Titin N2A Domain and Its Interactions at the Sarcomere
Abstract
1. Introduction
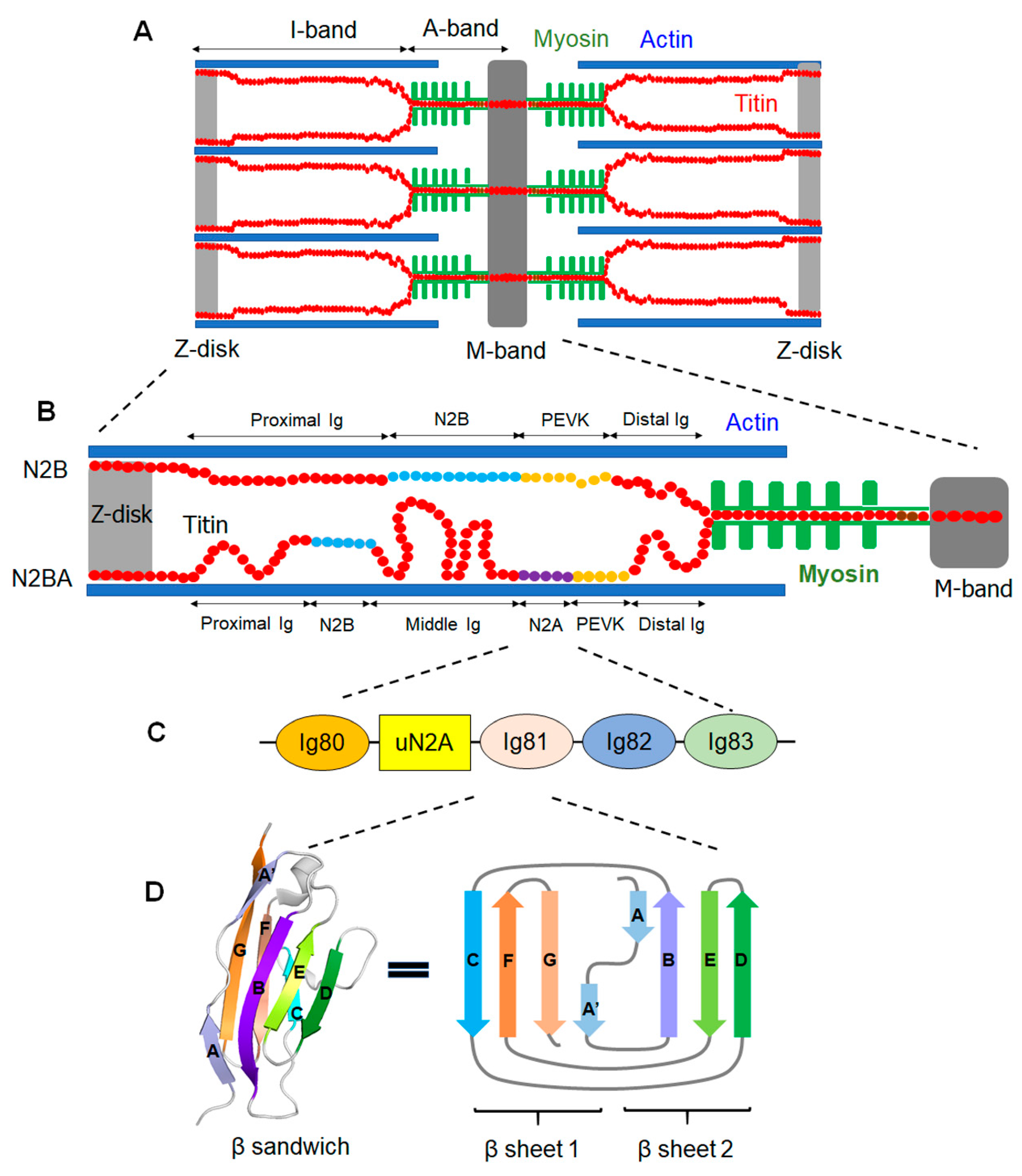
2. Structural Conformations of Titin’s N2A Domain
3. N2A Domain—An Interaction Node in Titin
3.1. Ca2+ and F-Actin to Modulate Contraction
3.2. Ca2+ to Regulate the Stability of N2A’s Ig Domains
3.3. HSP90 and SMYD2
3.4. Other Small HSPs
3.5. CARP
3.6. Calpain
4. N2A Domain’s Susceptibility to Proteolysis
4.1. Proteolysis by Calpains
4.2. Proteolysis by MMP-2
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Shadrin, I.Y.; Khodabukus, A.; Bursac, N. Striated muscle function, regeneration, and repair. Cell. Mol. life Sci. 2016, 73, 4175–4202. [Google Scholar] [CrossRef]
- Sweeney, H.L.; Hammers, D.W. Muscle Contraction. Cold Spring Harb. Perspect. Biol. 2018, 10, a023200. [Google Scholar] [CrossRef]
- Myhre, J.L.; Pilgrim, D. A Titan but not Necessarily a Ruler: Assessing the Role of Titin during Thick Filament Patterning and Assembly. Anat. Rec. 2014, 297, 1604–1614. [Google Scholar] [CrossRef] [PubMed]
- Koser, F.; Loescher, C.; Linke, W.A. Posttranslational modifications of titin from cardiac muscle: How, where, and what for? FEBS J. 2019, 286, 2240–2260. [Google Scholar] [CrossRef] [PubMed]
- Krüger, M.; Linke, W.A. The Giant Protein Titin: A Regulatory Node That Integrates Myocyte Signaling Pathways. J. Biol. Chem. 2011, 286, 9905–9912. [Google Scholar] [CrossRef]
- Voelkel, T.; Linke, W.A. Conformation-regulated mechanosensory control via titin domains in cardiac muscle. Pflügers Arch. Eur. J. Physiol. 2011, 462, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Krüger, M.; Kötter, S. Titin, a Central Mediator for Hypertrophic Signaling, Exercise-Induced Mechanosignaling and Skeletal Muscle Remodeling. Front. Physiol. 2016, 7, 76. [Google Scholar] [CrossRef]
- Eckels, E.C.; Tapia-Rojo, R.; Rivas-Pardo, J.A.; Fernández, J.M. The Work of Titin Protein Folding as a Major Driver in Muscle Contraction. Annu. Rev. Physiol. 2018, 80, 327–351. [Google Scholar] [CrossRef]
- Linke, W.A.; Hamdani, N. Gigantic business: Titin properties and function through thick and thin. Circ. Res. 2014, 114, 1052–1068. [Google Scholar] [CrossRef]
- Schappacher-Tilp, G.; Leonard, T.; Desch, G.; Herzog, W. A Novel Three-Filament Model of Force Generation in Eccentric Contraction of Skeletal Muscles. PLoS ONE 2015, 10, e0117634. [Google Scholar] [CrossRef]
- Rode, C.; Siebert, T.; Blickhan, R. Titin-induced force enhancement and force depression: A ’sticky-spring’ mechanism in muscle contractions? J. Theor. Biol. 2009, 259, 350–360. [Google Scholar] [CrossRef]
- Tahir, U.; Monroy, J.A.; Rice, N.A.; Nishikawa, K.C. Effects of a titin mutation on force enhancement and force depression in mouse soleus muscles. J. Exp. Biol. 2020, 223. [Google Scholar] [CrossRef] [PubMed]
- Tomalka, A.; Rode, C.; Schumacher, J.; Siebert, T. The active force-length relationship is invisible during extensive eccentric contractions in skinned skeletal muscle fibres. Proc. R. Soc. B Boil. Sci. 2017, 284, 20162497. [Google Scholar] [CrossRef] [PubMed]
