Analysis of Phytohormone Signal Transduction in Sophora alopecuroides under Salt Stress
Abstract
:1. Introduction
2. Results
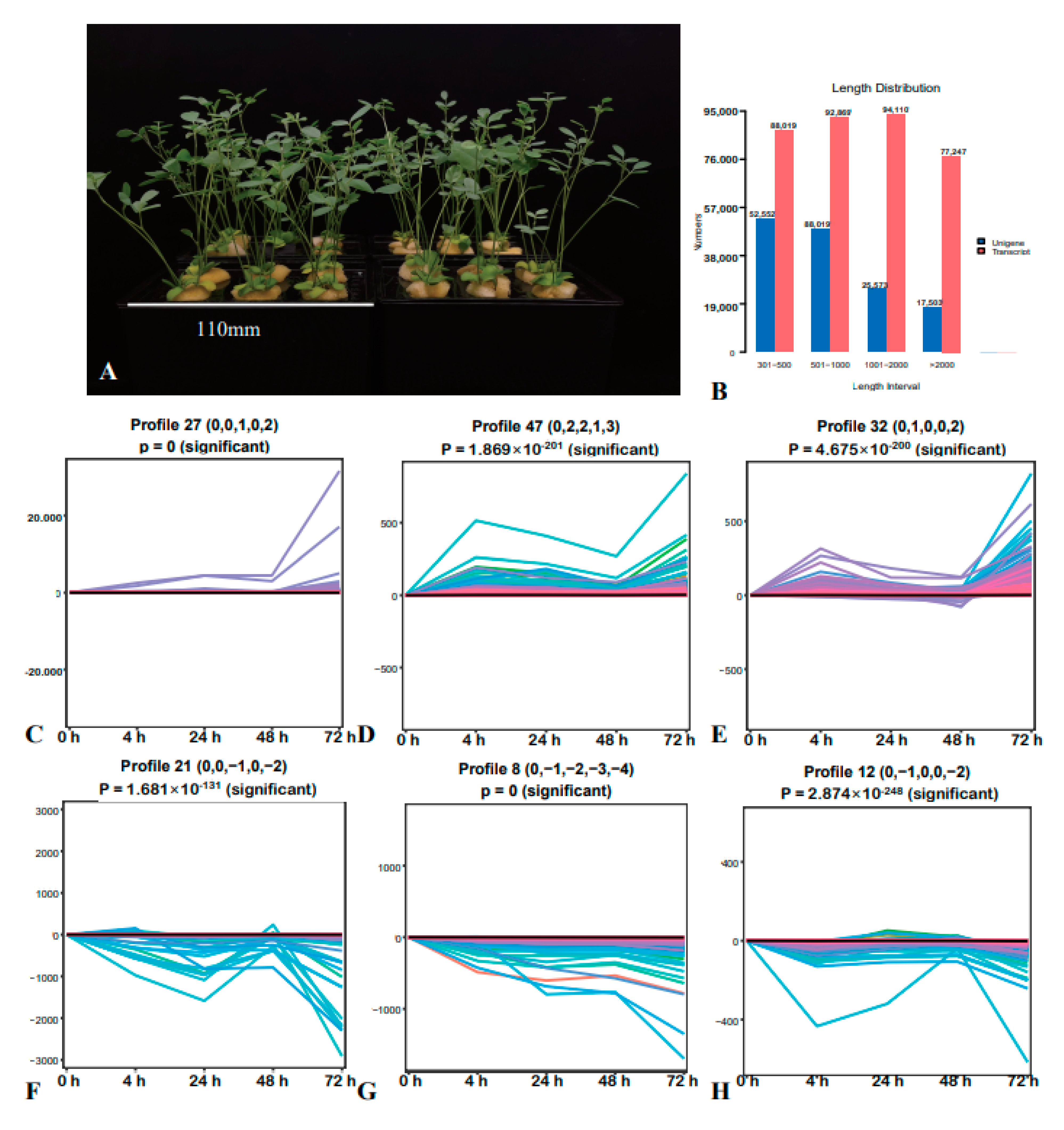
2.1. Transcriptome Analysis and STEM Analysis of DEGs
2.2. STEM Analysis of DMs
2.3. DEGs Were Significantly Enriched in Plant Hormone Signal Transduction
2.4. AUX, CKs, GA, and BRs Regulated S. alopecuroides Growth under Salt Stress
2.5. JA and ETH Signals Negatively Regulate S. alopecuroides in Response to Salt Stress
2.6. ABA and SA Levels Increased to Enhance S. alopecuroides Tolerance to Salt Stress
2.7. Phytohormones Regulated the Balance of S. alopecuroides Growth and Tolerance under Salt Stress
3. Discussion
4. Material and Methods
4.1. Growth of Plants and Salt Stress
4.2. Transcriptome Sequencing Analysis and STEM Analysis of DEGs
4.3. Non-Targeted Metabolite Detection and STEM Analysis of DMs
4.4. KEGG Enrichment Analysis of DEGs
4.5. Joint Analysis of DEGs and DMs of Phytohormone Signal Transduction Pathways
4.6. Analysis of the PPI Network of the DEGs
4.7. qRT-PCR Quantitative Verification of DEGs and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [Green Version]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.Y.; Li, J.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wang, Q.; Wang, Y.; Xu, Y.; Li, J.; Zhao, S.; Wang, D.; Ma, Z.; Yan, F.; Liu, Y. Combined Transcriptomic and Metabolomic Analysis Reveals the Role of Phenylpropanoid Biosynthesis Pathway in the Salt Tolerance Process of Sophora alopecuroides. Int. J. Mol. Sci. 2021, 22, 2399. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Zhu, Y.; Zhao, Y.; Wang, Y.; Li, J.; Wang, Q.; Liu, Y. De novo transcriptome sequencing and analysis of salt-, alkali-, and drought-responsive genes in Sophora alopecuroides. BMC Genom. 2020, 21, 423. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [Green Version]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Yu, Z.; Gao, L.; Zheng, C. SnRK2s at the Crossroads of Growth and Stress Responses. Trends Plant Sci. 2019, 24, 672–676. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, D.; Xu, Y.; Jin, S.; Zhang, L.; Zhang, S.; Yang, G.; Huang, J.; Yan, K.; Wu, C.; et al. CEPR2 phosphorylates and accelerates the degradation of PYR/PYLs in Arabidopsis. J. Exp. Bot. 2019, 70, 5457–5469. [Google Scholar] [CrossRef] [Green Version]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef]
- Ribba, T.; Garrido-Vargas, F.; O’Brien, J.A. Auxin-mediated responses under salt stress: From developmental regulation to biotechnological applications. J. Exp. Bot. 2020, 71, 3843–3853. [Google Scholar] [CrossRef]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, M.J.; Terrile, M.C.; Bartoli, C.G.; D’Ippólito, S.; Casalongué, C.A. Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis. Plant Mol. Biol. 2010, 74, 215–222. [Google Scholar] [CrossRef]
