Haplotype Analysis of BADH1 by Next-Generation Sequencing Reveals Association with Salt Tolerance in Rice during Domestication
Abstract
:1. Introduction
2. Results
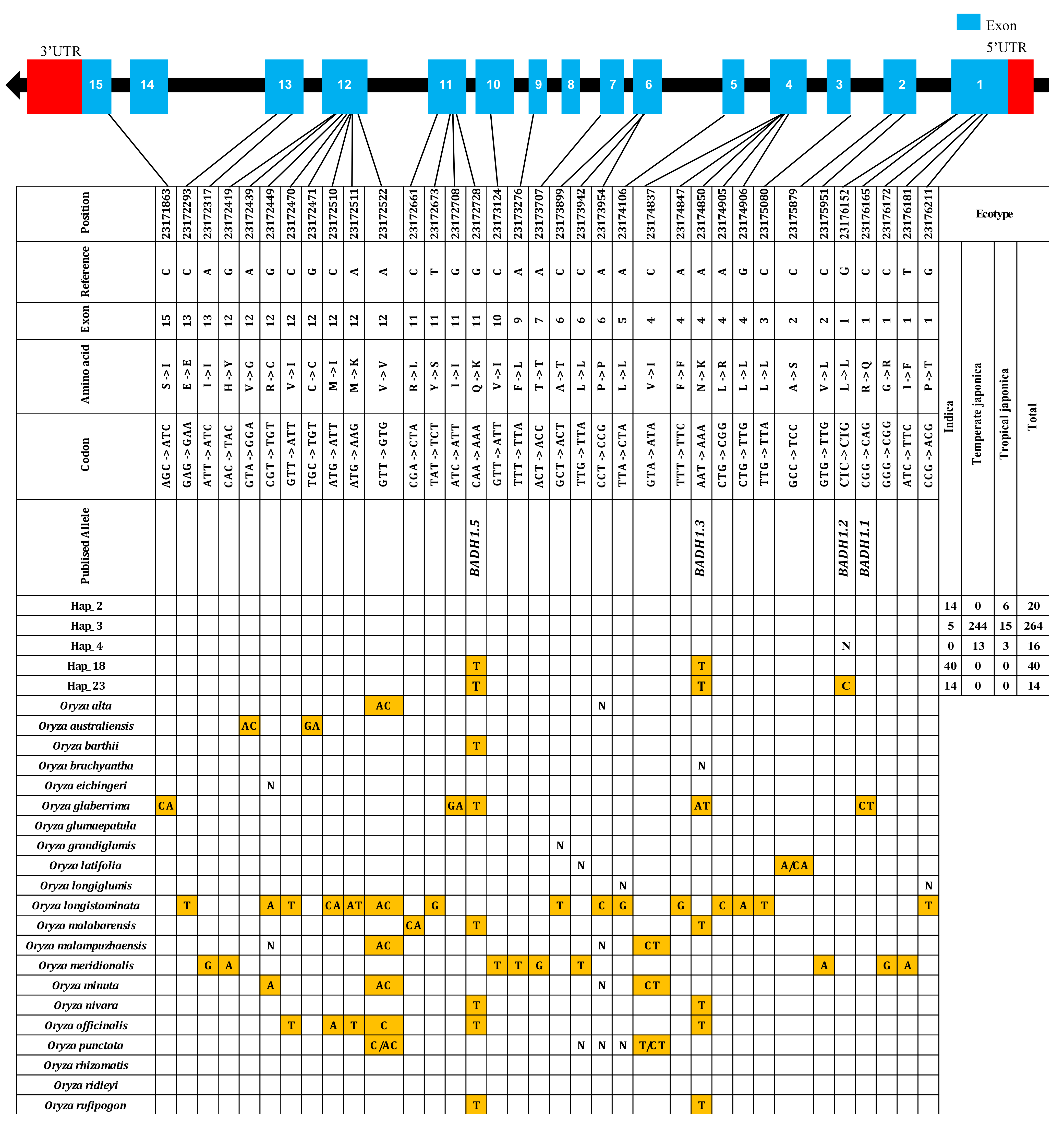
2.1. Discovery of Genetic Variations in BADH1
2.2. Population Structure, Principal Component Analysis (PCA), and Fixation Index (FST Test) of BADH1
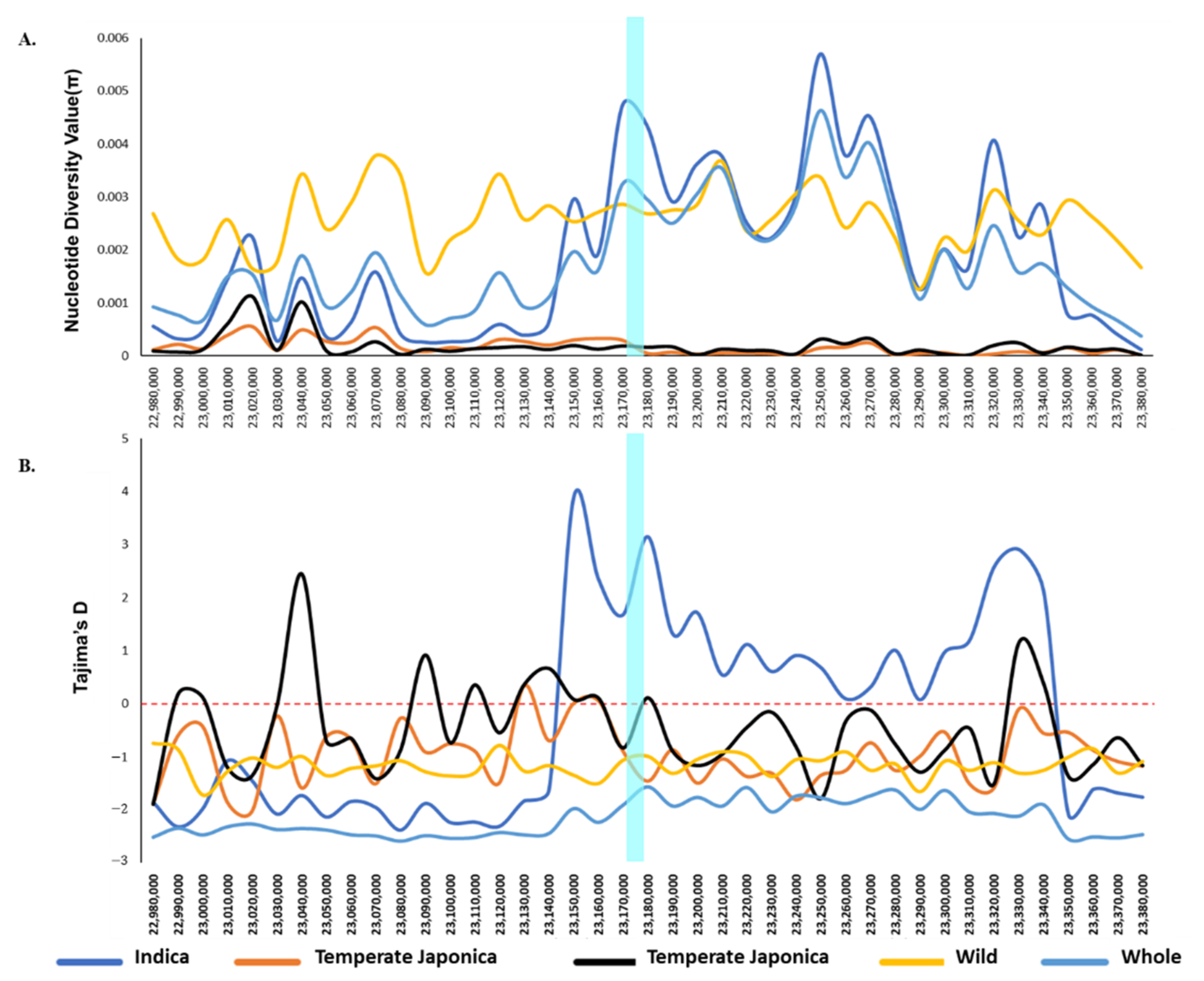
2.3. Genetic Diversity of BADH1
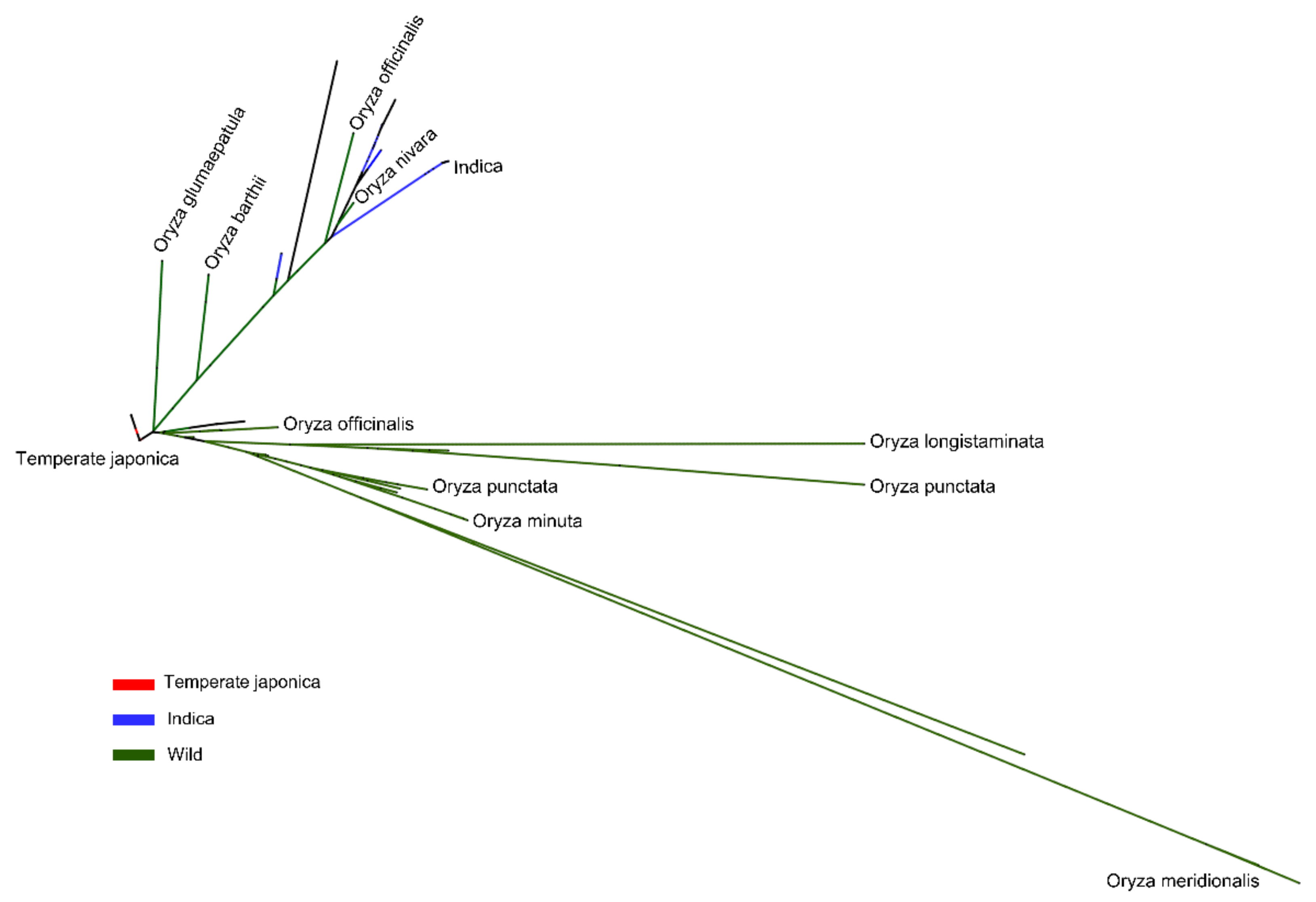
2.4. Phylogenetic Study of BADH1
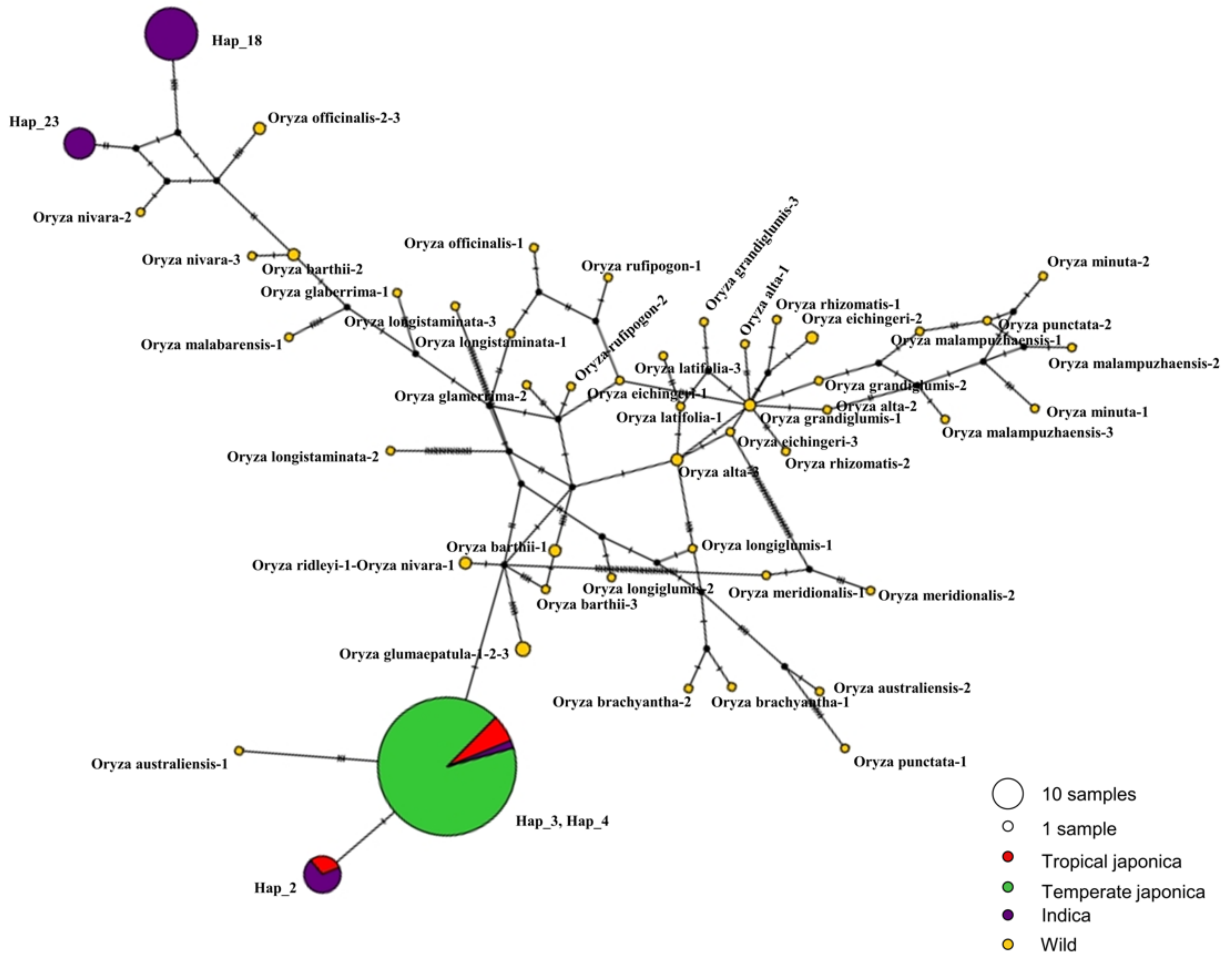
2.5. Haplotype Diversity
2.6. Screening and Evaluation of Salt Tolerance Phenotypes
2.7. Test/Control Ratio of Eight Major Plant Parameters
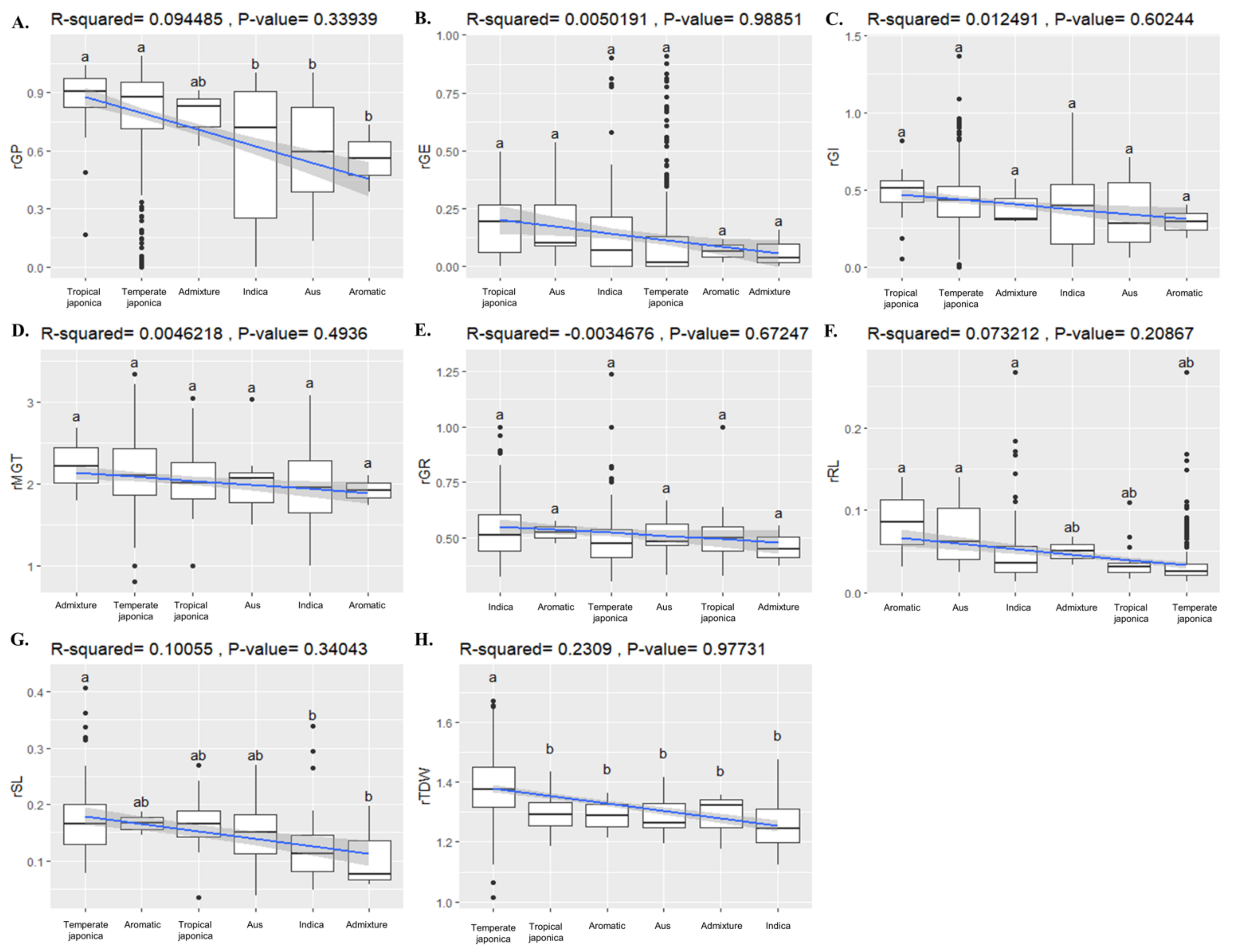
2.8. Association of BADH1 Haplotypes and Plant Parameters under Salt Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. DNA Extraction, Resequencing, and Variant Calling
4.3. Population Structure, Principal Component Analysis (PCA), and Phylogenetic Study