- Kötter, S.; Unger, A.; Hamdani, N.; Lang, P.; Vorgerd, M.; Nagel-Steger, L.; Linke, W.A. Human myocytes are protected from titin aggregation-induced stiffening by small heat shock proteins. J. Cell Biol. 2014, 204, 187–202. [Google Scholar] [CrossRef]
- Linke, W.A. Sense and stretchability: The role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc. Res. 2007, 77, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Kontrogianni-Konstantopoulos, A.; Ackermann, M.A.; Bowman, A.L.; Yap, S.V.; Bloch, R.J. Muscle Giants: Molecular Scaffolds in Sarcomerogenesis. Physiol. Rev. 2009, 89, 1217–1267. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.C.; Wright, N.T. Structure of giant muscle proteins. Front. Physiol. 2013, 4. [Google Scholar] [CrossRef]
- Hayashi, C.; Ono, Y.; Doi, N.; Kitamura, F.; Tagami, M.; Mineki, R.; Arai, T.; Taguchi, H.; Yanagida, M.; Hirner, S.; et al. Multiple Molecular Interactions Implicate the Connectin/Titin N2A Region as a Modulating Scaffold for p94/Calpain 3 Activity in Skeletal Muscle. J. Biol. Chem. 2008, 283, 14801–14814. [Google Scholar] [CrossRef]
- Dutta, S.; Tsiros, C.; Sundar, S.L.; Athar, H.; Moore, J.; Nelson, B.; Gage, M.J.; Nishikawa, K. Calcium increases titin N2A binding to F-actin and regulated thin filaments. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- van der Pijl, R.J.; van den Berg, M.; van de Locht, M.; Shen, S.; Bogaards, S.J.P.; Conijn, S.; Langlais, P.; Hooijman, P.E.; Labeit, S.; Heunks, L.M.A. Muscle ankyrin repeat protein 1 (MARP1) locks titin to the sarcomeric thin filament and is a passive force regulat r. J. Gen. Physiol. 2021, 153, e202112925. [Google Scholar] [CrossRef]
- Zhou, T.; Fleming, J.R.; Lange, S.; Hessel, A.L.; Bogomolovas, J.; Stronczek, C.; Grundei, D.; Ghassemian, M.; Biju, A.; Börgeson, E.; et al. Molecular Characterisation of Titin N2A and Its Binding of CARP Reveals a Titin/Actin Cross-linking Mechanism. J. Mol. Biol. 2021, 433, 166901. [Google Scholar] [CrossRef]
- Miller, M.K.; Bang, M.-L.; Witt, C.C.; Labeit, D.; Trombitas, C.; Watanabe, K.; Granzier, H.; McElhinny, A.S.; Gregorio, C.C.; Labeit, S. The Muscle Ankyrin Repeat Proteins: CARP, ankrd2/Arpp and DARP as a Family of Titin Filament-based Stress Response Molecules. J. Mol. Biol. 2003, 333, 951–964. [Google Scholar] [CrossRef] [PubMed]
- Donlin, L.T.; Andresen, C.; Just, S.; Rudensky, E.; Pappas, C.; Krufcger, M.; Jacobs, E.Y.; Unger, A.; Zieseniss, A.; Dobenecker, M.-W.; et al. Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev. 2012, 26, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Godage, D.N.P.M.; Vanhecke, G.C.; Samarasinghe, K.; Feng, H.-Z.; Hiske, M.; Holcomb, J.; Yang, Z.; Jin, J.; Chung, C.S.; Ahn, Y.-H. SMYD2 glutathionylation contributes to degradation of sarcomeric proteins. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Voelkel, T.; Andresen, C.; Ungera, A.; Just, S.; Rottbauer, W.; Wolfgang, A. LinkeadLysine methyltransferase Smyd2 regulates Hsp90-mediated protection of the sarcomeric titin springs and cardiac function. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 812–822. [Google Scholar] [CrossRef]
