Abstract
In vitro tissue culture plant regeneration is a complicated process that requires stressful conditions affecting the cell functioning at multiple levels, including signaling pathways, transcriptome functioning, the interaction between cellular organelles (retro-, anterograde), compounds methylation, biochemical cycles, and DNA mutations. Unfortunately, the network linking all these aspects is not well understood, and the available knowledge is not systemized. Moreover, some aspects of the phenomenon are poorly studied. The present review attempts to present a broad range of aspects involved in the tissue culture-induced variation and hopefully would stimulate further investigations allowing a better understanding of the phenomenon and the cell functioning.
1. Introduction
Tissue culture-induced variation (TCIV) is a well-established phenomenon of any changes affecting regenerants phenotype or genotype during in vitro plant regeneration [1,2,3,4]. If the changes are stably inherited via the generative cycle, it is usually called somaclonal variation (SV) [5,6]. However, the TCIV and SV terms are often used mutually [7]. TCIV may be pronounced at the morphological [8], biochemical [9], genetic [10] and epigenetic [11] levels, which do not necessarily overlap [12]. Possibly the most investigated level is based on DNA markers.
The studies of molecular aspects of TCIV started at the beginning of the 1980s [13,14,15]. The development of the restriction fragment length polymorphism (RFLP) [16] and the randomly amplified polymorphic DNA (RAPD) [17] techniques (Table 1) resulted in a burst of experimental data suggesting that even using anther culture, plant regeneration was subjected to numerous mutations such as single nucleotide polymorphisms (SNPs) [18]. Some studies suggested that variation was due to the so-called pre-existing variation [19,20], the others addressed mutations to the activation of transposable elements [21] due to DNA methylation pattern alternations [22]. For a long time, the origin of changes was not apparent. It was speculated that they might have come from the degradation of the cells during tissue cultures [23], the elevated production of reactive oxygen species species (ROS) [24], due to stressful conditions [25]. The degradation hypothesis claimed that cell death in culture under stress and programmed cell death leads to secondary metabolites (pigments, tannins). Their release into culture can induce somaclonal variation in an unspecified way [23]. It was speculated that the callus phase and indirect embryogenesis could have contributed to the phenomenon [26]. For about a decade, it was thought that studies on TCIV were deficient. However, the development of new molecular tools and progress in epigenetics led to the revision of previous data and pushed a deeper understanding of the phenomenon.

Table 1.
The arrangement of molecular techniques used to study TCIV.
The introduction of the amplified fragment length polymorphism (AFLP) approach [27], followed by the methylation-sensitive amplified polymorphism (MSAP) one [28,29], allowed studies of DNA methylation changes addressed to in vitro tissue culture plant regeneration [30,31]. Although that changes are frequent, but may vary in wide spectrum, from 0.07% [32] to 52% [33]. While the MSAP approach was productive, it could identify only DNA methylation changes related to CG and CHG sequence contexts [34]. Moreover, the ways to quantify changes used distinct algorithms [35] that might have differed from study to study [36,37], making a comparison of results somewhat tricky. Thus, the semi-quantitative MSAP approach, allowing for quantifying DNA methylation changes, was suggested to overcome the limitation [38]. However, studies on sequence variation required utilization of additional marker techniques. Consequently, another AFLP based technique, the so-called methylation sensitive AFLP (metAFLP) approach, was developed [1,39], allowing quantification of sequence and DNA methylation changes during a single experiment. Further advances in the technique showed that one might study not only CG and CHG but also the CHH methylation context, an essential tool in studies on cereals [1,12].
The development of the new generation sequencing (NGS) opened innovative opportunities. The diversity arrays technology methylation analysis (DArTseqMet) techniques permitted for identifying a large number of markers that could be employed in quantification procedures evaluated for the semiquantitative MSAP approach [40]. MethylRAD [41] or MethylSeq [42] are the other alternatives. The MethylSeq method is an NGS variant of the bisulfite-based sequencing approach. It could be used to study different types of genomic DNA methylation, but its application requires at least 10X coverage. The MethylRAD approach uses Mrr-like enzymes to collect 32-bp methylated DNA fragments from the whole genome for high-throughput sequencing. It allows for de novo methylation analysis using low DNA input. The two approaches have incredible potential in DNA methylome studies; however, they were not yet used in tissue culture studies.
All the techniques mentioned above were exploited in studies on the whole genome. When it was of value to investigate DNA methylation pattern change of a specific sequence, the bisulfite approach was recommended [43,44]. It allowed the establishment of the extent to which the CG, CHG, and CHH sequences were methylated in the genome [45]. Thus, the tools to study molecular aspects of the TCIV were established. Moreover, by that time, the genetic model of the studies was also evaluated [46]. It became apparent that studies on TCIV should start from a well-defined, preferentially homozygous double haploid plant that served as a donor of tissue for the regenerants subjected to further analysis. Utilizing specific plant materials and sophisticated molecular marker techniques, TCIV was studied in barley [47], triticale [48], Polyscias filicifolia [49] and Gentiana pannonica Scop. [4]. It was shown that the phenomenon is common to plants [50,51,52] and that at least some DNA methylation changes could be transmitted to the progeny [53,54]. Furthermore, DNA methylation changes affecting regenerants may require several generative cycles to be stabilized [48]. Using a modification of the metAFLP approach that utilized primers directed towards TEs, their putative role in sequence variation was demonstrated [47,55]. However, the mechanisms of the TCIV are still being discussed.
It is becoming understood that analyses of DNA methylation changes affecting tissue culture due to plant regeneration are insufficient to have an image of the phenomenon. Interdisciplinary studies are needed to have species-specific models describing consecutive steps required to change the cell fate towards somatic development. In this context, studies of the cell wall, cell membrane, subintinal layer, their components, and the way signals transmitting stresses are involved in the process are needed. An interesting approach to study the role of the cell wall or biochemical pathways affected by in vitro tissue culture is attenuated total reflectance Fourier transform infrared (ATR-FTIR) spectroscopy [56,57], which allows for the identification of putative cell wall components [58] or biochemical pathways [59] participating in sensing stresses. It was documented that β-glucans being built of glucose units [60], and probably present in the subintinal layer of some microspores [61], may be sources of glucose for new anther-culture-derived regenerants [62]. Furthermore, stressful conditions might influence DNA methylation, probably disturbing the methionine cycle [62]. The role of copper and silver ions in sequence variation [63] and green plant development [40] was also documented in barley regenerants derived via anther culture. Similarly, in embryo-derived regenerants of barley copper ions participated in sequence variation, possibly via modification of methylated sequences [12]. The respective relationships could be evaluated using mediation and moderation analyses [64] applied to DNA methylation changes and the ATR-FTIR spectroscopy data. Such results could be used in structural equation modeling (SEM), resulting in a deeper insight into the phenomenon leading towards its practical applications. However, due to limited sample size, SEM analysis allowing construction of an in vitro plant regeneration either via andro- or embryogenesis was not evaluated. If not the most substantial, a further limitation is the understanding of the role of the way the cell senses and transmits stresses leading towards TCIV.
The review aims to present data related to the in vitro tissue culture-induced variation, how stressful conditions may affect tissue culture, the role of the cell wall, cell membrane, mucilage layer, signaling compounds, and biochemical pathways.
1.1. Stresses and Their Role in Plant Regeneration through Tissue Culture In Vitro
It is assumed that inductive stress treatment is necessary to initiate the cell reprogramming process required for the microspore to switch from gametophytic to embryogenic fate. The stress treatment needed to switch the developmental fate of microspores depends on the plant species and the species genotype. Osmotic, starvation, cold, heat, and many other stresses are employed to initiate andro- and embryogenesis [67,68] (Table 2).

Table 2.
Summation of stresses that efficiently switch the developmental fate of microspores.
Three phases of embryogenic development due to androgenesis could be distinguished. The first one reflects the acquisition of embryogenic potential as the reaction to stress. At this stage, which lasts about three days, repression of gametophytic development and dedifferentiation of the cells occurs. The second stage follows the induction and culture initiation step. The microspores divide and produce multicellular structures persisting within the microspore wall (exine) [74], indicating the initiation of embryogenesis. This stage, at least in barley, may take two days. Next, the exine breaks down, and embryos follow a similar zygotic embryogenesis pathway through globular, heart, torpedo, and cotyledonary embryos. The formation of tissue culture structures reflects the third phase of embryogenic development that begins around the 21st day of in vitro tissue cultures [74]. Interestingly, the third phase of embryogenic development corresponds to the point when under increased copper concentration present in induction medium, regeneration of new barley plants via anther culture results in the highest output [40].
In the model system of Hordeum vulgare (barley), the inductive cold stress (4 °C) is applied, whereas, in Brassica napus, 32 °C is used to initiate a switch from gameto- to sporophytic path. Stress-related heat-shock proteins (HSP70 and HSP90) were detected in microspore embryogenesis cultures of Brassica napus and Capsicum annum due to cold treatment [75]. Thus, a protective role for the chaperones was suggested; however, their role in microspore embryogenesis is not clear. Low temperatures may alter endogenous ethylene (ET) levels enhancing tolerance with higher ET concentrations [76].
During reprogramming stages, induced stresses result in increased cell death and oxidative stress. At this point, ROS arise in excess and are accumulated [77]. However, in the case of Medicago sativa leaf protoplasts, the application of oxidative stress-inducing agents resulted in acceleration of cell cycle re-entry that was accompanied by a lower level of ROS accumulation [78]. The equilibrium between ROS-scavenging and ROS-producing mechanisms administrates the cell’s level of damage and oxidative stress. Several enzymes of the antioxidative machinery of the cell increase their activity in microspore cultures. These stress-related proteins may have a protective role [79].
Abiotic stress factors may impact auxin homeostasis [80] probably resulting in the induction of somatic embryogenesis [81,82,83]. While microspore culture does not require exogenous auxin (2,4-dichlorophenoxyacetic acid), endogenously, the phytohormone may participate in microspore reprogramming in Brassica napus and in vitro embryo formation [84]. It was shown that induction of microspore embryogenesis resulted in de novo endogenous auxin biosynthesis and accumulation of indole-3-acetic acid (responsible for embryo patterning, polarization, and differentiation) in pre-embryo cells starting from the first embryogenic divisions [84]. Auxin activity, biosynthesis, and transport are essential for stress-induced microspore embryogenesis. There is a link between auxin biosynthesis, its perception, transport, gene expression, signaling, and nontranscriptional responses; however, the exact mechanisms remain elusive [85,86,87]. The other hormone vital for plant cells is cytokinin that acts antagonistically to auxin. Cytokinin participates in cell growth and may control the early stages of somatic embryogenesis [88]. Spatiotemporal localization of cytokinin and auxin responses during microspore embryogenesis was suggested [74]. However, data concerning the presence of endogenous cytokinin in microspore embryogenesis was not evidenced.
It is worth mentioning that stress conditions may induce or impact autophagy, a significant pathway for recycling cell materials [89,90]. Autophagy may promote plant cell survival under starvation and stress conditions [91]. Moreover, excretion of cytoplasmic material (occasionally containing whole organelles) via single membrane-bound autophagic bodies and those deposited in the cell wall (remnants from the digestion of cytoplasmic organelles) of embryogenic microspores in between the cell wall of embryogenic microspores and the plasma membrane was observed [92]. Some of the autophagosomes were transported out of the cell, creating extracytoplasmic fibrillar and membranous material deposits It was shown that embryogenic microspores are associated with autophagy and excretion of the removed material [92]. Thus, autophagy is a kind of cytoplasmic cleaning, whereas excretion is a mechanism of avoiding unnecessary vacuolar system growth [92]. The excretion was essential for proper microspore embryogenesis; however, in some cases, the material was redirected to the cell [92], possibly implying ‘recycling’ of the materials for cellular processes. It should also be mentioned that autophagy can involve converting the lytic vacuole to a storage one [93,94].
The connection between ROS and autophagy in plants was suggested [95]. In barley, the activation of autophagy after inductive stress at 4 °C with upregulation of HvATG5 and HvATG6 genes, and a rise in the number of autophagosomes was reported [96]. Autophagy is activated and involved in cell death with the participation of cathepsins the proteases that degrade proteins during stress-induced microspore embryogenesis in barley [96]. It was also demonstrated that stress treatment of Brassica napus also resulted in induction of autophagy [92] in parallel to cysteine-dependent proteases (metacaspase) involved in programmed cell death, stress and cell proliferation [97], and gene expression [98]. Interestingly, application of agents directed towards ROS (MnCl2), autophagy [3-methyladenine (3-MA), inhibitor], and protease activities, caspase 3-like, and metacaspase activities (E64, Ac-DEVD-CHO, and Ac-VRPR-FMK; inhibitors), reduces cell death levels, increasing embryogenesis in rapeseed and barley [96] opening up pathways reducing stress-induced cell death at the early stages of microspore embryogenesis.
Inductive stresses are responsible for cell reprogramming. The process involves DNA demethylation and de novo methylation [99]. A global DNA hypomethylation during the change of the gametophytic to sporophytic fate and first embryogenic divisions were observed in barley [100]. Among inductive stressful conditions, darkness is one of the most exploited in cereals [101]. When mannitol is applied, triggering microspores to dividing [73], the culture is subjected to osmotic, and carbon starvation discussed earlier [102]. Sometimes, instead of darkness, heat stress is applied [103]. It was suggested that ethylene participates in the response of plants to heat stress [76]. Stresses induce ethylene accumulation. The higher the ethylene concentration, the higher the chance of plant survival under stressful conditions [104]. Most probably, ethylene modulates gene expression via ethylene signaling mechanisms [105].