4.4. Nucleotide Diversity, Tajima’s D, and Fixation Index (FST)
4.5. Haplotype Diversity Analysis
4.6. Screening of Salt Tolerance Phenotypes
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ashraf, M.; Foolad, M. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Zhu, J.-K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Moghaieb, R.E.; Saneoka, H.; Fujita, K. Effect of salinity on osmotic adjustment, glycinebetaine accumulation and the betaine aldehyde dehydrogenase gene expression in two halophytic plants, Salicornia europaea and Suaeda maritima. Plant Sci. 2004, 166, 1345–1349. [Google Scholar] [CrossRef]
- McNeil, S.D.; Rhodes, D.; Russell, B.L.; Nuccio, M.L.; Shachar-Hill, Y.; Hanson, A.D. Metabolic modeling identifies key constraints on an engineered glycine betaine synthesis pathway in tobacco. Plant Physiol. 2000, 124, 153–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhodes, D.; Hanson, A. Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu. Rev. Plant Biol. 1993, 44, 357–384. [Google Scholar] [CrossRef]
- Shirasawa, K.; Takabe, T.; Takabe, T.; Kishitani, S. Accumulation of glycinebetaine in rice plants that overexpress choline monooxygenase from spinach and evaluation of their tolerance to abiotic stress. Ann. Bot. 2006, 98, 565–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Singh, P.K.; Singh, R.; Pandit, A.; Mahato, A.K.; Gupta, D.K.; Tyagi, K.; Singh, A.K.; Singh, N.K.; Sharma, T.R. SNP haplotypes of the BADH1 gene and their association with aroma in rice (Oryza sativa L.). Mol. Breed. 2010, 26, 325–338. [Google Scholar] [CrossRef]
- Hasthanasombut, S.; Ntui, V.; Supaibulwatana, K.; Mii, M.; Nakamura, I. Expression of Indica rice OsBADH1 gene under salinity stress in transgenic tobacco. Plant Biotechnol. Rep. 2010, 4, 75–83. [Google Scholar] [CrossRef]
- Nakamura, T.; Yokota, S.; Muramoto, Y.; Tsutsui, K.; Oguri, Y.; Fukui, K.; Takabe, T. Expression of a betaine aldehyde dehydrogenase gene in rice, a glycinebetaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J. 