Dibenzofuran, 4-Chromanone, Acetophenone, and Dithiecine Derivatives: Cytotoxic Constituents from Eupatorium fortunei
Abstract
:1. Introduction
2. Results and Discussion
2.1. General
2.2. Structure Elucidation of the New Compounds
2.3. Structure Identification of the Known Isolated Compounds
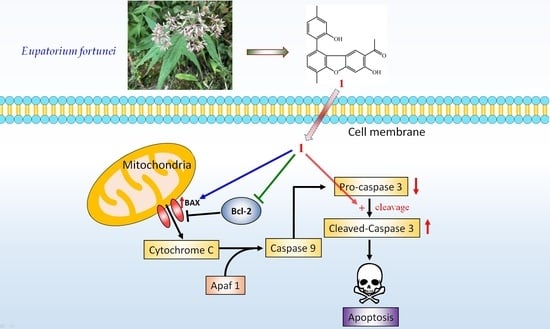
2.4. Biological Studies
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Biological Assay
3.4.1. Cell Culture
3.4.2. Cell Viability Assay
3.4.3. Colony-Formation Assay
3.4.4. Flow Cytometry
3.4.5. Western Blot
3.4.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Yang, F.F.; Chen, H.; Qi, Y.D.; Si, J.Y.; Wu, Q.; Liao, Y.H. Analysis of pyrrolizidine alkaloids in Eupatorium fortunei Turcz. and their in vitro neurotoxicity. Food Chem. Toxicol. 2021, 151, 112151. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Jiang, H.X.; Gao, K. One novel nortriterpene and other constituents from Eupatorium fortunei Turcz. Biochem. Syst. Ecol. 2013, 47, 1–4. [Google Scholar] [CrossRef]
- Pham, T.N.; Pham, H.D.; Dang, D.K.; Duong, T.T.; Le, T.P.Q.; Nguyen, Q.D.; Nguyen Tien, D. Anticyanobacterial phenolic constituents from the aerial parts of Eupatorium fortunei Turcz. Nat. Prod. Res. 2019, 33, 1345–1348. [Google Scholar] [CrossRef] [PubMed]
- Tori, M.; Ohara, Y.; Nakashima, K.; Sono, M. Thymol derivatives from Eupatorium fortunei. J. Nat. Prod. 2001, 64, 1048–1051. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Wang, H.; Jin, D.Q.; Chen, H.; Xu, J.; Ohizumi, Y.; Guo, Y. Thymol derivatives from Eupatorium fortunei and their inhibitory activities on LPS-induced NO production. Phytochem. Lett. 2014, 7, 190–193. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, Y.; Shi, R.; Zhang, D.; Li, C.; Shi, J. New thymol and isothymol derivatives from Eupatorium fortunei and their cytotoxic effects. Bioorg. Chem. 2020, 98, 103644. [Google Scholar] [CrossRef]
- Sekioka, T.; Shibano, M.; Kusano, G. Three trihydroxypiperidines, glycosidase inhibitors, from Eupatorium fortunei Turz. Nat. Med. 1995, 49, 332–335. [Google Scholar]
- Liu, K.; Roeder, E.; Chen, T.; Xiu, X. Pyrrolizidine alkaloids from Eupatorium fortunei. Phytochemistry 1992, 31, 2573–2574. [Google Scholar] [CrossRef]
- Jiang, H.X.; Liu, Q.; Gao, K. Benzofuran derivatives from Eupatorium fortunei. Nat. Prod. Res. 2008, 22, 937–941. [Google Scholar] [CrossRef]
- Perez-Vasquez, A.; Linares, E.; Bye, R.; Cerda-Garcia-Rojas, C.M.; Mata, R. Phytotoxic activity and conformational analysis of thymol analogs from Hofmeisteria schaffneri. Phytochemistry 2008, 69, 1339–1347. [Google Scholar] [CrossRef]