- Kelly, C.M.; Manukian, S.; Kim, E.; Gage, M.J. Differences in stability and calcium sensitivity of the Ig domains in titin’s N2A region. Protein Sci. 2020, 29, 1160–1171. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, X. The Molecular Mechanisms of Calpains Action on Skeletal Muscle Atrophy. Physiol. Res. 2016, 65, 547–560. [Google Scholar] [CrossRef]
- Wu, Y.; Cazorla, O.; Labeit, D.; Labeit, S.; Granzier, H. Changes in Titin and Collagen Underlie Diastolic Stiffness Diversity of Cardiac Muscle. J. Mol. Cell. Cardiol. 2000, 32, 2151–2161. [Google Scholar] [CrossRef]
- Bang, M.-L.; Centner, T.; Fornoff, F.; Geach, A.J.; Gotthardt, M.; McNabb, M.; Witt, C.C.; Labeit, D.; Gregorio, C.C.; Granzier, H.; et al. The Complete Gene Sequence of Titin, Expression of an Unusual ≈700-kDa Titin Isoform, and Its Interaction with Obscurin Identify a Novel Z-Line to I-Band Linking System. Circ. Res. 2001, 89, 1065–1072. [Google Scholar] [CrossRef]
- Cazorla, O.; Freiburg, A.; Helmes, M.; Centner, T.; McNabb, M.; Wu, Y.; Trombitas, K.; Labeit, S.; Granzier, H. Differential Expression of Cardiac Titin Isoforms and Modulation of Cellular Stiffness. Circ. Res. 2000, 86, 59–67. [Google Scholar] [CrossRef]
- Hessel, A.L.; Joumaa, V.; Eck, S.; Herzog, W.; Nishikawa, K.C. Optimal length, calcium sensitivity and twitch characteristics of skeletal muscles from mdm mice with a deletion in N2A titin. J. Exp. Biol. 2019, 222, jeb200840. [Google Scholar] [CrossRef] [PubMed]
- Tiffany, H.; Sonkar, K.; Gage, M.J. The insertion sequence of the N2A region of titin exists in an extended structure with helical characteristics. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hamdani, N.; Herwig, M.; Linke, W.A. Tampering with springs: Phosphorylation of titin affecting the mechanical function of cardiomyocytes. Biophys. Rev. 2017, 9, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Fleming, J.R.; Franke, B.; Bogomolovas, J.; Barsukov, I.; Rigden, D.J.; Labeit, S.; Mayans, O. CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex. FEBS Lett. 2016, 590, 3098–3110. [Google Scholar] [CrossRef] [PubMed]
- Marino, M.; Svergun, D.; Kreplak, L.; Konarev, P.; Maco, B.; Labeit, D.; Mayans, O. Poly-Ig tandems from I-band titin share extended domain arrangements irrespective of the distinct features of their modular constituents. J. Muscle Res. Cell Motil. 2005, 26, 355–365. [Google Scholar] [CrossRef]
- Bogomolovas, J.; Fleming, J.R.; Anderson, B.R.; Williams, R.; Lange, S.; Simon, B.; Khan, M.M.; Rudolf, R.; Franke, B.; Belinda, B.; et al. Exploration of pathomechanisms triggered by a single-nucleotide polymorphism in titin’s I-band: The cardiomyopathy-linked mutation T2580I. Open Biol. 2016, 6, 160114. [Google Scholar] [CrossRef]