The tissue culture medium itself may be treated as a stressful factor. The regeneration medium contains many ingredients such as 6-benzylaminopurine, 2,4-dichlorophenoxyacetic acid, alpha-naphthalene acetic acid, and indole-3-acetic acid not necessary are neutral to the cells may induce TCIV [101,106,107]. As such ingredients may be toxic, they may lead to genomic DNA degradation as indicated by comet analysis [101]. It was also demonstrated that copper and silver ions containing media might also affect DNA methylation patterns [12], resulting in DNA sequence changes [62]. Moreover, the two ions most probably change respiratory chain balance affecting the Yang cycle [62] and inducing sequence changes [63]. Interestingly, copper and silver may moderate relationships between de novo methylation/demethylation processes, leading to green plant regeneration [40]. For more details on that, see an excellent review published recently [74].
Assuming that different stresses may affect in vitro tissue culture, the mucilage layer, the cell wall, and the plasma membrane are reasonable candidates for sensing stresses [108,109] affecting the cell functioning and possibly promoting the TCIV. However, the information on how the mucilage layer, the cell wall, and plasma membrane perform due to in vitro tissue culture plant regeneration is not well addressed.
1.2. The Mucilage Layer
The mucilage layer is formed on the surface of cells. It consists of polysaccharides, proteins including histones, etc., and extracellular DNA (exDNA), known as danger-associated molecular patterns [110,111,112,113]. The components of the layer may activate the innate immune system. ExDNA is defined as “DNA located outside the cell and originating from intracellular DNA by active or passive extrusion mechanisms or by cell lysis” [114]. It is assumed that it participates in immune defense in plants and contributes to “damage-associated molecular patterns” (DAMPs). Depending on DNA origin (self-exDNA (endogenous) or non-self-exDNA (exogeneous)) may act differently. Non-self-exDNA in plants results in reactive oxygen species formation and callose deposition [115]. On the other hand, self-exDNA may inhibit root growth, the effect not identified in non-self-exDNA. It was also demonstrated that self-exDNA acts in a concentration-dependent manner [116,117,118] by reacting with pattern recognition receptors [119]. Its action depends on the self-damage level recognized by plants. The presence of methylated or non-methylated CpG sequences seems important in plant reaction and recognition of endo- and exogenous DNAs [120]. It was also demonstrated that self-exDNA participates in immune signaling [117], stimulating calcium signaling and plasma membrane decomposition in Zea mays [117]. Furthermore, endogenous DNA may inhibit seed germination and trigger H2O2 production following by mitogen-activation protein kinase (MAPK) in Phaseolus vulgaris [118]. The MAPK signaling pathway senses extracellular signals and transmitting them from the cell membrane to the nucleus in response to various environmental stimuli [121]. Still, the mechanism of exDNA action is not understood. Most shreds of evidence on exDNA role come from studies on plant roots [122,123]. The importance of protein constituents of the mucilage layer and its treatment with proteases was suggested [124]. It was also shown that DNase I may lead to necrosis under pathogen inoculation of root tips [125]. Still, the role of the mucilage layer decomposition and secretion of DNA molecules from surrounding cells remains uncovered.
Even less is known in the case of microspores. Microspores are cells surrounded by a primary and secondary wall with haploid nucleus specifically surrounded by mitochondria present in the vicinity of the nucleus’s pores. It was demonstrated that mitochondria might cover microspore surface and vegetative nuclei around postmeiotic mitosis I (a few days before flowering). The mitochondria were also detected apart nucleus (shortly after postmeiotic mitosis) in 30 nm from a nuclear pore [126]. It should be stressed that, under dark conditions, a callose subintinal layer is formed [127]. The presence of callose might be linked to microspores’ ability to switch from gameto- to sporophytic fate [128]. Furthermore, the autophagy phenomenon responsible for the secretion of damaged organelles outside the cell was observed in stressed microspores [96]. These phenomena are in line with the mucilage layer’s presence in the case of microspores. The layer may sense some abiotic stresses, inducing an innate immune response and allowing or enhancing the possibility of anther-culture-derived plant regeneration. Most probably, the layer may interact with the cell wall and or cell membrane.
1.3. Cell Wall and Plasma Membrane as Sensors of Stresses
Even less, the pollen cell wall protects male sperm from abiotic stresses (i.e., abrasion, desiccation, and UV radiation). A durable wall consists of gametophyte-derived intine and sporophyte-derived exine layers covered with lipid-rich pollen coat or trephine [129,130]. The intine resembles the primary cellulosic plant cell wall [129,131,132] whereas the exine is formed of heterogenous polymer composed of polyhydroxylated aliphatic constituents with aromatic or conjugated side chains containing ether and ester bonds [133,134,135] called sporopollenin synthesized from precursors in tapetum [129,130] and anchored on to microspores [136].
Pollen wall formation starts after microspore meiosis generating tetrads covered with callose. At the late tetrad stage, the microspore surface is formed of primexine, a transient cell wall matrix-shaped of polysaccharides outside of microspore plasma membrane inside callose. Primexine is composed of sporopollenin. This layer may contain cellulase [137]. The callose wall degrades after the tetrad phase allowing free microspore release into the locule and tecta. At this stage, a thin microspore-derived intine wall forming between the developing exine and a microspore-derived intine wall is evidenced [138].
As the composition of the microspore cell wall is not apparent, available information on the plant cell wall in general with emphasis on cellulose and wall matrix polysaccharides [139] are discussed. All of them form a rigid structure. The essential cell wall function relies on providing mechanical strength to resist turgor pressure. It forms a barrier against biotic and abiotic stresses that may alter the cell wall components’ deposition [140].
The cell wall cellulose may be present in amorphic, semi amorphic, or crystalline forms. Crystalline cellulose has significantly better stiffness than all other constituents. Amorphous cellulose is more penetrable and accessible to enzymes and has a higher enzyme binding capacity than its crystalline counterpart so that it has a higher hydrolysis rate. The ratio of these two regions characterizes crystallinity. The cellulose with high crystallinity usually has low enzymatic hydrolysis efficiency [141,142,143]. Cellulose synthase (CESA) complexes (CSCs) synthesize cellulose microfibrils assembled in the Golgi apparatus. CSCs actively synthesize cellulose when delivered to the plasma membrane (PM). CSCs move along cortical microtubule paths that define cellulose microfibrils synthesis. CSC traffic between the PM and various intracellular compartments plays a vital role in determining the level of cellulose synthesized [144]. Uridine diphosphate glucose (Glc) originates from the cytosolic invertase/uridine diphosphate Glc phosphorylase pathway [145]. It is the substrate for CESAs [146]. The CESA proteins are involved in the dimerization/oligomerization of CESA subunits [147] due to a cytosolic N-terminal region. Moreover, the proteins consist of two transmembrane domains, a large cytoplasmic central loop that contains the substrate binding and catalytic regions, six additional transmembrane domains [148,149], and an intracellular C-terminal domain. The CSC rosettes contain 18 CESA subunits synthetizing 18 glucans [150].
Phytohormones may influence cell wall composition. For example, the amount of cellulose increases in methyl jasmonate, whereas lignin content decreases after salicylic acid application. The profile of gene expression involved in cell wall biosynthesis is also modified [151]. Phytohormone signaling pathways regulate stress perception at the primary cell wall, followed by cellulose synthesis and microtubule arrangement [140]. Alternatively, ROS and peroxidases arising in response to stresses in the cell wall may cross-link phenolic compounds and glycoproteins, resulting in stiffening. If ROS-levels remain high during continued stress, OH°-radicals lead to polymer cleavage [139]. Cellulosomes could accomplish the same, an extracellular supramolecular multienzyme complex that can efficiently degrade cellulose and hemicelluloses in plant cell walls [152].
The role of cellulose in sensing stresses was demonstrated on mutant plants. Cellulose-deficient mutants are typically more sensitive to abiotic stress than wild-type plants [140]. Thus, the cellulose synthesis machinery may be vital in abiotic stress responses [140]. The other option is that some cell wall components may sense stresses via cell wall integrity [153]. The cell wall proteins, i.e., the Arabidopsis thaliana leucine-rich repeat receptor kinase LRR-RK male discoverer 1-interacting receptor-like kinase 2 (MIK2) was suggested to play such a role [154]. The MIK2 regulates cell wall damage responses upon cellulose biosynthesis inhibition due to abiotic stress [153]. MIK2 has overlapping functions with receptor-like protein kinase THESEUS 1 (THE1), also proposed as a cell wall integrity sensor. Moreover, plants may coordinate stress responses by integrating phytohormones (auxins, cytokinins, gibberellins, abscisic acid, ethylene, brassinosteroids, salicylic acid, jasmonates, and strigolactones) and their pathways [140,155,156].
Recent studies have demonstrated that cellulose forms participate in a signaling pathway that links cellulose and mitochondria. Two cell wall maintainer1 (cwm1) and cwm2 pentatricopeptide repeat protein genes were shown to be involved in editing mitochondrial transcripts encoding subunits of respiratory complexes (i.e., complex III linked to the maintenance of cell wall integrity under stress) and activation of retrograde mitochondrial signaling via ANAC017, a transcription factor participating in retrograde signaling to the nucleus upon mitochondrial dysfunction [157,158]. A complex hierarchy of transcription factors exists downstream of ANAC017. These involve ANAC and WRKY transcription factors associated with organellar signaling and senescence; moreover, the network includes ethylene- and gibberellic acid-related transcription factors involved in stress responses [159].
The development of plants requires the biosynthesis, deposition, and degradation of cell wall matrix polysaccharides. Biosynthesis of polysaccharides is performed in the endoplasmic reticulum and Golgi of the plant secretory system by polysaccharide glycosyltransferases (GTs) transmembrane proteins. The synthesis of the cell wall matrix glycans (pectins, xylans, xyloglucans, mannans, mixed-linkage glucans (MLGs), and arabinogalactan components of arabinogalactan proteins (AGPs) proteoglycans) was mapped to multigene families [160]. Different stresses can perturb targeted genes to promote functional adaptation [161]. The polysaccharide fraction of the cell wall may participate in stress sensing [162]. The environmental (mechanical) signals affect the cell wall determining its phenotypic features. However, the way stress is sensed and transmitted into the cell is still under investigation.
The plasma membrane was suggested to participate in signal transduction because of its position at the interface between the cell’s interior and the cell wall. Thus, the crucial players that maintain cell status and respond to stresses are in the plasma membrane’s same vicinity [163]. Interestingly, the layer between the plasma membrane and cell wall, the so-called subintinal layer present in microspores with embryogenic fate and composed of callose, is also considered essential in stress sensing. As callose is formed of glucans, it may serve as a source of glucose [164] required by glycolysis which impacts the Krebs cycle and the electron transport chain.
Cereal β-glucans have a specific combination of β-(1→4) and β-(1→3) linkages into linear long-chain polysaccharides of high molecular weight. β-glucans were identified in barley and oats [165]. Under elongated dark conditions, β-glucans deliver glucose for glycolysis, supporting biosynthesis of, i.e., lipids [166] that may act as signaling compounds [167,168], affecting gene expression [169,170]. Thus, the subintinal layer components are reasonable candidates for signals [171] of stresses in microspore culture. It was also demonstrated that β-glucans might impact DNA methylation of the CHG context in barley [62]. Whatever is the sensor of abiotic stresses at the cell wall level, the signal needs to be transmitted to the cell. It is being accepted that calcium is a second messenger that facilitates responses to stresses by activating calcium-binding proteins [172,173,174]. Another candidate that can sense stress and may act as a cell wall mechanism protecting plants from external stresses is β-1,3-linked D-glucose [140]. Furthermore, evidence shows that massively glycosylated hydroxyproline-rich proteins called AGP improve plant regeneration in barley anther culture [175]. The AGP content changes in Brassica napus are related to the developmental fate of microspores [176]. The proteins are present in the mucilage layer, the cell wall, and the cell membrane and participate in microspore fate change towards sporophytic pattern and may be involved in stress signaling [177].
1.4. Stresses and How They Affect Nucleus at the DNA and Histone Levels and Gene Expression
The crosstalk between DNA methylation, histone methylation/acetylation, and genome-wide chromatin remodeling is required for microspore reprogramming, totipotency, and initiation embryogenesis. It was shown that low levels of H3K9 methylation favor embryogenesis initiation, while its increased level is needed for embryo differentiation progresses. The process of histone demethylation and its methylation correlates with the expression of BnHKMT SUVR4-like and BnLSD1-like genes. Interestingly, BIX01294 small molecule (an inhibitor of H3K9 methylation) may promote microspore reprogramming in Brassica napus and Hordeum vulgare [92]. If used at advanced stages, the same molecule impairs embryo formation. Thus, the initiation of embryogenesis requires low H3K9 methylation while embryo differentiation its high levels [68]. Moreover, demethylation of histone H3K27me3 is critical for acquisition of callus formation (see review by Pasternak and Dudits [178]). The other histone modification, namely the acetylation of lysine residues within the N-terminal tail of histones H3 and H4 (associated with actively transcribed genes and open chromatin state), promotes microspore embryogenesis. The increased histone acetylation promotes microspore reprogramming and cell totipotency [179]. For sure, such multilevel machinery being unbalanced may affect genome functioning and induction of variation under in vitro tissue culture conditions.