- Chen, J.J.; Tsai, Y.C.; Hwang, T.L.; Wang, T.C. Thymol, benzofuranoid, and phenylpropanoid derivatives: Anti-inflammatory constituents from Eupatorium cannabinum. J. Nat. Prod. 2011, 74, 1021–1027. [Google Scholar] [CrossRef]
- Trang, N.D.; Wanner, M.J.; Koomen, G.J.; Dung, N.X. New acetophenone and thymol derivatives from Eupatorium stoechadosmum. Planta Med. 1993, 59, 480–481. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Tang, Q.; Liu, J.; Zhang, Z.; Liu, W. Preparative isolation and purification of phenolic acids from Smilax china by high-speed counter-current chromatography. Sep. Purif. Technol. 2008, 61, 474–478. [Google Scholar] [CrossRef]
- Bohlmann, F.; Bühmann, U. Synthese von natürlich vorkommenden Hydroxyacetophenon-Derivaten. Chem. Ber. 1972, 105, 863–873. [Google Scholar] [CrossRef]
- Wu, C.; Du, C.; Gubbens, J.; Choi, Y.H.; van Wezel, G.P. Metabolomics-driven discovery of a prenylated isatin antibiotic produced by Streptomyces species MBT28. J. Nat. Prod. 2015, 78, 2355–2363. [Google Scholar] [CrossRef]
- Sabui, S.K.; Venkateswaran, R.V. Synthesis of O-methyl epi-heliannuol E. Tetrahedron 2003, 59, 8375–8381. [Google Scholar] [CrossRef]
- Metternich, J.B.; Gilmour, R. One photocatalyst, n activation modes strategy for cascade catalysis: Emulating coumarin biosynthesis with (−)-riboflavin. J. Am. Chem. Soc. 2016, 138, 1040–1045. [Google Scholar] [CrossRef]
- Reynolds, W.F.; McLean, S.; Poplawski, J.; Enriquez, R.G.; Escobar, L.I.; Leon, I. Total assignment of 13C and 1H spectra of three isomeric triterpenol derivatives by 2D NMR: An investigation of the potential utility of 1H chemical shifts in structural investigations of complex natural products. Tetrahedron 1986, 42, 3419–3428. [Google Scholar] [CrossRef]
- Warr, W.A. Scientific workflow systems: Pipeline Pilot and KNIME. J. Comput. Aided Mol. Des. 2012, 26, 801–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Linghu, K.; Huang, S.; Battino, M.; Georgiev, M.I.; Zengin, G.; Li, D.; Deng, Y.; Wang, Y.; Cao, H. Flaxseed extract induces apoptosis in human breast cancer MCF-7 cells. Food Chem. Toxicol. 2019, 127, 188–196. [Google Scholar] [CrossRef]
- Zheng, L.W.; Wu, L.L.; Zhao, B.X.; Dong, W.L.; Miao, J.Y. Synthesis of novel substituted pyrazole-5-carbohydrazide hydrazone derivatives and discovery of a potent apoptosis inducer in A549 lung cancer cells. Bioorg. Med. Chem. 2009, 17, 1957–1962. [Google Scholar] [CrossRef] [PubMed]
- Li, X.H.; He, X.R.; Zhou, Y.Y.; Zhao, H.Y.; Zheng, W.X.; Jiang, S.T.; Zhou, Q.; Li, P.P.; Han, S.Y. Taraxacum mongolicum extract induced endoplasmic reticulum stress associated-apoptosis in triple-negative breast cancer cells. J. Ethnopharmacol. 2017, 206, 55–64. [Google Scholar] [CrossRef]