- Iglesias, M.J.; Terrile, M.C.; Windels, D.; Lombardo, M.C.; Bartoli, C.G.; Vazquez, F.; Estelle, M.; Casalongué, C.A. MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS ONE 2014, 9, e107678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyoung, S.; Cho, S.H.; Chung, J.H.; So, W.M.; Cui, M.H.; Shin, J.S. Cytokinin oxidase PpCKX1 plays regulatory roles in development and enhances dehydration and salt tolerance in Physcomitrella patens. Plant Cell Rep. 2020, 39, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Romanov, G.A.; Schmülling, T. Opening Doors for Cytokinin Trafficking at the ER Membrane. Trends Plant Sci. 2021, 26, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavlů, J.; Novák, J.; Koukalová, V.; Luklová, M.; Brzobohatý, B.; Černý, M. Cytokinin at the Crossroads of Abiotic Stress Signalling Pathways. Int. J. Mol. Sci. 2018, 19, 2450. [Google Scholar] [CrossRef] [Green Version]
- Schachtman, D.P.; Goodger, J.Q. Chemical root to shoot signaling under drought. Trends Plant Sci. 2008, 13, 281–287. [Google Scholar] [CrossRef]
- Wei, L.J.; Deng, X.G.; Zhu, T.; Zheng, T.; Li, P.X.; Wu, J.Q.; Zhang, D.W.; Lin, H.H. Ethylene is Involved in Brassinosteroids Induced Alternative Respiratory Pathway in Cucumber (Cucumis sativus L.) Seedlings Response to Abiotic Stress. Front. Plant Sci. 2015, 6, 982. [Google Scholar] [CrossRef] [Green Version]
- Zhu, T.; Deng, X.; Zhou, X.; Zhu, L.; Zou, L.; Li, P.; Zhang, D.; Lin, H. Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci. Rep. 2016, 6, 35392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davière, J.M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [Green Version]
- Bao, S.; Hua, C.; Shen, L.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Qin, H.; Zhou, S.; Wei, P.; Zhang, H.; Zhou, Y.; Miao, Y.; Huang, R. The Ubiquitin-Binding Protein OsDSK2a Mediates Seedling Growth and Salt Responses by Regulating Gibberellin Metabolism in Rice. Plant Cell 2020, 32, 414–428. [Google Scholar] [CrossRef] [Green Version]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.F.; Etheridge, N.; Schaller, G.E. Ethylene signal transduction. Ann. Bot. 2005, 95, 901–915. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Alonso, J.M. From Ethylene-Auxin Interactions to Auxin Biosynthesis and Signal Integration. Plant Cell 2019, 31, 1393–1394. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Smith, J.A.; Harberd, N.P.; Jiang, C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016, 91, 651–659. [Google Scholar] [CrossRef]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H.; Guo, H. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 2014, 10, e1004664. [Google Scholar] [CrossRef] [Green Version]
- Awan, S.A.; Khan, I.; Rizwan, M.; Zhang, X.; Brestic, M.; Khan, A.; El-Sheikh, M.A.; Alyemeni, M.N.; Ali, S.; Huang, L. Exogenous abscisic acid and jasmonic acid restrain polyethylene glycol-induced drought by improving the growth and antioxidative enzyme activities in pearl millet. Physiol. Plant. 2020, 172, 809–819. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edel, K.H.; Kudla, J. Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 2016, 33, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Motomura, Y.; Yagi, Y.; Nomura, H.; Kikuchi, S.; Nakai, M.; Shiina, T. Chloroplast envelope localization of EDS5, an essential factor for salicylic acid biosynthesis in Arabidopsis thaliana. Plant. Signal. Behav. 2013, 8, e23603. [Google Scholar] [CrossRef] [Green Version]