1997, 11, 1115–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, T.L.; Waters, D.L.E.; Henry, R.J. The effect of salt on betaine aldehyde dehydrogenase transcript levels and 2-acetyl-1-pyrroline concentration in fragrant and non-fragrant rice (Oryza sativa). Plant Sci. 2008, 175, 539–546. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43. [Google Scholar] [CrossRef]
- Wang, M.; Yu, Y.; Haberer, G.; Marri, P.R.; Fan, C.; Goicoechea, J.L.; Zuccolo, A.; Song, X.; Kudrna, D.; Ammiraju, J.S. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 2014, 46, 982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-S.; He, Q.; Kim, K.-W.; Yoon, M.-Y.; Ra, W.-H.; Li, F.P.; Tong, W.; Yu, J.; Oo, W.H.; Choi, B. Genome-wide resequencing of KRICE_CORE reveals their potential for future breeding, as well as functional and evolutionary studies in the post-genomic era. BMC Genom. 2016, 17, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapuz, R.R.; Javier, S.; Aquino, J.D.C.; Undan, J. Gene Expression and Sequence Analysis of BADH1 Gene in CLSU Aromatic Rice (Oryza sativa L.) Accessions Subjected to Drought and Saline Condition. J. Nutr. Sci. Vitaminol. 2019, 65, S196–S199. [Google Scholar] [CrossRef]
- Hasthanasombut, S.; Paisarnwipatpong, N.; Triwitayakorn, K.; Kirdmanee, C.; Supaibulwatana, K. Expression of OsBADH1 gene in Indica rice (Oryza sativa L.) in correlation with salt, plasmolysis, temperature and light stresses. Plant Omics 2011, 4, 75–81. [Google Scholar]
- Niu, X.; Zheng, W.; Lu, B.-R.; Ren, G.; Huang, W.; Wang, S.; Liu, J.; Tang, Z.; Luo, D.; Wang, Y. An unusual posttranscriptional processing in two betaine aldehyde dehydrogenase loci of cereal crops directed by short, direct repeats in response to stress conditions. Plant Physiol. 2007, 143, 1929–1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cubry, P.; Tranchant-Dubreuil, C.; Thuillet, A.-C.; Monat, C.; Ndjiondjop, M.