1.5. Chromosome Doubling
The development of a diploid plant from microspores requires a chromosome doubling stage. The process may proceed spontaneously or may be induced artificially by chromosome segregation blocking mitotic inhibitors such as colchicine [180]. In cereals, the haploid genome undergoes spontaneous diploidization with the frequency ranging from 40 to 50% in Brassica napus and 90% in barley and rye [102,181,182,183]. Chromosome spontaneous doubling mechanism assumes nuclear fusion in cereal species [184]. It is considered a preferred way of obtaining double haploid plants as this mechanism avoids induction of mutagenesis, usually associated with artificial chromosome doubling. Artificial chromosome doubling may involve colchicine, a poisonous and mutagenic alkaloid compound extracted from bulbs and seeds of autumn crocus or meadow saffron (Colchicum autumnale). Its action relies on preventing the microtubule formation and chromosome segregation resulting in chromosome doubling [180]. The process may induce large chromosomal rearrangements [185,186] resulting in somaclonal variation.
1.6. Transcriptome Changes
Many studies have shown changes in the plant transcriptome under the influence of stress factors [187,188,189]. Plant regeneration via anther culture requires the application of several stresses, including cold treatment [190]. The stress treatment allows induction of embryogenesis in the haploid uninucleate microspore [191]. Under such conditions, haploid microspores need to switch their gametophytic fate [192]. In barley before and after applying stress, significant transcriptome changes (3382 genes’ expression changes) are indicated by RNA-seq analysis. Among genes with altered expression, glutathione S-transferases (check for Section 1.8 Biochemical Aspects), heat shock proteins, and ribosomal subunit proteins were indicated. Moreover, activation transcription factors in early embryogenesis, hormone biosynthesis, hormonal signaling, and genes involved in secondary metabolism were shown [193]. Interestingly, differential expression of genes involved in plastid transcription, translation, and starch synthesis was observed in barley capable of regenerating preferentially green plants vs. that regenerating albino plants. The expression of granule-bound starch synthase I (GBSSI) gene in early-mid microspores correlated with a genotype ability to regenerate green plants via androgenesis [194]. Results concerning the role of nuclear and plastic-encoded polymerases in albino and green plant regeneration indicating antero- and retrograde phenomenon in barley were presented from the same group [195]. Research conducted on Arabidopsis thaliana has shown that the transcriptome composition changes significantly during callus induction in plant tissue culture, proving the high dynamics of DNA methylation [196]. It has been found that hypomethylation is characteristic of active transcription, while transcriptional repression is associated with increased DNA methylation level resulting from changes in the chromatin structure [197,198]. Protein transcriptional regulators mediate the regulation of transcription. Transcription regulators alter nucleosome structure via acetylation, deacetylation, or ATP-dependent nucleosome structure changes [199]. Among regulators studied by now, the VIP1 protein present in the nucleus [197] exhibited overexpression in tobacco cells inhibiting shoot formation, not affecting callus development and root formation. In turn, the work of Pischke et al. [200] knowledge on transcriptome alteration in the synthesis of cytokinins, compounds necessary for the proper growth and development of plants. Cytokinins participate in signaling in callus tissues [201,202,203,204]. The other particles that exhibited a change in the expression levels are small RNAs (namely, a unique microRNA set). Some of these particles are being highly expressed, influencing multicopper oxidases (laccases). When suppressed, laccases prevent lignification and cell wall thickening, keeping the cells in an undifferentiated, meristematic state. The microRNAs expression correlated with the transition from the undifferentiated to the differentiated stage of cells [205]. The study demonstrates that copper ions may influence the cell’s biochemical pathways but in complexes that may affect epigenetic machinery (see review [206]). Recent studies in barley have also demonstrated that different micro RNAs are involved during microspore development. Some of them appear due to stress responses. Furthermore, miRNA-directed regulation of several transcription factor families (ARF, SPL, GRF, and HD-ZIPIII) was needed for the transition of microspores toward the embryogenesis pathway [207]. Differential expression of small RNAs was also denoted in the early development of broccoli (Brassica oleracea var. italica) pollen [208]. Differences in expression of some miRNAs were shown depending on the developmental stages of broccoli pollen. They were involved in the developmental transition from uni- to bi- and trinucleate pollen grains. Some of the miRNAs were specific for the phases. Based on gene ontology functional annotation, the miRNAs targeted plant organ formation, morphogenesis, and male reproduction development [209].
1.7. Retrograde and Anterograde Signaling
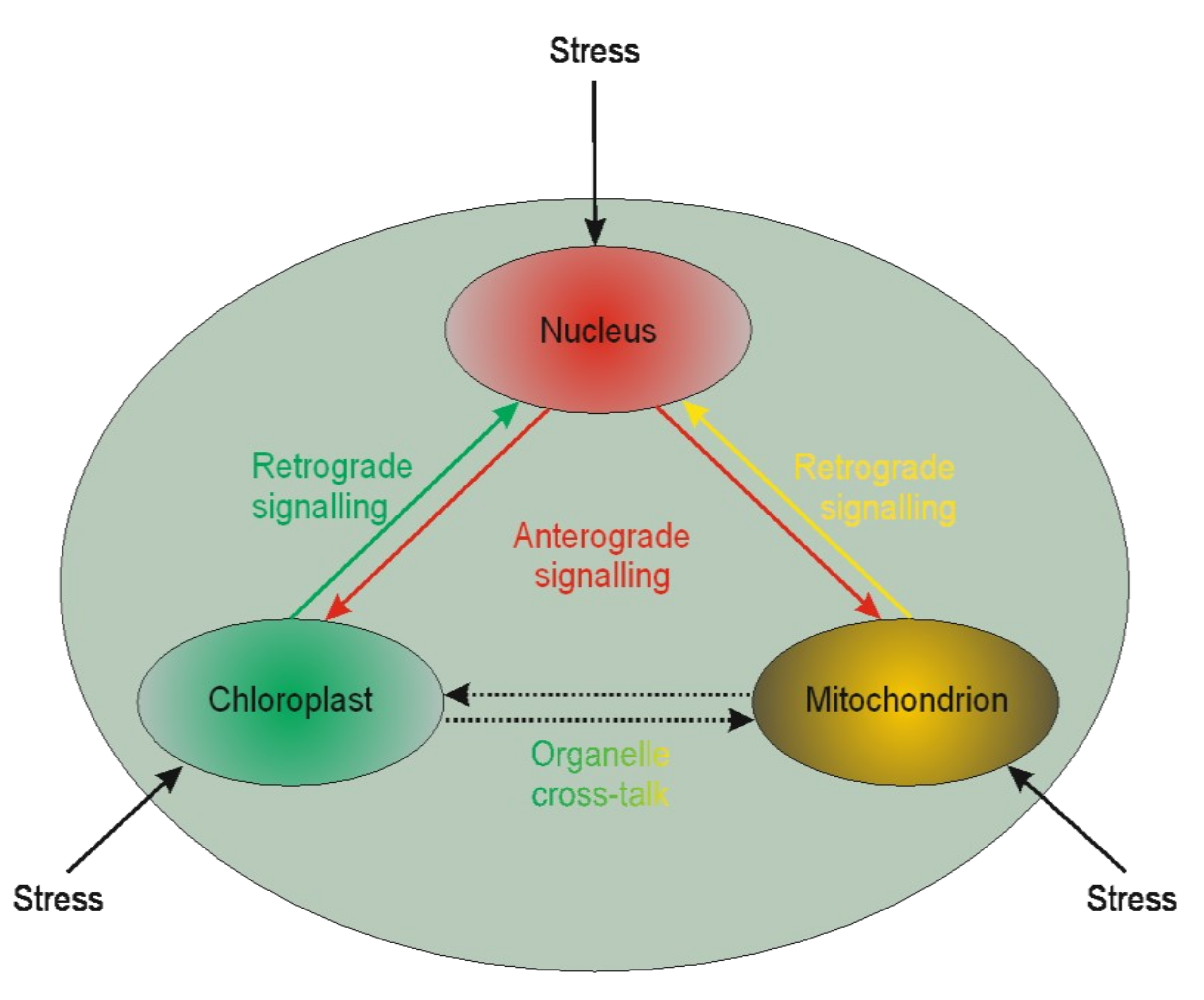
It is becoming evident that nuclear genes’ transcription regulation may be modulated by plastid gene expression [210,211,212]. Such regulation is called retrograde signaling. The reversed interaction when nucleus genes influence plastids’ genome functioning is called anterograde [213] (Figure 1).
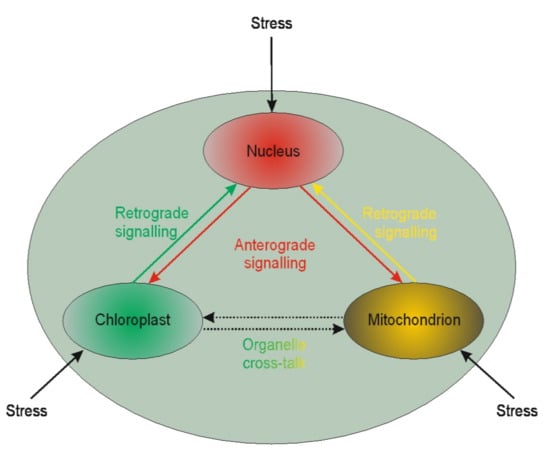
Figure 1.
Schematic illustration of interplay between cellular organelles. Anterograde is the signaling from the nucleus towards chloroplasts or mitochondria, whereas the reversed direction is called retrograde signaling. The communication between mitochondria and chloroplast (and other organelles except but nucleus) is called cross-talk.
Little is known of the two interactions in the context of microspores. The available literature is devoted chiefly to chloroplasts. Based on chloroplast biogenesis, retrograde signaling could be divided into “biogenic” and “operational” signals. The “biogenic signals” are the signals exhibited during early chloroplast development, whereas the “operational signals” are related to chloroplasts’ normal function in mature plants. Based on the signals’ origin, the retrograde signaling pathways in chloroplasts may include tetrapyrroles biosynthesis, redox state, chloroplast gene expression, reactive oxygen species, and protein import into plastids. The retrograde signaling pathway was described in barley mutant albostrians, which lacks plastid ribosomes and shows reduced amounts and/or activities of nuclear-encoded plastid proteins (the small subunit of ribulose-1,5-bisphosphate carboxylase⁄oxygenase (Rubisco), ferredoxin NADP+ reductase, and enzymes of the Calvin cycle) [214]. Inhibition of tetrapyrrole biosynthesis and plastid gene expression activity resulted in the moderated expression of nuclear starch biosynthesis genes in tobacco [215]. It is well known that in vitro plant regeneration involves stressful conditions that induce chloroplast proteome remodeling. Nuclear factors regulate chloroplast gene expression, and this form of anterograde regulation has roles in plant adaptation to abiotic stress [216]. Furthermore, retrograde and anterograde signaling was demonstrated in barley in the case of albino plant regeneration via androgenesis [193,194]. The other line of evidence suggests that retrograde mitochondrial signaling may also be essential for plants. For instance, mitochondria have been proposed to play a vital role in programmed cell death (PCD) in anther tapetum cells. PCD is crucial during anther development providing lipids coating pollen exines [217]. The role of PCD in the tapetum was also demonstrated in sunflower [218]. However, it could not be excluded that such similar signaling may act for anther cultures. Thus, the retro and anterograde signaling pathways may affect organelle functioning’s biochemical level, linked to epigenetic aspects of the in vitro tissue culture-induced variation.