- Shakirova, F.M.; Sakhabutdinova, A.R.; Bezrukova, M.V.; Fatkhutdinova, R.A.; Fatkhutdinova, D.R. Changes in the hormonal status of wheat seedlings induced by salicylic acid and salinity. Plant Sci. 2003, 164, 317–322. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: A New Plant Hormone and/or a Plant Master Regulator? Trends Plant Sci. 2019, 24, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, K.; Soga, K.; Hoson, T.; Kotake, T.; Kojima, M.; Sakakibara, H.; Yamazaki, T.; Higashibata, A.; Ishioka, N.; Shimazu, T.; et al. Persistence of plant hormone levels in rice shoots grown under microgravity conditions in space: Its relationship to maintenance of shoot growth. Physiol. Plant. 2017, 161, 285–293. [Google Scholar] [CrossRef]
- Ding, Y.; Murphy, K.M.; Poretsky, E.; Mafu, S.; Yang, B.; Char, S.N.; Christensen, S.A.; Saldivar, E.; Wu, M.; Wang, Q.; et al. Multiple genes recruited from hormone pathways partition maize diterpenoid defences. Nat. Plants 2019, 5, 1043–1056. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J.; Zhou, Y. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2020, 18, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Choi, W.G.; Toyota, M.; Kim, S.H.; Hilleary, R.; Gilroy, S. Salt stress–induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 2014, 111, 6497–6502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvan-Ampudia, C.S.; Julkowska, M.M.; Darwish, E.; Gandullo, J.; Korver, R.A.; Brunoud, G.; Haring, M.A.; Munnik, T.; Vernoux, T.; Testerink, C. Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol. 2013, 23, 2044–2050. [Google Scholar] [CrossRef] [Green Version]
- Barkla, B.J.; Castellanos-Cervantes, T.; de León, J.L.; Matros, A.; Mock, H.P.; Perez-Alfocea, F.; Salekdeh, G.H.; Witzel, K.; Zörb, C. Elucidation of salt stress defense and tolerance mechanisms of crop plants using proteomics—Current achievements and perspectives. Proteomics 2013, 13, 1885–1900. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zolla, G.; Heimer, Y.M.; Barak, S. Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J. Exp. Bot. 2010, 61, 211–224. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Li, R.J.; Han, T.T.; Cai, W.; Fu, Z.W.; Lu, Y.T. Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, R.; Watanabe, Y.; Fujita, Y.; Le, D.T.; Kojima, M.; Werner, T.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Kakimoto, T.; et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 2011, 23, 2169–2183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Shen, W.; Chan, Z.; Wu, Y. Endogenous Cytokinin Overproduction Modulates ROS Homeostasis and Decreases Salt Stress Resistance in Arabidopsis Thaliana. Front. Plant Sci. 2015, 6, 1004. [Google Scholar] [CrossRef] [Green Version]
- Divi, U.K.; Rahman, T.; Krishna, P. Brassinosteroid-Mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 2010, 10, 151. [Google Scholar] [CrossRef] [Green Version]
- Su, Q.; Zheng, X.; Tian, Y.; Wang, C. Exogenous Brassinolide Alleviates Salt Stress in Malus hupehensis Rehd. by Regulating the Transcription of NHX-Type Na+(K+)/H+ Antiporters. Front. Plant Sci. 2020, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Guan, L.; Sun, Y.; Zhu, Y.; Liu, L.; Lu, R.; Jiang, M.; Tan, M.; Zhang, A. Calcium and ZmCCaMK are involved in brassinosteroid-induced antioxidant defense in maize leaves. Plant Cell Physiol. 2015, 56, 883–896. [Google Scholar] [CrossRef] [Green Version]