- Kelly, C.; Pace, N.; Gage, M.; Pfuhl, M. Solution NMR Structure of Titin N2A Region Ig Domain I83 and Its Interaction with Metal Ions. J. Mol. Biol. 2021, 433, 166977. [Google Scholar] [CrossRef]
- Stronczek, C.; Lange, S.; Bullard, B.; Wolniak, S.; Börgeson, E.; Mayans, O.; Fleming, J.R. The N2A region of titin has a unique structural configuration. J. Gen. Physiol. 2021, 153. [Google Scholar] [CrossRef]
- von Castelmur, E.; Marino, M.; Svergun, D.I.; Kreplak, L.; Ucurum-Fotiadis, Z.; Konarev, P.V.; Urzhumtsev, A.; Labeit, D.; Labeit, S.; Mayans, O. A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain. Proc. Natl. Acad. Sci. USA 2008, 105, 1186–1191. [Google Scholar] [CrossRef]
- Sun, X.; Jones, W.T.; Uversky, V.N. Applications of Bioinformatics and Experimental Methods to Intrinsic Disorder-Based Protein-Protein Interactions. In Protein Engineering; Kaumaya, P., Ed.; InTech: Rijeka, Croatia, 2012; Chapter 9; pp. 181–206. [Google Scholar]
- Xue, B.; Dunbrack, R.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 996–1010. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Linke, W.; Ivemeyer, M.; Labeit, S.; Hinssen, H.; Rüegg, J.; Gautel, M. Actin-titin interaction in cardiac myofibrils: Probing a physiological role. Biophys. J. 1997, 73, 905–919. [Google Scholar] [CrossRef]
- Kulke, M.; Fujita-Becker, S.; Rostkova, E.; Neagoe, C.; Labeit, D.; Manstein, D.; Gautel, M.; Linke, W. Interaction Between PEVK-Titin and Actin Filaments. Circ. Res. 2001, 89, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.A.; Carland, C.R.; Guo, Y.; Bernstein, S.I. Getting folded: Chaperone proteins in muscle development, maintenance and disease. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2014, 297, 1637–1649. [Google Scholar] [CrossRef]
- Nagy, A.; Cacciafesta, P.; Grama, L.; Kengyel, A.; Málnási-Csizmadia, A.; Kellermayer, M.S.Z. Differential actin binding along the PEVK domain of skeletal muscle titin. J. Cell Sci. 2004, 117 Pt 24, 5781–5789. [Google Scholar] [CrossRef]
- Tomalka, A.; Röhrle, O.; Han, J.-C.; Pham, T.; Taberner, A.J.; Siebert, T. Extensive eccentric contractions in intact cardiac trabeculae: Revealing compelling differences in contractile behaviour compared to skeletal muscles. Proc. R. Soc. B Boil. Sci. 2019, 286, 20190719. [Google Scholar] [CrossRef]
- Bianco, P.; Nagy, A.; Kengyel, A.; Szatmari, D.; Mártonfalvi, Z.; Huber, T.; Kellermayer, M.S. Interaction Forces between F-Actin and Titin PEVK Domain Measured with Optical Tweezers. Biophys. J. 2007, 93, 2102–2109. [Google Scholar] [CrossRef]
- Linke, W.A.; Rudy, D.E.; Centner, T.; Gautel, M.; Witt, C.; Labeit, S.; Gregorio, C.C. I-Band Titin in Cardiac Muscle Is a Three-Element Molecular Spring and Is Critical for Maintaining Thin Filament Structure. J. Cell Biol. 1999, 146, 631–644. [Google Scholar] [CrossRef]
