet J, Bulteau A-L, Touat-Hamici Z, Mosca M, Bierla K, Mounicou S, Lobinski R, Chavatte L. Selenoproteome Expression Studied by Non-Radioactive Isotopic Selenium-Labeling in Human Cell Lines. International Journal of Molecular Sciences. 2021; 22(14):7308. https://doi.org/10.3390/ijms22147308
Chicago/Turabian StyleSonet, Jordan, Anne-Laure Bulteau, Zahia Touat-Hamici, Maurine Mosca, Katarzyna Bierla, Sandra Mounicou, Ryszard Lobinski, and Laurent Chavatte. 2021. "Selenoproteome Expression Studied by Non-Radioactive Isotopic Selenium-Labeling in Human Cell Lines" International Journal of Molecular Sciences 22, no. 14: 7308. https://doi.org/10.3390/ijms22147308
APA StyleSonet, J., Bulteau, A.-L., Touat-Hamici, Z., Mosca, M., Bierla, K., Mounicou, S., Lobinski, R., & Chavatte, L. (2021). Selenoproteome Expression Studied by Non-Radioactive Isotopic Selenium-Labeling in Human Cell Lines. International Journal of Molecular Sciences, 22(14), 7308. https://doi.org/10.3390/ijms22147308