- Duan, F.; Ding, J.; Lee, D.; Lu, X.; Feng, Y.; Song, W. Overexpression of SoCYP85A1, a Spinach Cytochrome p450 Gene in Transgenic Tobacco Enhances Root Development and Drought Stress Tolerance. Front. Plant Sci. 2017, 8, 1909. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M.; Zhou, D.X.; Yang, W.; Zhao, Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Mei, Z.; Duan, J.; Chen, H.; Feng, H.; Cai, W. OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS ONE 2014, 9, e87110. [Google Scholar] [CrossRef] [PubMed]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008, 56, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, Z.; Xiao, G.; Zhai, M.; Pan, X.; Huang, R.; Zhang, H. CYP71D8L is a key regulator involved in growth and stress responses by mediating gibberellin homeostasis in rice. J. Exp. Bot. 2020, 71, 1160–1170. [Google Scholar]
- Yang, C.; Ma, B.; He, S.J.; Xiong, Q.; Duan, K.X.; Yin, C.C.; Chen, H.; Lu, X.; Chen, S.Y.; Zhang, J.S. MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 Regulate Ethylene Response of Roots and Coleoptiles and Negatively Affect Salt Tolerance in Rice. Plant Physiol. 2015, 169, 148–165. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Duan, J.; Shan, C.; Mei, Z.; Chen, H.; Feng, H.; Zhu, J.; Cai, W. Ethylene insensitive3-like2 (OsEIL2) confers stress sensitivity by regulating OsBURP16, the β subunit of polygalacturonase (PG1β-like) subfamily gene in rice. Plant Sci. 2020, 292, 110353. [Google Scholar] [CrossRef]
- An, F.; Zhao, Q.; Ji, Y.; Li, W.; Jiang, Z.; Yu, X.; Zhang, C.; Han, Y.; He, W.; Liu, Y.; et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 2010, 22, 2384–2401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.; Belfield, E.J.; Cao, Y.; Smith, J.A.; Harberd, N.P. An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 2013, 25, 3535–3552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, R.L.; Kim, H.; Bakshi, A.; Binder, B.M. The Ethylene Receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 Have Contrasting Roles in Seed Germination of Arabidopsis during Salt Stress. Plant Physiol. 2014, 165, 1353–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liu, C.; Li, K.; Sun, F.; Hu, H.; Li, X.; Zhao, Y.; Han, C.; Zhang, W.; Duan, Y.; et al. Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol. Biol. 2007, 64, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dong, W.; Zhang, N.; Ai, X.; Wang, M.; Huang, Z.; Xiao, L.; Xia, G. A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 2014, 164, 1068–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Abellan, J.O.; Fernandez-Garcia, N.; Lopez-Berenguer, C.; Egea, I.; Flores, F.B.; Angosto, T.; Capel, J.; Lozano, R.; Pineda, B.; Moreno, V.; et al. The tomato res mutant which accumulates JA in roots in non-stressed conditions restores cell structure alterations under salinity. Physiol. Plant. 2015, 155, 296–314. [Google Scholar] [CrossRef] [Green Version]
- Hazman, M.; Hause, B.; Eiche, E.; Nick, P.; Riemann, M. Increased tolerance to salt stress in OPDA-deficient rice ALLENE OXIDE CYCLASE mutants is linked to an increased ROS-scavenging activity. J. Exp. Bot. 2015, 66, 3339–3352. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, R.M.; Cheng, C.; Sheng, J.; Wang, W.; Ren, H.; Aslam, M.; Yan, Y. Interruption of Jasmonic Acid Biosynthesis Causes Differential Responses in the Roots and Shoots of Maize Seedlings against Salt Stress. Int. J. Mol. Sci. 2019, 20, 6202. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [Green Version]