- Al-Qusairi, L.; Laporte, J. T-tubule biogenesis and triad formation in skeletal muscle and implication in human diseases. Skelet. Muscle 2011, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Barone, V.; Giacomello, E.; Cusimano, V.; Sorrentino, V. The Sarcoplasmic Reticulum: An Organized Patchwork of Specialized Domains. Traffic 2008, 9, 1044–1049. [Google Scholar] [CrossRef] [PubMed]
- Borgia, M.B.; Borgia, A.; Best, R.; Steward, A.; Nettels, D.; Wunderlich, B.; Schuler, B.; Clarke, J. Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins. Nat. Cell Biol. 2011, 474, 662–665. [Google Scholar] [CrossRef]
- Duvall, M.M.; Gifford, J.L.; Amrein, M.; Herzog, W. Altered mechanical properties of titin immunoglobulin domain 27 in the presence of calcium. Eur. Biophys. J. 2012, 42, 301–307. [Google Scholar] [CrossRef]
- Kojic, S.; Radojkovic, D.; Faulkner, G. Muscle ankyrin repeat proteins: Their role in striated muscle function in health and disease. Crit. Rev. Clin. Lab. Sci. 2011, 48, 269–294. [Google Scholar] [CrossRef] [PubMed]
- Loescher, C.M.; Breitkreuz, M.; Li, Y.; Nickel, A.; Unger, A.; Dietl, A.; Schmidt, A.; Mohamed, B.A.; Kötter, S.; Schmitt, J.P.; et al. Regulation of titin-based cardiac stiffness by unfolded domain oxidation (UnDOx). Proc. Natl. Acad. Sci. USA 2020, 117, 24545–24556. [Google Scholar] [CrossRef] [PubMed]
- Rice, M.; Jiang, Y.; Holcomb, J.; Trescott, L.; Spellmon, N.; Sirinupong, N.; Yang, Z. SMYD2 Structure and Function: A Multispecificity Protein Lysine Methyltransferase. J. Cytol. Molecul. Biol. 2014, 1, 7. [Google Scholar]
- Zhang, N.; Xie, X.-J.; Wang, J.-A. Multifunctional protein: Cardiac ankyrin repeat protein. J. Zhejiang Univ. Sci. B 2016, 17, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Evans, S.; Chen, J.; Kuo, H.; Harvey, R.; Chien, K. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development 1997, 124, 793–804. [Google Scholar] [CrossRef]
- Bang, M.-L.; Gu, Y.; Dalton, N.D.; Peterson, K.L.; Chien, K.R.; Chen, J. The Muscle Ankyrin Repeat Proteins CARP, Ankrd2, and DARP Are Not Essential for Normal Cardiac Development and Function at Basal Conditions and in Response to Pressure Overload. PLoS ONE 2014, 9, e93638. [Google Scholar] [CrossRef]
- Lun, A.S.; Chen, J.; Lange, S. Probing Muscle Ankyrin-Repeat Protein (MARP) Structure and Function. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2014, 297, 1615–1629. [Google Scholar] [CrossRef]
- Moulik, M.; Vatta, M.; Witt, S.H.; Arola, A.M.; Murphy, R.T.; McKenna, W.J.; Boriek, A.M.; Oka, K.; Labeit, S.; Bowles, N.E.; et al. ANKRD1, the Gene Encoding Cardiac Ankyrin Repeat Protein, Is a Novel Dilated Cardiomyopathy Gene. J. Am. Coll. Cardiol. 2009, 54, 325–333. [Google Scholar] [CrossRef]
- Lanzicher, T.; Zhou, T.; Saripalli, C.; Keschrumrus, V.; Iii, J.E.S.; Mayans, O.; Sbaizero, O.; Granzier, H. Single-Molecule Force Spectroscopy on the N2A Element of Titin: Effects of Phosphorylation and CARP. Front. Physiol. 2020, 11, 173. [Google Scholar] [CrossRef]