- Huyen, C.T.T.; Luyen, B.T.T.; Khan, G.J.; Oanh, H.V.; Hung, T.M.; Li, H.J.; Li, P. Chemical constituents from Cimicifuga dahurica and their anti-proliferative effects on MCF-7 breast cancer cells. Molecules 2018, 23, 1083. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.Y.; Chang, T.C.; Wu, Y.J.; Chen, Y.; Chen, J.J. Benzophenone and Benzoylphloroglucinol Derivatives from Hypericum sampsonii with Anti-Inflammatory Mechanism of Otogirinin A. Molecules 2020, 25, 4463. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.L.; Liu, Y.H.; Liu, C.; Qi, M.P.; Liu, R.N.; Zhu, X.F.; Zhou, Q.G.; Chen, Y.Y.; Guo, A.Z.; Hu, C.M. Indirubin inhibits LPS-induced inflammation via TLR4 abrogation mediated by the NF-kB and MAPK signaling pathways. Inflammation 2017, 40, 1–12. [Google Scholar] [CrossRef] [PubMed]










| 1 | 2 | |||
|---|---|---|---|---|
| Position | δH (J in Hz) a | δC b | δH (J in Hz) a | δC b |
| 1 | 7.65, s | 125.4 | 7.59, s | 125.1 |
| 2 | 116.6 | 116.0 | ||
| 3 | 163.2 | 162.9 | ||
| 4 | 7.07, s | 99.8 | 7.05, s | 99.6 |
| 4a | 161.2 | 161.3 | ||
| 5a | 155.9 | 155.4 | ||
| 6 | 121.8 | 120.5 | ||
| 7 | 7.33, br d (7.5) | 128.6 | 7.27, br d (7.6) | 127.9 |
| 8 | 7.22, d (7.5) | 125.1 | 7.17, d (7.6) | 125.0 |
| 9 | 127.7 | 130.6 | ||
| 9a | 121.9 | 121.7 | ||
| 9b | 116.3 | 117.8 | ||
| 1′ | 122.4 | 125.4 | ||
| 2′ | 152.7 | 156.7 | ||
| 3′ | 6.96, br s | 116.1 | 6.91, br s | 111.5 |
| 4′ | 140.4 | 139.8 | ||
| 5′ | 6.93, br d (7.5) | 121.7 | 6.95, br d (7.5) | 121.4 |
| 6′ | 7.25, d (7.5) | 130.2 | 7.28, d (7.5) | 131.0 |
| COMe-2 | 203.9 | 203.7 | ||
| COMe-2 | 2.41, s | 26.2 | 2.40, s | 26.2 |
| OH-3 | 12.65, s | 12.62, s | ||
| Me-6 | 2.62, s | 15.1 | 2.60, s | 15.1 |
| OH-2′ | 4.97, s | |||
| OMe-2′ | 3.70, s | 55.5 | ||
| Me-4′ | 2.44, s | 21.4 | 2.49, s | 21.7 |
| Position | δH J (Hz) a | δC b |
|---|---|---|
| 2 | 81.0 | |
| 3 | 2.79, s | 48.5 |
| 4 | 191.7 | |
| 4a | 119.6 | |
| 5 | 8.08, d (2.0) | 120.0 |
| 6 | 129.6 | |
| 7 | 7.70, d (2.0) | 114.3 |
| 8 | 149.8 | |
| 8a | 154.0 | |
| Me-2 | 1.55, s | 26.5 |
| COMe-6 | 196.6 | |
| COMe-6 | 2.60, s | 26.2 |
| OMe-8 | 3.95, s | 56.4 |
| Position | δH J (Hz) a | δC b |
|---|---|---|
| 1 | 149.3 | |
| 2 | 128.3 | |
| 3 | 7.74, d (8.0) | 130.4 |
| 4 | 7.12, br d (8.0) | 126.7 |
| 5 | 144.7 | |
| 6 | 6.94, br s | 124.4 |
| 1′ | 166.1 | |
| 2′ | 127.0 | |
| 3′ | 6.30, qq (7.3, 1.8) | 141.4 |
| 4′ | 2.08, dq (7.3, 1.3) | 16.0 |
| 5′ | 2.08, qd (1.8, 1.3) | 20.6 |
| COMe-2 | 197.1 | |
| COMe-2 | 2.52, s | 29.4 |
| Me-5 | 2.40, s | 21.4 |
| Position | δH J (Hz) a | δC b |
|---|---|---|
| 2 | 122.6 | |