1.8. Biochemical Aspects
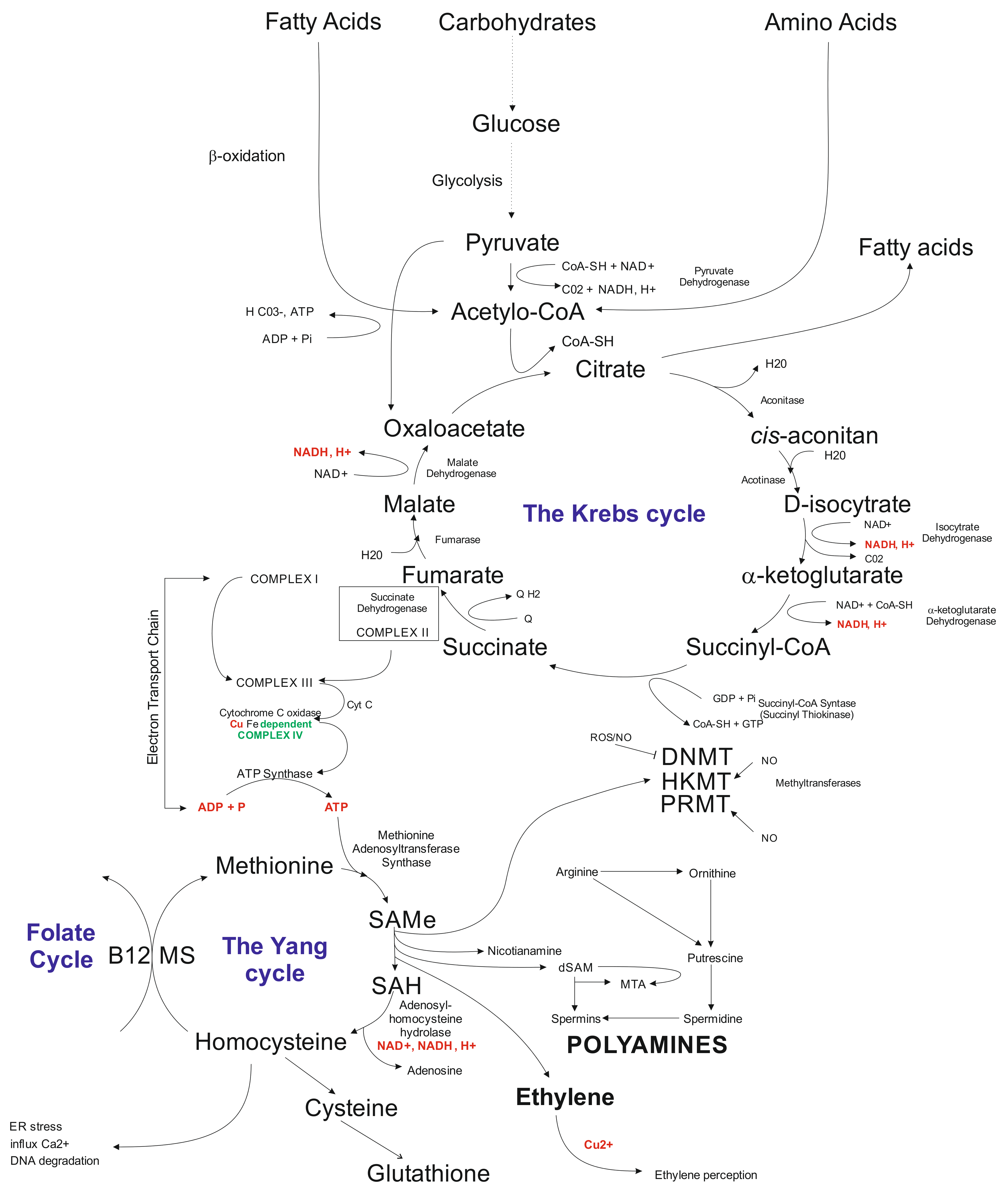
Another layer that is being affected by in vitro tissue culture plant regeneration relates to biochemical pathways. The available data indicate that callose present in the subintinal layer [128] of microspores may affect the Krebs’ cycle via complex II involved in electron transport chain (ETC) [62]. Problems with ATP synthesis may disturb the Yang cycle [219], followed by DNA methylation problems [220] and induction of mutations [221], and possibly activation of transposable elements (TEs) [222]. Indirectly, the Yang cycle is responsible for spermine and spermidine synthesis [223]. The compounds may participate in gene expression regulation [224]. Furthermore, the Yang cycle is essential for the cell’s glutathione synthesis as an antioxidant reagent preventing modification of, for example, methylated cytosines. It was documented that glutathione significantly improved plant regeneration via anther culture in rye [102]. Possibly, that disturbances in the Krebs’ cycle may change fatty acid synthesis (Figure 2). Fatty acids may influence, for example, gene transcription [225] and glycolysis. Furthermore, glycolysis could be disturbed under carbon starvation stress [226]. Thus, in vitro tissue culture plant regeneration is a complex, multidimensional process affecting all cell functioning levels. Understanding the relationships among all of the system’s components is vital for the elaboration of knowledge-based approaches of in vitro plant regeneration and regulation of the levels of somaclonal variation.
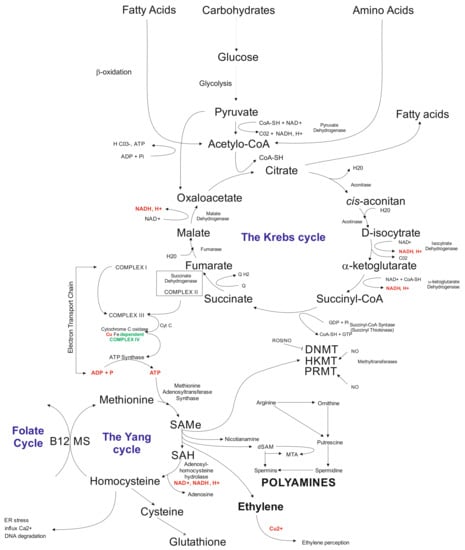
Figure 2.
Schematic illustration of biochemical cycles that may participate in the induction of in vitro tissue culture variation. Briefly, under cold treatment and darkness and maltose presence, carbon starvation stress results in disfunction of glycolysis affecting the Krebs cycle. Disbalance in the Krebs cycle could be sensed at the electron transfer chain level via complex II and problems with ATP synthesis, which influence the Yang cycle. The latter is responsible for producing SAM involved in 80% of the cell compounds’ methylation processes, including genomic cytosines. The Yang cycle’s mall functioning may also influence glutathione production (antioxidant) used but the cell during oxidative stress. Oxidative stress may modify methylated cytosine inducing point mutations. Furthermore, byproducts of the Yang cycle may regulate transcriptome as the result of abiotic stresses. Abbreviations: B12 (vitamin B12); CoA (Coenzyme A (acyl-CoA); CoASH (coenzyme A not attached to acyl group); DNMT (DNA methyltransferase); dSAM (decarboxylated SAM); ER stress (endoplasmic reticulum stress); HKMT (histone lysine methyltransferase); PRMT; MS (methionine synthesis); MTA (5′-methyl thioadenosine); NAD (nicotinamide adenine dinucleotide); NADH (1,4-dihydro-nicotinamide adenine dinucleotide); PRMT (protein arginine N-methyltransferase); ROS (radical oxygen species); SAMe (S-adenosyl-L-methionine); SAH (S-adenosylhomocysteine).
2. Conclusions
The available literature demonstrates that TCIV is a complex phenomenon, including the mucilage layer, the cell wall, the cell membrane, and the interactions of cellular organelles. The other layer involves transcriptome functioning changes. Furthermore, all these alterations may be linked to biochemical pathways responsible for DNA methylation and sequence variation.
The crucial factors influencing the balance between the different aspects of cell organization are abiotic stresses affecting in vitro tissue cultures and pretreatment stages.
The establishment of all the linkages involved in the TCIV requires further interdisciplinary studies that would extend our knowledge of cell functioning under in vitro tissue culture conditions. Thus, the presented review was an attempt to gather the scattered data in a single place.
Author Contributions
Conceptualization, P.T.B.; resources, P.T.B.; R.O.; writing—original draft preparation, P.T.B.; K.A.P.; W.M.D.; J.M.; R.O.; writing—review and editing, P.T.B.; R.O.; visualization, P.T.B.; supervision, P.T.B.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We thank Jan J. Rybczyński (Polish Academy of Sciences Botanical Garden –Center for Biological Diversity Conservation, Poland) for critical reading of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 3-MA | 3-methyladenine |
| AFLP | amplified fragment length polymorphism |
| ATR-FTIR | attenuated total reflectance Fourier transform infrared |
| B12 | vitamin B12 |
| CESA | cellulose synthase |
| CoA | coenzyme A (acyl-CoA) |
| CoASH | coenzyme A not attached to acyl group |
| CSC | cellulose synthase complex |
| cwm1 | cell wall maintainer1 |
| cwm2 | cell wall maintainer2 |
| DAMPs | damage-associated molecular pattern |
| DArTseqMet | diversity arrays technology methylation analysis |
| DNMT | DNA methyltransferase |
| dSAM | decarboxylated SAM |
| ER stress | endoplasmic reticulum stress |
| ET | endogenous ethylene |
| ETC | electron transport chain ETC |
| exDNA | extracellular DNA |
| GBSSI | granule-bound starch synthase I |
| Glc | glucose |
| GTs | glycosyltransferases |
| HKMT | histone lysine methyl-transferase |
| MAPK | mitogen-activation protein kinase |
| metAFLP | methylation sensitive amplified fragment length polymorphism |
| MIK2 | leucine-rich repeat receptor kinase LRR-RK male discoverer 1-interacting receptor-like kinase 2 |
| MLGs | mixed-linkage glucans MLGs |
| MS | methionine synthesis |
| MSAP | methylation sensitive amplification polymorphism |
| MTA | 5′-methyl thioadenosine |
| NAD | nicotinamide adenine dinucleotide |
| NADH | 1,4-dihydronicotinamide adenine dinucleotide |
| NGS | next generation sequencing |
| PCD | programmed cell death |
| PM | plasma membrane |
| PRMT | protein arginine N-methyltransferase |
| RAPD | randomly amplified polymorphic DNA |
| RFLP | restriction fragment length polymorphisms |
| ROS | reactive oxygen species |
| Rubisco | ribulose-1,5-bisphosphate carboxylase⁄oxygenase (Rubisco) |
| SAH | S-adenosylhomocysteine |
| SAMe | S-adenosyl-L-methionine |
| SEM | structural equation modeling |
| SNPs | single nucleotide polymorphisms |
| SV | somaclonal variation |
| TCIV | tissue-culture-induced variation |
| THE1 | receptor-like protein kinase THESEUS 1 |
References
- Orłowska, R.; Bednarek, P.T. Precise evaluation of tissue culture-induced variation during optimisation of in vitro regeneration regime in barley. Plant Mol. Biol. 2020, 103, 33–50. [Google Scholar] [CrossRef]
- Machczyńska, J.; Zimny, J.; Bednarek, P. Tissue culture-induced genetic and epigenetic variation in triticale (× Triticosecale spp. Wittmack ex A. Camus 1927) regenerants. Plant Mol. Biol. 2015, 89, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Mikuła, A.; Tomiczak, K.; Rybczynski, J.J. Cryopreservation enhances embryogenic capacity of Gentiana cruciata (L.) suspension culture and maintains (epi)genetic uniformity of regenerants. Plant Cell Rep. 2011, 30, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Fiuk, A.; Bednarek, P.T.; Rybczyński, J.J. Flow Cytometry, HPLC-RP, and metAFLP Analyses to Assess Genetic Variability in Somatic Embryo-Derived Plantlets of Gentiana pannonica Scop. Plant Mol. Biol. Rep. 2010, 28, 413–420. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Bhojwani, S.S. Genetic variability in the progeny of androgenic dihaploid plants and selection of high agronomic performing lines in Brassica juncea. Biol. Plant. 2004, 48, 503–508. [Google Scholar] [CrossRef]
- Zehr, B.E.; Williams, M.E.; Duncan, D.R.; Widholm, J.M. Somaclonal variation in the progeny of plants regenerated from callus cultures of seven inbred lines of maize. Can. J. Bot. 1987, 65, 491–499. [Google Scholar] [CrossRef]
- Ghosh, A.; Igamberdiev, A.U.; Debnath, S.C. Tissue culture-induced DNA methylation in crop plants: A review. Mol. Biol. Rep. 2021, 48, 823–841. [Google Scholar] [CrossRef]
- Cao, Z.; Sui, S.; Cai, X.; Yang, Q.; Deng, Z. Somaclonal variation in ‘Red Flash’ caladium: Morphological, cytological and molecular characterization. Plant Cell Tissue Organ Cult. 2016, 126, 269–279. [Google Scholar] [CrossRef]
- Deepthi, V.P. Somaclonal variation in micro propagated bananas. Adv. Plants Agric. Res. 2018, 8, 624–627. [Google Scholar] [CrossRef]
- Qin, Y.; Shin, K.-S.; Woo, H.-J.; Lim, M.-H. Genomic Variations of Rice Regenerants from Tissue Culture Revealed by Whole Genome Re-Sequencing. Plant Breed. Biotechnol. 2018, 6, 426–433. [Google Scholar] [CrossRef]
- Azizi, P.; Hanafi, M.M.; Sahebi, M.; Harikrishna, J.A.; Taheri, S.; Yassoralipour, A.; Nasehi, A. Epigenetic changes and their relationship to somaclonal variation: A need to monitor the micropropagation of plantation crops. Funct. Plant Biol. 2020, 47, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Orłowska, R. Barley somatic embryogenesis-an attempt to modify variation induced in tissue culture. J. Biol. Res. Thessalon. 2021, 28, 9. [Google Scholar] [CrossRef]
- Skirvin, R.M.; Janick, J. Tissue culture-induced variation in scented Pelargonium spp. J. Am. Soc. Hortic. Sci. 1976, 101, 281–290. [Google Scholar]
- Scowcroft, W.R. Somaclonal variation: The myth of clonal uniformity. In Genetic Flux in Plants; Hohn, B., Dennis, E.B., Eds.; Springer: Vienna, Austria, 1985; pp. 217–245. [Google Scholar]
- Larkin, P.J.; Scowcroft, W.R. Somaclonal variation—A novel source of variability from cell cultures for plant improvment. Theor. Appl. Genet. 1981, 60, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polimorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Jeong, I.S.; Yoon, U.H.; Lee, G.S.; Ji, H.S.; Lee, H.J.; Han, C.D.; Hahn, J.H.; An, G.; Kim, T.H. SNP-based analysis of genetic diversity in anther-derived rice by whole genome sequencing. Rice 2013, 6, 6. [Google Scholar] [CrossRef]
- Peschke, V.M.; Phillips, R.L.; Genggenbach, B.G. Discovery of transposable element activity among progeny of tissue-culture derived plants. Science 1987, 238, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Hosokawa, M.; Doi, M. Somaclonal Variation Is Induced De Novo via the Tissue Culture Process: A Study Quantifying Mutated Cells in Saintpaulia. PLoS ONE 2011, 6, e23541. [Google Scholar] [CrossRef] [PubMed]
- Barret, P.; Brinkman, M.; Beckert, M. A sequence related to rice Pong transposable element displays transcriptional activation by in vitro culture and reveals somaclonal variations in maize. Genome 2006, 49, 1399–1407. [Google Scholar] [CrossRef]
- Tanurdzic, M.; Vaughn, M.W.; Jiang, H.; Lee, T.-J.; Slotkin, R.K.; Sosinski, B.; Thompson, W.F.; Doerge, R.W.; Martienssen, R.A. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 2008, 6, e302. [Google Scholar] [CrossRef] [PubMed]
- Kaeppler, S.M.; Phillips, R.L.; Olhoft, P. Molecular basis of heritable tissue culture-induced variation in plants. In Somaclonal Variation and Induced Mutation in Crop Improvement; Jain, S.M., Brar, D.S., Ahloowalia, B.S., Eds.; Kluwer Academic: Dordrecht, The Netherlands, 1998; pp. 465–484. [Google Scholar]