- Cenni, V.; Kojic, S.; Capanni, C.; Faulkner, G.; Lattanzi, G. Ankrd2 in Mechanotransduction and Oxidative Stress Response in Skeletal Muscle: New Cues for the Pathogenesis of Muscular Laminopathies. Oxidative Med. Cell. Longev. 2019, 2019, 1–15. [Google Scholar] [CrossRef]
- Pandurangan, M.; Hwang, I. The role of calpain in skeletal muscle. Anim. Cells Syst. 2012, 16, 431–437. [Google Scholar] [CrossRef]
- Prill, K.; Dawson, J.F. Assembly and Maintenance of Sarcomere Thin Filaments and Associated Diseases. Int. J. Mol. Sci. 2020, 21, 542. [Google Scholar] [CrossRef] [PubMed]
- Huebsch, K.A.; Kudryashova, E.; Wooley, C.M.; Sher, R.B.; Seburn, K.L.; Spencer, M.J.; Cox, G.A. Mdm muscular dystrophy: Interactions with calpain 3 and a novel functional role for titin’s N2A domain. Hum. Mol. Genet. 2005, 14, 2801–2811. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nishikawa, K.; Lindstedt, S.L.; Hessel, A.; Mishra, D. N2A Titin: Signaling Hub and Mechanical Switch in Skeletal Muscle. Int. J. Mol. Sci. 2020, 21, 3974. [Google Scholar] [CrossRef]
- Purintrapiban, J.; Wang, M.C.; Forsberg, N.E. Degradation of sarcomeric and cytoskeletal proteins in cultured skeletal muscle cells. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003, 136, 393–401. [Google Scholar] [CrossRef]
- Willis, M.S.; Schisler, J.C.; Portbury, A.L.; Patterson, C. Build it up-Tear it down: Protein quality control in the cardiac sarcomere. Cardiovasc. Res. 2009, 81, 439–448. [Google Scholar] [CrossRef]
- Lyon, R.C.; Lange, S.; Sheikh, F. Breaking down protein degradation mechanisms in cardiac muscle. Trends Mol. Med. 2013, 19, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, A.; Prado, A.; Antonio, R.C.; Issa, J.P.; Gerlach, R.F. Matrix Metalloproteinases are Involved in Cardiovascular Diseases. Basic Clin. Pharmacol. Toxicol. 2014, 115, 301–314. [Google Scholar] [CrossRef]
- Barta, J.; Tóth, A.; Édes, I.; Vaszily, M.; Papp, J.G.; Varró, A.; Papp, Z. Calpain-1-sensitive myofibrillar proteins of the human myocardium. Mol. Cell. Biochem. 2005, 278, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, R.; Maeda, K.; Hattori, A.; Takahashi, K. Calcium binding to an elastic portion of connectin/titin filaments. J. Muscle Res. Cell Motil. 2001, 22, 149–162. [Google Scholar] [CrossRef]
- Unger, A.; Beckendorf, L.; Böhme, P.; Kley, R.; Von Frieling-Salewsky, M.; Lochmüller, H.; Schröder, R.; Fürst, D.O.; Vorgerd, M.; Linke, W.A. Translocation of molecular chaperones to the titin springs is common in skeletal myopathy patients and affects sarcomere function. Acta Neuropathol. Commun. 2017, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Labeit, S.; Kolmerer, B. Titins: Giant proteins in charge of muscle ultrastructure and elasticity. Science 1995, 270, 293–296. [Google Scholar] [CrossRef]
- Raynaud, F.; Fernandez, E.; Coulis, G.; Aubry, L.; Vignon, X.; Bleimling, N.; Gautel, M.; Benyamin, Y.; Ouali, A. Calpain 1-titin interactions concentrate calpain 1 in the Z-band edges and in the N2-line region within the skeletal myofibril. FEBS J. 2005, 272, 2578–2590. [Google Scholar] [CrossRef] [PubMed]