- Cai, S.; Chen, G.; Wang, Y.; Huang, Y.; Marchant, D.B.; Wang, Y.; Yang, Q.; Dai, F.; Hills, A.; Blatt, M.R. Evolutionary Conservation of ABA Signaling for Stomatal Closure. Plant Physiol. 2017, 174, 732–747. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Xie, J.; Chen, C.; Cao, H.; Sun, J.; Kong, Q.; Shabala, S.; Shabala, L.; Huang, Y.; Bie, Z. An early ABA-induced stomatal closure, Na+ sequestration in leaf vein and K+ retention in mesophyll confer salt tissue tolerance in Cucurbita species. J. Exp. Bot. 2018, 69, 4945–4960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahanger, M.A.; Aziz, U.; Alsahli, A.A.; Alyemeni, M.N.; Ahmad, P. Influence of Exogenous Salicylic Acid and Nitric Oxide on Growth, Photosynthesis, and Ascorbate-Glutathione Cycle in Salt Stressed Vigna angularis. Biomolecules 2019, 10, 42. [Google Scholar] [CrossRef] [Green Version]
- Yuan-Yuan, W.; Yu, W.; Guang-Zhe, L.; Lin, H. Salicylic acid-altering Arabidopsis plant response to cadmium exposure: Underlying mechanisms affecting antioxidation and photosynthesis-related processes. Ecotoxicol. Environ. Saf. 2019, 169, 645–653. [Google Scholar]
- Jayakannan, M.; Bose, J.; Babourina, O.; Shabala, S.; Massart, A.; Poschenrieder, C.; Rengel, Z. The NPR1-dependent salicylic acid signalling pathway is pivotal for enhanced salt and oxidative stress tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 1865–1875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quilis, J.; Peñas, G.; Messeguer, J.; Brugidou, C.; San Segundo, B. The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol. Plant Microbe Interact. 2008, 21, 1215–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, K.J.P.; Mahna, N.; Mou, Z.; Folta, K.M. NPR1 as a transgenic crop protection strategy in horticultural species. Hortic. Res. 2018, 5, 15. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Chen, M.X.; Liu, R.; Zhang, L.; Hou, X.; Liu, S.; Ding, X.; Jiang, Y.; Xu, J.; Zhang, J.; et al. Abscisic Acid Regulates Auxin Distribution to Mediate Maize Lateral Root Development Under Salt Stress. Front. Plant Sci. 2019, 10, 716. [Google Scholar] [CrossRef]
- Albacete, A.; Ghanem, M.E.; Martínez-Andújar, C.; Acosta, M.; Sánchez-Bravo, J.; Martínez, V.; Lutts, S.; Dodd, I.C.; Pérez-Alfocea, F. Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 2008, 59, 4119–4131. [Google Scholar] [CrossRef]
- Ghanem, M.E.; Albacete, A.; Martínez-Andújar, C.; Acosta, M.; Romero-Aranda, R.; Dodd, I.C.; Lutts, S.; Pérez-Alfocea, F. Hormonal changes during salinity-induced leaf senescence in tomato (Solanum lycopersicum L.). J. Exp. Bot. 2008, 59, 3039–3050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Zhang, X.; Gong, Z.; Yang, S.; Shi, Y. ABI4 represses the expression of type-A ARRs to inhibit seed germination in Arabidopsis. Plant J. 2017, 89, 354–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Gan, S. AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 2011, 156, 1612–1619. [Google Scholar] [CrossRef] [Green Version]