- Cassels, A.C.; Curry, R.F. Oxidative stress and physiological, epigenetic and genetic variability in plant tissue culture: Implications for micropropagators and genetic engineers. Plant Cell Tissue Organ Cult. 2001, 64, 145–157. [Google Scholar] [CrossRef]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Dey, T.; Saha, S.; Ghosh, P.D. Somaclonal variation among somatic embryo derived plants—Evaluation of agronomically important somaclones and detection of genetic changes by RAPD in Cymbopogon winterianus. S. Afr. J. Bot. 2015, 96, 112–121. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef]
- Reyna-Lopez, G.E.; Simpson, J.; Ruiz-Herrera, J. Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol. Gen. Genet. 1997, 253, 703–710. [Google Scholar] [CrossRef]
- Xiong, L.Z.; Xu, C.G.; Saghai Maroof, M.A.; Zhang, Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplifcation polymorphism technique. Mol. Genet. Genom. 1999, 261, 439–446. [Google Scholar] [CrossRef]
- Baranek, M.; Cechova, J.; Kovacs, T.; Eichmeier, A.; Wang, S.; Raddova, J.; Necas, T.; Ye, X. Use of Combined MSAP and NGS Techniques to Identify Differentially Methylated Regions in Somaclones: A Case Study of Two Stable Somatic Wheat Mutants. PLoS ONE 2016, 11, e0165749. [Google Scholar] [CrossRef]
- Francischini, J.H.M.B.; Kemper, E.L.; Costa, J.B.; Manechini, J.R.V.; Pinto, L.R. DNA methylation in sugarcane somaclonal variants assessed through methylation-sensitive amplified polymorphism. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef]
- Bobadilla Landey, R.; Cenci, A.; Georget, F.; Bertrand, B.; Camayo, G.; Dechamp, E.; Herrera, J.C.; Santoni, S.; Lashermes, P.; Simpson, J.; et al. High Genetic and Epigenetic Stability in Coffea arabica Plants Derived from Embryogenic Suspensions and Secondary Embryogenesis as Revealed by AFLP, MSAP and the Phenotypic Variation Rate. PLoS ONE 2013, 8, e56372. [Google Scholar] [CrossRef]
- Dann, A.L.; Wilson, C.R. Comparative assessment of genetic and epigenetic variation among regenerants of potato (Solanum tuberosum) derived from long-term nodal tissue-culture and cell selection. Plant Cell Rep. 2011, 30, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Guevara, M.; de María, N.; Sáez-Laguna, E.; Vélez, M.D.; Cervera, M.T.; Cabezas, J.A. Analysis of DNA Cytosine Methylation Patterns Using Methylation-Sensitive Amplification Polymorphism (MSAP). Methods Mol. Biol. 2017, 1456, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Fulneček, J.; Kovařík, A. How to interpret Methylation Sensitive Amplified Polymorphism (MSAP) profiles? BMC Genet. 2014, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Balao, F.; Bazaga, P.; Perez, R. Epigenetic contribution to successful polyploidizations: Variation in global cytosine methylation along an extensive ploidy series in Dianthus broteri (Caryophyllaceae). New Phytol. 2016, 212, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Eckstein, R.L.; Durka, W. Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol. Ecol. Resour. 2013, 13, 642–653. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R.; Niedziela, A. A relative quantitative Methylation-Sensitive Amplified Polymorphism (MSAP) method for the analysis of abiotic stress. BMC Plant Biol. 2017, 17, 79. [Google Scholar] [CrossRef]
- Machczyńska, J.; Orłowska, R.; Zimny, J.; Bednarek, P.T. Extended metAFLP approach in studies of the tissue culture induced variation (TCIV) in case of triticale. Mol. Breed. 2014, 34, 845–854. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R. Time of In Vitro Anther Culture May Moderate Action of Copper and Silver Ions that Affect the Relationship between DNA Methylation Change and the Yield of Barley Green Regenerants. Plants 2020, 9, 1064. [Google Scholar] [CrossRef]
- Wang, S.; Lv, J.; Zhang, L.; Dou, J.; Sun, Y.; Li, X.; Fu, X.; Dou, H.; Mao, J.; Hu, X.; et al. MethylRAD: A simple and scalable method for genome-wide DNA methylation profiling using methylation-dependent restriction enzymes. Open Biol. 2015, 5, 150130. [Google Scholar] [CrossRef]
- Brunner, A.L.; Johnson, D.S.; Kim, S.W.; Valouev, A.; Reddy, T.E.; Neff, N.F.; Anton, E.; Medina, C.; Nguyen, L.; Chiao, E.; et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009, 19, 1044–1056. [Google Scholar] [CrossRef]
- Haque, N.; Nishiguchi, M. Bisulfite sequencing for cytosine-methylation analysis in plants. Methods Mol. Biol. 2011, 744, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rohde, C.; Tierling, S.; Stamerjohanns, H.; Reinhardt, R.; Walter, J.; Jeltsch, A. DNA Methylation Analysis by Bisulfite Conversion, Cloning, and Sequencing of Individual Clones. In DNA Methylation: Methods and Protocols; Tost, J., Ed.; Humana Press: Totowa, NY, USA, 2009; pp. 177–187. [Google Scholar] [CrossRef]
- Sun, Q.; Qiao, J.; Zhang, S.; He, S.; Shi, Y.; Yuan, Y.; Zhang, X.; Cai, Y. Changes in DNA methylation assessed by genomic bisulfite sequencing suggest a role for DNA methylation in cotton fruiting branch development. PeerJ 2018, 6, e4945. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P.T.; Orłowska, R.; Koebner, R.M.D.; Zimny, J. Quantification of the tissue-culture induced variation in barley (Hordeum vulgare L.). BMC Plant Biol. 2007, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Orłowska, R.; Machczyńska, J.; Oleszczuk, S.; Zimny, J.; Bednarek, P.T. DNA methylation changes and TE activity induced in tissue cultures of barley (Hordeum vulgare L.). J. Biol. Res. 2016, 23, 19. [Google Scholar] [CrossRef]
- Machczyńska, J.; Orłowska, R.; Mańkowski, D.R.; Zimny, J.; Bednarek, P.T. DNA methylation changes in triticale due to in vitro culture plant regeneration and consecutive reproduction. Plant Cell Tissue Organ Cult. 2014, 119, 289–299. [Google Scholar] [CrossRef]
- Śliwińska, A.A.; Białek, A.; Orłowska, R.; Mańkowski, D.; Sykłowska-Baranek, K.; Pietrosiuk, A. Comparative Study of the Genetic and Biochemical Variability of Polyscias filicifolia (Araliaceae) Regenerants Obtained by Indirect and Direct Somatic Embryogenesis as a Source of Triterpenes. Int. J. Mol. Sci. 2021, 22, 5752. [Google Scholar] [CrossRef] [PubMed]
- Mikuła, A.; Tomiczak, K.; Wójcik, A.; Rybczynski, J.J. Encapsulation-dehydration method elevates embryogenic abilities of Gentiana kurroo cell suspension and carrying on genetic stability of its regenerants after cryopreservation. In Acta Horticulturae; International Society for Horticultural Science: Leuven, Belgium, 2011; Volume 908, pp. 143–154. [Google Scholar]
- Oleszczuk, S.; Zimny, J.; Bednarek, P.T. The application of the AFLP method to determine the purity of homozygous lines of barley (Hordeum vulgare L.). Cell. Mol. Biol. Lett. 2002, 7, 777–783. [Google Scholar]
- Coronel, C.J.; González, A.I.; Ruiz, M.L.; Polanco, C. Analysis of somaclonal variation in transgenic and regenerated plants of Arabidopsis thaliana using methylation related metAFLP and TMD markers. Plant Cell Rep. 2018, 37, 137–152. [Google Scholar] [CrossRef]
- Lukens, L.N.; Zhan, S. The plant genome’s methylation status and response to stress: Implications for plant improvement. Curr. Opin. Plant Biol. 2007, 10, 317–322. [Google Scholar] [CrossRef]
- Jiang, C.; Mithani, A.; Gan, X.; Belfield, E.J.; Klingler, J.P.; Zhu, J.K.; Ragoussis, J.; Mott, R.; Harberd, N.P. Regenerant Arabidopsis lineages display a distinct genome-wide spectrum of mutations conferring variant phenotypes. Curr. Biol. 2011, 21, 1385–1390. [Google Scholar] [CrossRef]
- Orłowska, R.; Pachota, K.A.; Dynkowska, W.M.; Niedziela, A.; Bednarek, P.T. Androgenic-Induced Transposable Elements Dependent Sequence Variation in Barley. Int. J. Mol. Sci. 2021, 22, 6783. [Google Scholar] [CrossRef]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef]
- Kumar, S.; Lahlali, R.; Liu, X.; Karunakaran, C. Infrared spectroscopy combined with imaging: A new developing analytical tool in health and plant science. Appl. Spectrosc. Rev. 2016, 51, 466–483. [Google Scholar] [CrossRef]
- Kačuráková, M.; Capek, P.; Sasinková, V.; Wellner, N.; Ebringerová, A. FT-IR study of plant cell wall model compounds: Pectic polysaccharides and hemicelluloses. Carbohydr. Polym. 2000, 43, 195–203. [Google Scholar] [CrossRef]
- Kazarian, S.G.; Chan, K.L.A. ATR-FTIR spectroscopic imaging: Recent advances and applications to biological systems. Analyst 2013, 138, 1940–1951. [Google Scholar] [CrossRef] [PubMed]
- Synytsya, A.; Novak, M. Structural analysis of glucans. Ann. Transl. Med. 2014, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Zhu, J.; Yang, Z. Molecular Cell Biology of Pollen Walls. In Applied Plant Cell Biology: Cellular Tools and Approaches for Plant Biotechnology; Nick, P., Opatrny, Z., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 179–205. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Zebrowski, J.; Orłowska, R. Exploring the Biochemical Origin of DNA Sequence Variation in Barley Plants Regenerated via in Vitro Anther Culture. Int. J. Mol. Sci. 2020, 21, 5770. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R. CG Demethylation Leads to Sequence Mutations in an Anther Culture of Barley Due to the Presence of Cu, Ag Ions in the Medium and Culture Time. Int. J. Mol. Sci. 2020, 21, 4401. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.F. Introduction to Mediation, Moderation, and Conditional Process Analysis. A Regression Bases Approach; A Division of Guilford Publications, Inc.: New York, NY, USA, 2018; p. 507. [Google Scholar]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef]
- Pereira, W.J.; Pappas, M.d.C.R.; Grattapaglia, D.; Pappas, G.J., Jr. A cost-effective approach to DNA methylation detection by Methyl Sensitive DArT sequencing. PLoS ONE 2020, 15, e0233800. [Google Scholar] [CrossRef]
- Maraschin, S.F.; de Priester, W.; Spaink, H.P.; Wang, M. Androgenic switch: An example of plant embryogenesis from the male gametophyte perspective. J. Exp. Bot. 2005, 56, 1711–1726. [Google Scholar] [CrossRef]
- Galán-Ávila, A.; García-Fortea, E.; Prohens, J.; Herraiz, F.J. Microgametophyte Development in Cannabis sativa L. and First Androgenesis Induction Through Microspore Embryogenesis. Front. Plant Sci. 2021, 12, 669424. [Google Scholar] [CrossRef] [PubMed]
- Touraev, A.; Indrianto, A.; Wratschko, I.; Vicente, O.; Heberle-Bors, E. Efficient microspore embryogenesis in wheat (Triticum aestivum L.) induced by starvation at high temperature. Sex. Plant Reprod. 1996, 9, 209–215. [Google Scholar] [CrossRef]
- Hoekstra, S.; Hoekstra, S.; Hoekstra, I.R.; Hoekstra, R.A.; Hoekstra, E. The Interaction of 2,4-D Application and Mannitol Pretreatment in Anther and Microspore Culture of Hordeum vulgare L. cv. Igri. J. Plant Physiol. 1996, 148, 696–700. [Google Scholar] [CrossRef]
- Binarova, P.; Hause, G.; Cenklová, V.; Cordewener, J.H.; Campagne, M.L. A short severe heat shock is required to induce embryogenesis in late bicellular pollen of Brassica napus L. Sex. Plant Reprod. 1997, 10, 200–208. [Google Scholar] [CrossRef]
- Popova, T.; Grozeva, S.; Todorova, V.; Stankova, G.; Anachkov, N.; Rodeva, V. Effects of low temperature, genotype and culture media on in vitro androgenic answer of pepper (Capsicum annuum L.). Acta Physiol. Plant. 2016, 38, 273. [Google Scholar] [CrossRef]
- Tenhola-Roininen, T.; Tanhuanpää, P.; Immonen, S. The effect of cold and heat treatments on the anther culture response of diverse rye genotypes. Euphytica 2005, 145, 1–9. [Google Scholar] [CrossRef]