- Neuhof, C.; Neuhof, H. Calpain system and its involvement in myocardial ischemia and reperfusion injury. World J. Cardiol. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Ojima, K.; Ono, Y.; Doi, N.; Yoshioka, K.; Kawabata, Y.; Labeit, S.; Sorimachi, H. Myogenic Stage, Sarcomere Length, and Protease Activity Modulate Localization of Muscle-specific Calpain. J. Biol. Chem. 2007, 282, 14493–14504. [Google Scholar] [CrossRef] [PubMed]
- Jobin, P.G.; Butler, G.S.; Overall, C.M. New intracellular activities of matrix metalloproteinases shine in the moonlight. Biochim. Biophys. Acta Bioenerg. 2017, 1864, 2043–2055. [Google Scholar] [CrossRef]
- Kandasamy, A.D.; Chow, A.K.; Ali, M.A.M.; Schulz, R. Matrix metalloproteinase-2 and myocardial oxidative stress injury: Beyond the matrix. Cardiovasc. Res. 2009, 85, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Stepanko, A.; Fan, X.; Holt, A.; Schulz, R. Calpain inhibitors exhibit matrix metalloproteinase-2 inhibitory activity. Biochem. Biophys. Res. Commun. 2012, 423, 1–5. [Google Scholar] [CrossRef]
- DeCoux, A.; Lindsey, M.; Villarreal, F.; Garcia, R.A.; Schulz, R. Myocardial matrix metalloproteinase-2: Inside out and upside down. J. Mol. Cell. Cardiol. 2014, 77, 64–72. [Google Scholar] [CrossRef]
- Hughes, B.; Schulz, R. Targeting MMP-2 to treat ischemic heart injury. Basic Res. Cardiol. 2014, 109, 424. [Google Scholar] [CrossRef]
- Ali, M.; Fan, X.; Schulz, R. Cardiac Sarcomeric Proteins: Novel Intracellular Targets of Matrix Metalloproteinase-2 in Heart Disease. Trends Cardiovasc. Med. 2011, 21, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.M.; Cho, W.J.; Hudson, B.; Kassiri, Z.; Granzier, H.; Schulz, R. Titin is a Target of Matrix Metalloproteinase-2Clinical Perspective. Implications in Myocardial Ischemia/Reperfusion Injury. Circulation 2010, 122, 2039–2047. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.B.; Hryshko, L.; Freed, D.; Dhalla, N.S. Activation of proteolytic enzymes and depression of the sarcolemmal Na+/K+-ATPase in ischemia–reperfused heart may be mediated through oxidative stress. Can. J. Physiol. Pharmacol. 2012, 90, 249–260. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adewale, A.O.; Ahn, Y.-H. Titin N2A Domain and Its Interactions at the Sarcomere. Int. J. Mol. Sci. 2021, 22, 7563. https://doi.org/10.3390/ijms22147563
Adewale AO, Ahn Y-H. Titin N2A Domain and Its Interactions at the Sarcomere. International Journal of Molecular Sciences. 2021; 22(14):7563. https://doi.org/10.3390/ijms22147563
Chicago/Turabian StyleAdewale, Adeleye O., and Young-Hoon Ahn. 2021. "Titin N2A Domain and Its Interactions at the Sarcomere" International Journal of Molecular Sciences 22, no. 14: 7563. https://doi.org/10.3390/ijms22147563
APA StyleAdewale, A. O., & Ahn, Y.-H. (2021). Titin N2A Domain and Its Interactions at the Sarcomere. International Journal of Molecular Sciences, 22(14), 7563. https://doi.org/10.3390/ijms22147563