- Tran, L.S.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [Green Version]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Hou, L.; Meng, J.; You, H.; Li, Z.; Gong, Z.; Yang, S.; Shi, Y. The Antagonistic Action of Abscisic Acid and Cytokinin Signaling Mediates Drought Stress Response in Arabidopsis. Mol. Plant 2018, 11, 970–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.; Tang, Q.; Hua, X. Arabidopsis Brassinosteroid Mutants det2-1 and bin2-1 Display Altered Salt Tolerance. J. Plant Growth Regul. 2010, 29, 44–52. [Google Scholar] [CrossRef]
- Li, Z.Y.; Xu, Z.S.; He, G.Y.; Yang, G.X.; Chen, M.; Li, L.C.; Ma, Y.Z. A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Commun. 2012, 426, 522–527. [Google Scholar] [CrossRef]
- Sun, F.; Yu, H.; Qu, J.; Cao, Y.; Ding, L.; Feng, W.; Khalid, M.H.B.; Li, W.; Fu, F. Maize ZmBES1/BZR1-5 Decreases ABA Sensitivity and Confers Tolerance to Osmotic Stress in Transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 996. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Yang, R.; Huo, Y.; Liu, S.; Yang, G.; Huang, J.; Zheng, C.; Wu, C. Expression of cotton PLATZ1 in transgenic Arabidopsis reduces sensitivity to osmotic and salt stress for germination and seedling establishment associated with modification of the abscisic acid, gibberellin, and ethylene signalling pathways. BMC Plant Biol. 2018, 18, 218. [Google Scholar] [CrossRef] [Green Version]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, P.; Li, Y.; Wang, S.; Tang, S.; Liu, C. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2016, 85, 348–361. [Google Scholar] [CrossRef] [Green Version]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Zhang, P.; Li, C.; Xia, G. The moss jasmonate ZIM-domain protein PnJAZ1 confers salinity tolerance via crosstalk with the abscisic acid signalling pathway. Plant Sci. 2019, 280, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ma, X.; Zhang, X.; Hu, Q.; Qian, R. Salicylic acid promotes plant growth and salt-related gene expression in Dianthus superbus L. (Caryophyllaceae) grown under different salt stress conditions. Physiol. Mol. Biol. Plants 2018, 24, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, L.; Yuan, F.; Chen, M. Exogenous salicylic acid improves the germination of Limonium bicolor seeds under salt stress. Plant Signal. Behav. 2019, 14, e1644595. [Google Scholar] [CrossRef] [PubMed]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Wang, Q.; Gao, Z.; Wang, Y.; Liu, Y.; Ma, Z.; Chen, Y.; Zhang, Y.; Yan, F.; Li, J. Analysis of Phytohormone Signal Transduction in Sophora alopecuroides under Salt Stress. Int. J. Mol. Sci. 2021, 22, 7313. https://doi.org/10.3390/ijms22147313
Zhu Y, Wang Q, Gao Z, Wang Y, Liu Y, Ma Z, Chen Y, Zhang Y, Yan F, Li J. Analysis of Phytohormone Signal Transduction in Sophora alopecuroides under Salt Stress. International Journal of Molecular Sciences. 2021; 22(14):7313. https://doi.org/10.3390/ijms22147313
Chicago/Turabian StyleZhu, Youcheng, Qingyu Wang, Ziwei Gao, Ying Wang, Yajing Liu, Zhipeng Ma, Yanwen Chen, Yuchen Zhang, Fan Yan, and Jingwen Li. 2021. "Analysis of Phytohormone Signal Transduction in Sophora alopecuroides under Salt Stress" International Journal of Molecular Sciences 22, no. 14: 7313. https://doi.org/10.3390/ijms22147313