- Testillano, P.S. Microspore embryogenesis: Targeting the determinant factors of stress-induced cell reprogramming for crop improvement. J. Exp. Bot. 2019, 70, 2965–2978. [Google Scholar] [CrossRef]
- Seguí-Simarro, J.M.; Bárány, I.; Suárez, R.; Fadón, B.; Testillano, P.S.; Risueño, M.C. Nuclear bodies domain changes with microspore reprogramming to embryogenesis. Eur. J. Histochem. 2006, 50, 35–44. [Google Scholar]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176. [Google Scholar] [CrossRef]
- Rodríguez-Serrano, M.; Bárány, I.; Prem, D.; Coronado, M.-J.; Risueño, M.C.; Testillano, P.S. NO, ROS, and cell death associated with caspase-like activity increase in stress-induced microspore embryogenesis of barley. J. Exp. Bot. 2012, 63, 2007–2024. [Google Scholar] [CrossRef]
- Pasternak, T. Oxidative stress inducing agents’ copper and alloxan accelerate cell cycle re-entering of somatic plant cells in the presence of suboptimal exogenous auxin. bioRxiv 2020. preprint. [Google Scholar] [CrossRef]
- Uváčková, L.; Takáč, T.; Boehm, N.; Obert, B.; Samaj, J. Proteomic and biochemical analysis of maize anthers after cold pretreatment and induction of androgenesis reveals an important role of anti-oxidative enzymes. J. Proteom. 2012, 75, 1886–1894. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, C.; De Almeida, M.; Ruedell, C.; Schwambach, J.; Maraschin, F.; Fett-Neto, A. When stress and development go hand in hand: Main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133. [Google Scholar] [CrossRef]
- Fehér, A. Somatic embryogenesis—Stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta 2015, 1849, 385–402. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, T.P.; Prinsen, E.; Ayaydin, F.; Miskolczi, P.l.; Potters, G.; Asard, H.; Van Onckelen, H.A.; Dudits, D.n.; Fehér, A. The Role of Auxin, pH, and Stress in the Activation of Embryogenic Cell Division in Leaf Protoplast-Derived Cells of Alfalfa. Plant Physiol. 2002, 129, 1807–1819. [Google Scholar] [CrossRef]
- Pasternak, T.P.; Ötvös, K.; Domoki, M.; Fehér, A. Linked activation of cell division and oxidative stress defense in alfalfa leaf protoplast-derived cells is dependent on exogenous auxin. Plant Growth Regul. 2007, 51, 109–117. [Google Scholar] [CrossRef]
- Rodríguez-Sanz, H.; Solís, M.T.; López, M.F.; Gómez-Cadenas, A.; Risueño, M.C.; Testillano, P.S. Auxin Biosynthesis, Accumulation, Action and Transport are Involved in Stress-Induced Microspore Embryogenesis Initiation and Progression in Brassica napus. Plant Cell Physiol. 2015, 56, 1401–1417. [Google Scholar] [CrossRef]
- Weijers, D.; Nemhauser, J.; Yang, Z. Auxin: Small molecule, big impact. J. Exp. Bot. 2018, 69, 133–136. [Google Scholar] [CrossRef]
- Li, S.-B.; Xie, Z.-Z.; Hu, C.-G.; Zhang, J.-Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef]
- Mironova, V.; Teale, W.; Shahriari, M.; Dawson, J.; Palme, K. The Systems Biology of Auxin in Developing Embryos. Trends Plant Sci. 2017, 22, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Liu, Y.B.; Bai, B.; Zhang, X.S. Establishment of embryonic shoot–root axis is involved in auxin and cytokinin response during Arabidopsis somatic embryogenesis. Front. Plant Sci. 2015, 5, 792. [Google Scholar] [CrossRef] [PubMed]
- Hofius, D.; Li, L.; Hafrén, A.; Coll, N.S. Autophagy as an emerging arena for plant-pathogen interactions. Curr. Opin. Plant Biol. 2017, 38, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Chen, Q.; Havé, M. Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol. 2017, 39, 8–17. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T.; Baluška, F.; Bozhkov, P.V.; Elander, P.H.; Fernie, A.R.; Galili, G.; Hassan, A.; Hofius, D.; Isono, E.; Le Bars, R.; et al. Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J. Experimrntal Bot. 2018, 69, 1335–1353. [Google Scholar] [CrossRef]
- Corral-Martínez, P.; Parra-Vega, V.; Seguí-Simarro, J.M. Novel features of Brassica napus embryogenic microspores revealed by high pressure freezing and freeze substitution: Evidence for massive autophagy and excretion-based cytoplasmic cleaning. J. Exp. Bot. 2013, 64, 3061–3075. [Google Scholar] [CrossRef]
- Michaeli, S.; Avin-Wittenberg, T.; Galili, G. Involvement of autophagy in the direct ER to vacuole protein trafficking route in plants. Front. Plant Sci. 2014, 5, 134. [Google Scholar] [CrossRef]
- Tan, X.; Li, K.; Wang, Z.; Zhu, K.; Tan, X.; Cao, J. A Review of Plant Vacuoles: Formation, Located Proteins, and Functions. Plants 2019, 8, 327. [Google Scholar] [CrossRef]
- Pérez-Pérez, M.E.; Lemaire, S.D.; Crespo, J.L. Reactive Oxygen Species and Autophagy in Plants and Algae. Plant Physiol. 2012, 160, 156–164. [Google Scholar] [CrossRef]
- Bárány, I.; Berenguer, E.; Solís, M.T.; Pérez-Pérez, Y.; Santamaría, M.E.; Crespo, J.L.; Risueño, M.C.; Díaz, I.; Testillano, P.S. Autophagy is activated and involved in cell death with participation of cathepsins during stress-induced microspore embryogenesis in barley. J. Exp. Bot. 2018, 69, 1387–1402. [Google Scholar] [CrossRef] [PubMed]
- Tsiatsiani, L.; Van Breusegem, F.; Gallois, P.; Zavialov, A.; Lam, E.; Bozhkov, P.V. Metacaspases. Cell Death Differ. 2011, 18, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Berenguer, E.; Bárány, I.; Solís, M.-T.; Pérez-Pérez, Y.; Risueño, M.C.; Testillano, P.S. Inhibition of Histone H3K9 Methylation by BIX-01294 Promotes Stress-Induced Microspore Totipotency and Enhances Embryogenesis Initiation. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [PubMed]
- Solís, M.-T.; El-Tantawy, A.-A.; Cano, V.; Risueño, M.C.; Testillano, P.S. 5-azacytidine promotes microspore embryogenesis initiation by decreasing global DNA methylation, but prevents subsequent embryo development in rapeseed and barley. Front. Plant Sci. 2015, 6, 472. [Google Scholar] [CrossRef] [PubMed]
- El-Tantawy, A.A.; Solís, M.T.; Risueño, M.C.; Testillano, P.S. Changes in DNA Methylation Levels and Nuclear Distribution Patterns after Microspore Reprogramming to Embryogenesis in Barley. Cytogenet. Genome Res. 2014, 143, 200–208. [Google Scholar] [CrossRef]
- Çakmak, E.; Uncuoğlu, A.A.; Aydın, Y. Evaluation of in vitro genotoxic effects induced by in vitro anther culture conditions in sunflower. Plant Signal. Behav. 2019, 14, 1633885. [Google Scholar] [CrossRef]
- Zieliński, K.; Krzewska, M.; Żur, I.; Juzoń, K.; Kopeć, P.; Nowicka, A.; Moravčiková, J.; Skrzypek, E.; Dubas, E. The effect of glutathione and mannitol on androgenesis in anther and isolated microspore cultures of rye (Secale cereale L.). Plant Cell Tissue Organ Cult. 2020, 140, 577–592. [Google Scholar] [CrossRef]
- Castander-Olarieta, A.; Montalbán, I.A.; De Medeiros Oliveira, E.; Dell’Aversana, E.; D’Amelia, L.; Carillo, P.; Steiner, N.; Fraga, H.P.D.F.; Guerra, M.P.; Goicoa, T.; et al. Effect of Thermal Stress on Tissue Ultrastructure and Metabolite Profiles During Initiation of Radiata Pine Somatic Embryogenesis. Front. Plant Sci. 2019, 9, 4. [Google Scholar] [CrossRef]
- Gamalero, E.; Glick, B.R. Ethylene and Abiotic Stress Tolerance in Plants. In Environmental Adaptations and Stress Tolerance of Plants in the Era of Climate Change; Ahmad, P., Prasad, M.N.V., Eds.; Springer New York: New York, NY, USA, 2012; pp. 395–412. [Google Scholar] [CrossRef]
- Klay, I.; Pirrello, J.; Riahi, L.; Bernadac, A.; Cherif, A.; Bouzayen, M.; Bouzid, S. Ethylene response factor Sl-ERF. B. 3 is responsive to abiotic stresses and mediates salt and cold stress response regulation in tomato. Sci. World J. 2014, 2014, 167681. [Google Scholar] [CrossRef]
- Mu, C.; Wang, S.; Zhang, S.; Pan, J.; Chen, N.; Li, X.; Wang, Z.; Liu, H. Small heat shock protein LimHSP16.45 protects pollen mother cells and tapetal cells against extreme temperatures during late zygotene to pachytene stages of meiotic prophase I in David Lily. Plant Cell Rep. 2011, 30, 1981. [Google Scholar] [CrossRef]
- Orłowska, R.; Pachota, K.A.; Machczyńska, J.; Niedziela, A.; Makowska, K.; Zimny, J.; Bednarek, P.T. Improvement of anther cultures conditions using the Taguchi method in three cereal crops. Electron. J. Biotechnol. 2020, 43, 8–15. [Google Scholar] [CrossRef]
- Vaahtera, L.; Schulz, J.; Hamann, T. Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 2019, 5, 924–932. [Google Scholar] [CrossRef]
- De Lorenzo, G.; Ferrari, S.; Giovannoni, M.; Mattei, B.; Cervone, F. Cell wall traits that influence plant development, immunity, and bioconversion. Plant J. 2019, 97, 134–147. [Google Scholar] [CrossRef]
- Bacic, A.; Moody, S.F.; Clarke, A.E. Structural Analysis of Secreted Root Slime from Maize (Zea mays L.). Plant Physiol. 1986, 80, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Moody, S.F.; Clarke, A.E.; Bacic, A. Structural analysis of secreted slime from wheat and cowpea roots. Phytochemistry 1988, 27, 2857–2861. [Google Scholar] [CrossRef]
- Chaboud, A.; Rougier, M. Comparison of maize root mucilages isolated from root exudates and root surface extracts by complementary cytological and biochemical investigations. Protoplasma 1990, 156, 163–173. [Google Scholar] [CrossRef]
- Hawes, M.C.; Curlango-Rivera, G.; Xiong, Z.; Kessler, J.O. Roles of root border cells in plant defense and regulation of rhizosphere microbial populations by extracellular DNA ‘trapping’. Plant Soil 2012, 355, 1–16. [Google Scholar] [CrossRef]
- Ceccherini, M.T.; Ascher, J.; Agnelli, A.; Borgogni, F.; Pantani, O.L.; Pietramellara, G. Experimental discrimination and molecular characterization of the extracellular soil DNA fraction. Antonie Van Leeuwenhoek 2009, 96, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Yakushiji, S.; Ishiga, Y.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Bacterial DNA activates immunity in Arabidopsis thaliana. J. Gen. Plant Pathol. 2009, 75, 227–234. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Bonanomi, G.; Incerti, G.; Chiusano, M.L.; Termolino, P.; Mingo, A.; Senatore, M.; Giannino, F.; Cartenì, F.; Rietkerk, M. Inhibitory and toxic effects of extracellular self-DNA in litter: A mechanism for negative plant–soil feedbacks? New Phytol. 2015, 205, 1195–1210. [Google Scholar] [CrossRef]
- Barbero, F.; Guglielmotto, M.; Capuzzo, A.; Maffei, M.E. Extracellular self-DNA (esDNA), but not heterologous plant or insect DNA (etDNA), induces plasma membrane depolarization and calcium signaling in lima bean (Phaseolus lunatus) and maize (Zea mays). Int. J. Mol. Sci. 2016, 17, 1659. [Google Scholar] [CrossRef]
- Duran-Flores, D.; Heil, M. Extracellular self-DNA as a damage-associated molecular pattern (DAMP) that triggers self-specific immunity induction in plants. Brain Behav. Immun. 2018, 72, 78–88. [Google Scholar] [CrossRef]
- Roh, J.S.; Sohn, D.H. Damage-Associated Molecular Patterns in Inflammatory Diseases. Immune Netw. 2018, 18, e27. [Google Scholar] [CrossRef] [PubMed]
- Vega-Muñoz, I.; Feregrino-Pérez, A.A.; Torres-Pacheco, I.; Guevara-González, R.G. Exogenous fragmented DNA acts as a damage-associated molecular pattern (DAMP) inducing changes in CpG DNA methylation and defence-related responses in Lactuca sativa. Funct. Plant Biol. 2018, 45, 1065–1072. [Google Scholar] [CrossRef]
- Fang, Y.-L.; Xia, L.-M.; Wang, P.; Zhu, L.-H.; Ye, J.-R.; Huang, L. The MAPKKK CgMck1 Is Required for Cell Wall Integrity, Appressorium Development, and Pathogenicity in Colletotrichum gloeosporioides. Genes 2018, 9, 543. [Google Scholar] [CrossRef] [PubMed]