| 3 | 130.6 | |
| 4 | 6.84, s | 119.1 |
| 5 | 159.3 | |
| 7 | 159.3 | |
| 8 | 6.84, s | 119.1 |
| 9 | 130.6 | |
| 10 | 122.6 | |
| COMe-2 | 190.2 | |
| COMe-2 | 2.50, s | 29.4 |
| C≡CMe-3 | 73.2 | |
| C≡CMe-3 | 94.8 | |
| C≡CMe-3 | 2.09, s | 4.8 |
| OMe-5 | 3.94, s | 58.8 |
| OMe-7 | 3.94, s | 58.8 |
| C≡CMe-9 | 73.2 | |
| C≡CMe-9 | 94.8 | |
| C≡CMe-9 | 2.09, s | 4.8 |
| COMe-10 | 190.2 | |
| COMe-10 | 2.50, s | 29.4 |
| Compound | IC50 (μM) a | |
|---|---|---|
| A549 | MCF-7 | |
| Eupatodibenzofuran A (1) | 5.95 ± 0.89 ** | 5.55 ± 0.23 ** |
| Eupatodibenzofuran B (2) | 93.37 ± 1.14 | 85.91 ± 3.94 |
| 6-Acetyl-8-methoxy-2,2-dimethylchroman-4-one (3) | >100 | >100 |
| Eupatofortunone (4) | 86.63 ± 10.89 * | 82.15 ± 8.26 ** |
| Eupatodithiecine (5) | 39.44 ± 2.81 * | 31.20 ± 4.23 * |
| Thymyl angelate (6) | >100 | >100 |
| 8,9-Dehydrothymol 3-O-tiglate (7) | >100 | >100 |
| 9-Angeloyloxythymol (8) | 60.08 ± 3.39 * | 52.11 ± 2.16 * |
| 9-O-Angeloyl-8,10-dehydrothymol (9) | 55.36 ± 0.80 * | 51.70 ± 0.48 * |
| 2-Hydroxy-4-methylacetophenone (10) | >100 | >100 |
| trans-o-Coumaric acid (11) | >100 | >100 |
| 6-Hydroxy-7-methoxy-2-isopropenyl-5- acetylcumaran (12) | 73.97 ± 2.88 * | 72.67 ± 3.51 * |
| 2,4-Di-tert-butylphenol (13) | >100 | >100 |
| 1-(2-Hydroxy-5-methoxy-4-methylphenyl)ethanone (14) | >100 | >100 |
| Coumarin (15) | >100 | >100 |
| Taraxasterol (16) | 94.79 ± 10.23 * | 93.59 ± 6.11 * |
| 5-Fluorouracil (5FU) b | 10.57 ± 1.89 ** | 8.59 ± 1.03 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, C.-H.; Wu, S.; Hsu, K.-C.; Huang, W.-J.; Chen, J.-J. Dibenzofuran, 4-Chromanone, Acetophenone, and Dithiecine Derivatives: Cytotoxic Constituents from Eupatorium fortunei. Int. J. Mol. Sci. 2021, 22, 7448. https://doi.org/10.3390/ijms22147448
Chang C-H, Wu S, Hsu K-C, Huang W-J, Chen J-J. Dibenzofuran, 4-Chromanone, Acetophenone, and Dithiecine Derivatives: Cytotoxic Constituents from Eupatorium fortunei. International Journal of Molecular Sciences. 2021; 22(14):7448. https://doi.org/10.3390/ijms22147448
Chicago/Turabian StyleChang, Chun-Hao, Semon Wu, Kai-Cheng Hsu, Wei-Jan Huang, and Jih-Jung Chen. 2021. "Dibenzofuran, 4-Chromanone, Acetophenone, and Dithiecine Derivatives: Cytotoxic Constituents from Eupatorium fortunei" International Journal of Molecular Sciences 22, no. 14: 7448. https://doi.org/10.3390/ijms22147448
APA StyleChang, C.-H., Wu, S., Hsu, K.-C., Huang, W.-J., & Chen, J.-J. (2021). Dibenzofuran, 4-Chromanone, Acetophenone, and Dithiecine Derivatives: Cytotoxic Constituents from Eupatorium fortunei. International Journal of Molecular Sciences, 22(14), 7448. https://doi.org/10.3390/ijms22147448