- Ferrusquía-Jiménez, N.I.; Chandrakasan, G.; Torres-Pacheco, I.; Rico-Garcia, E.; Feregrino-Perez, A.A.; Guevara-González, R.G. Extracellular DNA: A Relevant Plant Damage-Associated Molecular Pattern (DAMP) for Crop Protection Against Pests—A Review. J. Plant Growth Regul. 2020, 40, 451–463. [Google Scholar] [CrossRef]
- Monticolo, F.; Palomba, E.; Termolino, P.; Chiaiese, P.; de Alteriis, E.; Mazzoleni, S.; Chiusano, M.L. The Role of DNA in the Extracellular Environment: A Focus on NETs, RETs and Biofilms. Front. Plant Sci. 2020, 11, 589837. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, T.; Satoh, H.; Yamada, Y.; Hashimoto, T. A maize glycine-rich protein is synthesized in the lateral root cap and accumulates in the mucilage. Plant Physiol. 1999, 120, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Hawes, M.C.; Curlango-Rivera, G.; Wen, F.; White, G.J.; VanEtten, H.D.; Xiong, Z. Extracellular DNA: The tip of root defenses? Plant Sci. 2011, 180, 741–745. [Google Scholar] [CrossRef]
- Nagata, N.; Saito, C.; Sakai, A.; Kuroiwa, H.; Kuroiwa, T. Unique positioning of mitochondria in developing microspores and pollen grains in Pharbitis nil: Mitochondria cover the nuclear surface at specific developmental stages. Protoplasma 2000, 213, 74–82. [Google Scholar] [CrossRef]
- Parra-Vega, V.; Corral-Martínez, P.; Rivas-Sendra, A.; Seguí-Simarro, J.M. Induction of Embryogenesis in Brassica Napus Microspores Produces a Callosic Subintinal Layer and Abnormal Cell Walls with Altered Levels of Callose and Cellulose. Front. Plant Sci. 2015, 6, 1018. [Google Scholar] [CrossRef]
- Rivas-Sendra, A.; Corral-Martínez, P.; Porcel, R.; Camacho-Fernández, C.; Calabuig-Serna, A.; Seguí-Simarro, J.M. Embryogenic competence of microspores is associated with their ability to form a callosic, osmoprotective subintinal layer. J. Exp. Bot. 2019, 70, 1267–1281. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Toriyama, K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 2011, 62, 437–460. [Google Scholar] [CrossRef] [PubMed]
- Quilichini, T.D.; Grienenberger, E.; Douglas, C.J. The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 2015, 113, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Heslop-Harrison, J. Pollen wall development. Science 1968, 161, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Blackmore, S.; Wortley, A.H.; Skvarla, J.J.; Rowley, J.R. Pollen wall development in flowering plants. New Phytol. 2007, 174, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Ahlers, F.; Lambert, J.; Wiermann, R. Acetylation and silylation of piperidine solubilized sporopollenin from pollen of Typha angustifolia L. Z. Für Nat. C 2003, 58, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Blokker, P.; Boelen, P.; Broekman, R.; Rozema, J. The occurrence of p-coumaric acid and ferulic acid in fossil plant materials and their use as UV-proxy. In Plants and Climate Change; Springer: Berlin/Heidelberg, Germany, 2006; pp. 197–208. [Google Scholar]
- Bubert, H.; Lambert, J.; Steuernagel, S.; Ahlers, F.; Wiermann, R. Continuous decomposition of sporopollenin from pollen of Typha angustifolia L. by acidic methanolysis. Verl. Der Z. Für Nat. 2002, 57, 1035–1041. [Google Scholar] [CrossRef]
- Li, W.L.; Liu, Y.; Douglas, C.J. Role of Glycosyltransferases in Pollen Wall Primexine Formation and Exine Patterning. Plant Physiol. 2017, 173, 167–182. [Google Scholar] [CrossRef]
- Heslop-Harrison, J. Wall development within the microspore tetrad of Lilium longiflorum. Can. J. Bot. 1968, 46, 1185–1192. [Google Scholar] [CrossRef]
- Quilichini, T.D.; Douglas, C.J.; Samuels, A.L. New views of tapetum ultrastructure and pollen exine development in Arabidopsis thaliana. Ann. Bot. 2014, 114, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef] [PubMed]
- Kesten, C.; Menna, A.; Sánchez-Rodríguez, C. Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 2017, 40, 106–113. [Google Scholar] [CrossRef]
- Lampugnani, E.R.; Khan, G.A.; Somssich, M.; Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Majda, M.; Robert, S. The role of auxin in cell wall expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef] [PubMed]
- Saffer, A.M. Expanding roles for pectins in plant development. J. Integr. Plant Biol. 2018, 60, 910–923. [Google Scholar] [CrossRef] [PubMed]
- Polko, J.K.; Kieber, J.J. The Regulation of Cellulose Biosynthesis in Plants. Plant Cell 2019, 31, 282–296. [Google Scholar] [CrossRef]
- Barnes, W.J.; Anderson, C.T. Cytosolic invertases contribute to cellulose biosynthesis and influence carbon partitioning in seedlings of Arabidopsis thaliana. Plant J. 2018, 94, 956–974. [Google Scholar] [CrossRef] [PubMed]
- Verbančič, J.; Lunn, J.E.; Stitt, M.; Persson, S. Carbon supply and the regulation of cell wall synthesis. Mol. Plant 2018, 11, 75–94. [Google Scholar] [CrossRef]
- Kurek, I.; Kawagoe, Y.; Jacob-Wilk, D.; Doblin, M.; Delmer, D. Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc. Natl. Acad. Sci. USA 2002, 99, 11109–11114. [Google Scholar] [CrossRef]
- Sethaphong, L.; Haigler, C.H.; Kubicki, J.D.; Zimmer, J.; Bonetta, D.; DeBolt, S.; Yingling, Y.G. Tertiary model of a plant cellulose synthase. Proc. Natl. Acad. Sci. USA 2013, 110, 7512–7517. [Google Scholar] [CrossRef]
- Slabaugh, E.; Davis, J.K.; Haigler, C.H.; Yingling, Y.G.; Zimmer, J. Cellulose synthases: New insights from crystallography and modeling. Trends Plant Sci. 2014, 19, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.L.; Hammudi, M.B.; Tien, M. The Arabidopsis cellulose synthase complex: A proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 2014, 26, 4834–4842. [Google Scholar] [CrossRef] [PubMed]
- Napoleão, T.A.; Soares, G.; Vital, C.E.; Bastos, C.; Castro, R.; Loureiro, M.E.; Giordano, A. Methyl jasmonate and salicylic acid are able to modify cell wall but only salicylic acid alters biomass digestibility in the model grass Brachypodium distachyon. Plant Sci. 2017, 263, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Leng, L.; Islam, M.K.; Liu, F.; Lin, C.S.K.; Leu, S.-Y. Substrate-Related Factors Affecting Cellulosome-Induced Hydrolysis for Lignocellulose Valorization. Int. J. Mol. Sci. 2019, 20, 3354. [Google Scholar] [CrossRef] [PubMed]
- Van der Does, D.; Boutrot, F.; Engelsdorf, T.; Rhodes, J.; McKenna, J.F.; Vernhettes, S.; Koevoets, I.; Tintor, N.; Veerabagu, M.; Miedes, E.; et al. The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet. 2017, 13, e1006832. [Google Scholar] [CrossRef]
- Humphrey, T.V.; Bonetta, D.T.; Goring, D.R. Sentinels at the wall: Cell wall receptors and sensors. New Phytol. 2007, 176, 7–21. [Google Scholar] [CrossRef]
- Kafkaletou, M.; Fasseas, C.; Tsantili, E. Increased firmness and modified cell wall composition by ethylene were reversed by the ethylene inhibitor 1-methylcyclopropene (1-MCP) in the non-climacteric olives harvested at dark green stage—Possible implementation of ethylene for olive quality. J. Plant Physiol. 2019, 238, 63–71. [Google Scholar] [CrossRef]
- Salazar, R.; Pollmann, S.; Morales-Quintana, L.; Herrera, R.; Caparrós-Ruiz, D.; Ramos, P. In seedlings of Pinus radiata, jasmonic acid and auxin are differentially distributed on opposite sides of tilted stems affecting lignin monomer biosynthesis and composition. Plant Physiol. Biochem. 2019, 135, 215–223. [Google Scholar] [CrossRef]
- Hu, Z.; Vanderhaeghen, R.; Cools, T.; Wang, Y.; De Clercq, I.; Leroux, O.; Nguyen, L.; Belt, K.; Millar, A.H.; Audenaert, D.; et al. Mitochondrial Defects Confer Tolerance against Cellulose Deficiency. Plant Cell 2016, 28, 2276–2290. [Google Scholar] [CrossRef]
- Hofmann, N.R. A Functional Link between Mitochondria and the Cell Wall in Stress Responses. Plant Cell 2016, 28, 1996. [Google Scholar] [CrossRef]
- Meng, X.; Li, L.; De Clercq, I.; Narsai, R.; Xu, Y.; Hartmann, A.; Claros, D.L.; Custovic, E.; Lewsey, M.G.; Whelan, J.; et al. ANAC017 Coordinates Organellar Functions and Stress Responses by Reprogramming Retrograde Signaling. Plant Physiol. 2019, 180, 634–653. [Google Scholar] [CrossRef] [PubMed]
- Amos, R.A.; Mohnen, D. Critical Review of Plant Cell Wall Matrix Polysaccharide Glycosyltransferase Activities Verified by Heterologous Protein Expression. Front. Plant Sci. 2019, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Uluisik, S.; Seymour, G.B. Pectate lyases: Their role in plants and importance in fruit ripening. Food Chem. 2020, 309, 125559. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, F.; Stanislas, T. The Plasma Membrane—An Integrating Compartment for Mechano-Signaling. Plants 2020, 9, 505. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Fincher, G.B. (1,3;1,4)-β-D-Glucans in Cell Walls of the Poaceae, Lower Plants, and Fungi: A Tale of Two Linkages. Mol. Plant 2009, 2, 873–882. [Google Scholar] [CrossRef]
- Marković, S.M.; Đukić, N.H.; Knežević, D.; Leković, S.V. Divergence of barley and oat varieties according to their content of β-glucan. J. Serb. Chem. Soc. 2017, 82, 379–388. [Google Scholar] [CrossRef]
- Zhai, Z.; Liu, H.; Xu, C.; Shanklin, J. Sugar Potentiation of Fatty Acid and Triacylglycerol Accumulation. Plant Physiol. 2017, 175, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Funck, D.; Winter, G.; Baumgarten, L.; Forlani, G. Requirement of proline synthesis during Arabidopsis reproductive development. BMC Plant Biol. 2012, 12, 191. [Google Scholar] [CrossRef] [PubMed]
- Price, J.; Laxmi, A.; St Martin, S.K.; Jang, J.-C. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 2004, 16, 2128–2150. [Google Scholar] [CrossRef]
- Jang, J.C.; Sheen, J. Sugar sensing in higher plants. Plant Cell 1994, 6, 1665–1679. [Google Scholar] [CrossRef]
- Pego, J.V.; Smeekens, S.C. Plant fructokinases: A sweet family get-together. Trends Plant Sci. 2000, 5, 531–536. [Google Scholar] [CrossRef]
- Díaz-Sala, C. Molecular Dissection of the Regenerative Capacity of Forest Tree Species: Special Focus on Conifers. Front. Plant Sci. 2019, 9, 1943. [Google Scholar] [CrossRef] [PubMed]
- Gravino, M.; Savatin, D.V.; Macone, A.; De Lorenzo, G. Ethylene production in Botrytis cinerea- and oligogalacturonide-induced immunity requires calcium-dependent protein kinases. Plant J. 2015, 84, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tergel, T.; Chen, J.; Yang, J.; Kang, Y.; Qi, Z. Arabidopsis transcriptional response to extracellular Ca2+ depletion involves a transient rise in cytosolic Ca2+. J. Integr. Plant Biol. 2015, 57, 138–150. [Google Scholar] [CrossRef]
- Knight, H. Calcium Signaling during Abiotic Stress in Plants. In International Review of Cytology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 1999; Volume 195, pp. 269–324. [Google Scholar]
- Makowska, K.; Kałużniak, M.; Oleszczuk, S.; Zimny, J.; Czaplicki, A.; Konieczny, R. Arabinogalactan proteins improve plant regeneration in barley (Hordeum vulgare L.) anther culture. Plant Cell Tissue Organ Cult. 2017, 131, 247–257. [Google Scholar] [CrossRef]
- Corral-Martínez, P.; Driouich, A.; Seguí-Simarro, J.M. Dynamic Changes in Arabinogalactan-Protein, Pectin, Xyloglucan and Xylan Composition of the Cell Wall During Microspore Embryogenesis in Brassica napus. Front. Plant Sci. 2019, 10, 332. [Google Scholar] [CrossRef]
- El-Tantawy, A.-A.; Solís, M.-T.; Da Costa, M.L.; Coimbra, S.; Risueño, M.-C.; Testillano, P.S. Arabinogalactan protein profiles and distribution patterns during microspore embryogenesis and pollen development in Brassica napus. Plant Reprod. 2013, 26, 231–243. [Google Scholar] [CrossRef]
- Pasternak, T.; Dudits, D. Epigenetic Clues to Better Understanding of the Asexual Embryogenesis in planta and in vitro. Front. Plant Sci. 2019, 10, 778. [Google Scholar] [CrossRef]
- Li, H.; Soriano, M.; Cordewener, J.; Muiño, J.M.; Riksen, T.; Fukuoka, H.; Angenent, G.C.; Boutilier, K. The histone deacetylase inhibitor trichostatin a promotes totipotency in the male gametophyte. Plant Cell 2014, 26, 195–209. [Google Scholar] [CrossRef]
- Manzoor, A.; Ahmad, T.; Bashir, M.A.; Hafiz, I.A.; Silvestri, C. Studies on Colchicine Induced Chromosome Doubling for Enhancement of Quality Traits in Ornamental Plants. Plants 2019, 8, 194. [Google Scholar] [CrossRef]
- Castillo, A.M.; Cistué, L.; Vallés, M.P.; Soriano, M. Chromosome doubling in monocots. In Advances in Haploid Production in Higher Plants; Touraev, A., Forster, B.P., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 329–338. [Google Scholar]
- Prem, D.; Solís, M.T.; Bárány, I.; Rodríguez-Sanz, H.; Risueño, M.C.; Testillano, P.S. A new microspore embryogenesis system under low temperature which mimics zygotic embryogenesis initials, expresses auxin and efficiently regenerates doubled-haploid plants in Brassica napus. BMC Plant Biol. 2012, 12, 127. [Google Scholar] [CrossRef] [PubMed]
- Abass, M.H.; Al-Utbi, S.D.; Al-Samir, E.A.R.H. Genotoxicity assessment of high concentrations of 2,4-D, NAA and Dicamba on date palm callus (Phoenix dactylifera L.) using protein profile and RAPD markers. J. Genet. Eng. Biotechnol. 2017, 15, 287–295. [Google Scholar] [CrossRef] [PubMed]
- González-Melendi, P.; Ramírez, C.; Testillano, P.S.; Kumlehn, J.; Risueño, M.C. Three dimensional confocal and electron microscopy imaging define the dynamics and mechanisms of diploidisation at early stages of barley microspore-derived embryogenesis. Planta 2005, 222, 47–57. [Google Scholar] [CrossRef]
- Oleszczuk, S.; Grzechnik, N.; Mason, A.S.; Zimny, J. Heritability of meiotic restitution and fertility restoration in haploid triticale. Plant Cell Rep. 2019, 38, 1515–1525. [Google Scholar] [CrossRef]
- Oleszczuk, S.; Lukaszewski, A.J. The origin of unusual chromosome constitutions among newly formed allopolyploids. Am. J. Bot. 2014, 101, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Kreps, J.A.; Wu, Y.; Chang, H.-S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome Changes for Arabidopsis in Response to Salt, Osmotic, and Cold Stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef]
- Pontin, M.A.; Piccoli, P.N.; Francisco, R.; Bottini, R.; Martinez-Zapater, J.M.; Lijavetzky, D. Transcriptome changes in grapevine (Vitis vinifera L.) cv. Malbec leaves induced by ultraviolet-B radiation. BMC Plant Biol. 2010, 10, 224. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.C.; Chen, S.L.; Osbourn, A.; Kontogianni, V.G.; Liu, L.W.; Jordán, M.J. Temporal transcriptome changes induced by methyl jasmonate in Salvia sclarea. Gene 2015, 558, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.D.; Nayyar, H. Regulatory Networks in Pollen Development under Cold Stress. Front. Plant Sci. 2016, 7, 402. [Google Scholar] [CrossRef]
- Sakata, T.; Oda, S.; Tsunaga, Y.; Shomura, H.; Kawagishi-Kobayashi, M.; Aya, K.; Saeki, K.; Endo, T.; Nagano, K.; Kojima, M. Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol. 2014, 164, 2011–2019. [Google Scholar] [CrossRef] [PubMed]
- Hale, B.; Phipps, C.; Rao, N.; Wijeratne, A.; Phillips, G.C. Differential Expression Profiling Reveals Stress-Induced Cell Fate Divergence in Soybean Microspores. Plants 2020, 9, 1510. [Google Scholar] [CrossRef]
- Bélanger, S.; Marchand, S.; Jacques, P.-É.; Meyers, B.; Belzile, F. Differential Expression Profiling of Microspores During the Early Stages of Isolated Microspore Culture Using the Responsive Barley Cultivar Gobernadora. G3 2018, 8, 1603–1614. [Google Scholar] [CrossRef]
- Gajecka, M.; Marzec, M.; Chmielewska, B.; Jelonek, J.; Zbieszczyk, J.; Szarejko, I. Plastid differentiation during microgametogenesis determines green plant regeneration in barley microspore culture. Plant Sci. 2020, 291, 110321. [Google Scholar] [CrossRef] [PubMed]
- Gajecka, M.; Marzec, M.; Chmielewska, B.; Jelonek, J.; Zbieszczyk, J.; Szarejko, I. Changes in plastid biogenesis leading to the formation of albino regenerants in barley microspore culture. BMC Plant Biol. 2021, 21, 22. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Liu, J.; Fan, M.; Xin, W.; Hu, Y.; Xu, C. A genome-wide transcriptome profiling reveals the early molecular events during callus initiation in Arabidopsis multiple organs. Genomics 2012, 100, 116–124. [Google Scholar] [CrossRef]
- Avivi, Y.; Morad, V.; Ben-Meir, H.; Zhao, J.; Kashkush, K.; Tzfira, T.; Citovsky, V.; Grafi, G. Reorganization of specific chromosomal domains and activation of silent genes in plant cells acquiring pluripotentiality. Dev. Dyn. 2004, 230, 12–22. [Google Scholar] [CrossRef]
- Jones, P.L.; Wolffe, A.P. Relationships between chromatin organization and DNA methylation in determining gene expression. Semin. Cancer Biol. 1999, 9, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Workman, J.L.; Kingston, R.E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998, 67, 545–579. [Google Scholar] [CrossRef] [PubMed]
- Pischke, M.S.; Huttlin, E.L.; Hegeman, A.D.; Sussman, M.R. A transcriptome-based characterization of habituation in plant tissue culture. Plant Physiol. 2006, 140, 1255–1278. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kakimoto, T. CKI1, a Histidine Kinase Homolog Implicated in Cytokinin Signal Transduction. Science 1996, 274, 982–985. [Google Scholar] [CrossRef]
- Hwang, I.; Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001, 413, 383–389. [Google Scholar] [CrossRef]
- Sakai, H.; Honma, T.; Aoyama, T.; Sato, S.; Kato, T.; Tabata, S.; Oka, A. ARR1, a Transcription Factor for Genes Immediately Responsive to Cytokinins. Science 2001, 294, 1519–1521. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Miyata, S.; Urao, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Overexpression of Arabidopsis response regulators, ARR4/ATRR1/IBC7 and ARR8/ATRR3, alters cytokinin responses differentially in the shoot and in callus formation. Biochem. Biophys. Res. Commun. 2002, 293, 806–815. [Google Scholar] [CrossRef]
- Rodriguez-Enriquez, J.; Dickinson, H.G.; Grant-Downton, R.T. MicroRNA misregulation: An overlooked factor generating somaclonal variation? Trends Plant Sci. 2011, 16, 242–248. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R. Plant tissue culture environment as a switch-key of (epi)genetic changes. Plant Cell Tissue Organ Cult. 2020, 140, 245–257. [Google Scholar] [CrossRef]
- Bélanger, S.; Baldrich, P.; Lemay, M.-A.; Marchand, S.; Esteves, P.; Meyers, B.C.; Belzile, F. The commitment of barley microspores into embryogenesis correlates with miRNA-directed regulation of members of the SPL, GRF and HD-ZIPIII transcription factor families. Plant Direct 2020, 4, e00289. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Wu, M.; Li, L.; Jin, C.; Zhang, Q.; Chen, C.; Song, W.; Wang, C. Small RNA Sequencing Reveals Differential miRNA Expression in the Early Development of Broccoli (Brassica oleracea var. italica) Pollen. Front. Plant Sci. 2017, 8, 404. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Yang, J.; Pan, F.; Jin, B. Differential expression of microRNAs may regulate pollen development in Brassica oleracea. Genet. Mol. Res. 2015, 14, 15024–15034. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Verdeja, T.; Strand, Å. Retrograde Signals Navigate the Path to Chloroplast Development. Plant Physiol. 2017, 176, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Singh, S.; Parihar, P.; Singh, V.P.; Prasad, S.M. Retrograde signaling between plastid and nucleus: A review. J. Plant Physiol. 2015, 181, 55–66. [Google Scholar] [CrossRef]
- Pfalz, J.; Oelmüller, R. Plastid Retrograde Signals: More to Discover. In Sensory Biology of Plants; Sopory, S., Ed.; Springer: Singapore, 2019; pp. 477–507. [Google Scholar] [CrossRef]
- Fujii, S.; Toriyama, K. Genome Barriers between Nuclei and Mitochondria Exemplified by Cytoplasmic Male Sterility. Plant Cell Physiol. 2008, 49, 1484–1494. [Google Scholar] [CrossRef] [PubMed]
- Börner, T. The discovery of plastid-to-nucleus retrograde signaling—a personal perspective. Protoplasma 2017, 254, 1845–1855. [Google Scholar] [CrossRef]
- Enami, K.; Ozawa, T.; Motohashi, N.; Nakamura, M.; Tanaka, K.; Hanaoka, M. Plastid-to-Nucleus Retrograde Signals Are Essential for the Expression of Nuclear Starch Biosynthesis Genes during Amyloplast Differentiation in Tobacco BY-2 Cultured Cells. Plant Physiol. 2011, 157, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Watson, S.J.; Sowden, R.G.; Jarvis, P. Abiotic stress-induced chloroplast proteome remodelling: A mechanistic overview. J. Exp. Bot. 2018, 69, 2773–2781. [Google Scholar] [CrossRef]
- Kawanabe, T.; Ariizumi, T.; Kawai-Yamada, M.; Uchimiya, H.; Toriyama, K. Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol. 2006, 47, 784–787. [Google Scholar] [CrossRef] [PubMed]
- Balk, J.; Leaver, C.J. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 2001, 13, 1803–1818. [Google Scholar] [CrossRef] [PubMed]
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine salvage and S-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013, 451, 145–154. [Google Scholar] [CrossRef]
- Moffatt, B.A.; Weretilnyk, E.A. Sustaining S-adenosyl-l-methionine-dependent methyltransferase activity in plant cells. Physiol. Plant. 2001, 113, 435–442. [Google Scholar] [CrossRef]
- Yoshida, T.; Furihata, H.Y.; To, T.K.; Kakutani, T.; Kawabe, A. Genome defense against integrated organellar DNA fragments from plastids into plant nuclear genomes through DNA methylation. Sci. Rep. 2019, 9, 2060. [Google Scholar] [CrossRef]
- Ong-Abdullah, M.; Ordway, J.M.; Jiang, N.; Ooi, S.-E.; Kok, S.-Y.; Sarpan, N.; Azimi, N.; Hashim, A.T.; Ishak, Z.; Rosli, S.K.; et al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 2015, 525, 533. [Google Scholar] [CrossRef] [PubMed]
- Zierer, W.; Hajirezaei, M.R.; Eggert, K.; Sauer, N.; von Wirén, N.; Pommerrenig, B. Phloem-Specific Methionine Recycling Fuels Polyamine Biosynthesis in a Sulfur-Dependent Manner and Promotes Flower and Seed Development. Plant Physiol. 2016, 170, 790–806. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine Function in Plants: Metabolism, Regulation on Development, and Roles in Abiotic Stress Responses. Front. Plant Sci. 2019, 9, 1945. [Google Scholar] [CrossRef]
- Shang, B.; Xu, C.; Zhang, X.; Cao, H.; Xin, W.; Hu, Y. Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 5101–5106. [Google Scholar] [CrossRef] [PubMed]
- Contento, A.L.; Kim, S.-J.; Bassham, D.C. Transcriptome Profiling of the Response of Arabidopsis Suspension Culture Cells to Suc Starvation. Plant Physiol. 2004, 135, 2330–2347. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).