-N.; Labadie, K.; Cruaud, C.; Engelen, S.; Scarcelli, N.; Rhoné, B. The rise and fall of African rice cultivation revealed by analysis of 246 new genomes. Curr. Biol. 2018, 28, 2274–2282.e2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.Y.; Zaidem, M.; Gutaker, R.; Dorph, K.; Singh, R.K.; Purugganan, M.D. The complex geography of domestication of the African rice Oryza glaberrima. PLoS Genet. 2019, 15, e1007414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Q.; Yu, J.; Kim, T.-S.; Cho, Y.-H.; Lee, Y.-S.; Park, Y.-J. Resequencing reveals different domestication rate for BADH1 and BADH2 in rice (Oryza sativa). PLoS ONE 2015, 10, e0134801. [Google Scholar]
- Wambugu, P.W.; Ndjiondjop, M.-N.; Henry, R. Genetics and Genomics of African Rice (Oryza glaberrima Steud) Domestication. Rice 2021, 14, 6. [Google Scholar] [CrossRef]
- Hilbert, L.; Neves, E.G.; Pugliese, F.; Whitney, B.S.; Shock, M.; Veasey, E.; Zimpel, C.A.; Iriarte, J. Evidence for mid-Holocene rice domestication in the Americas. Nat. Ecol. Evol. 2017, 1, 1693–1698. [Google Scholar] [CrossRef] [PubMed]
- Cubry, P.; Vigouroux, Y. Population genomics of crop domestication: Current state and perspectives. Popul. Genom. 2018, 685–707. [Google Scholar]
- Doebley, J.F.; Gaut, B.S.; Smith, B. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; Sun, C.J.T.C.J. What happened during domestication of wild to cultivated rice. Crop. J. 2021, 9, 564–576. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, X.; Zhao, Y.; Khan, N.U.; Zhao, Z.; Zhang, Y.; Wen, X.; Tang, F.; Wang, F.; Li, Z. Genetic basis and identification of candidate genes for salt tolerance in rice by GWAS. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhou, Y.; Fan, J.; Fu, Y.; Shen, L.; Yao, Y.; Li, R.; Fu, S.; Duan, R.; Hu, X.; et al. SpBADH of the halophyte Sesuvium portulacastrum strongly confers drought tolerance through ROS scavenging in transgenic Arabidopsis. Plant Physiol. Biochem. 2015, 96, 377–387. [Google Scholar] [CrossRef]
- Kim, K.-W.; Chung, H.-K.; Cho, G.-T.; Ma, K.-H.; Chandrabalan, D.; Gwag, J.-G.; Kim, T.-S.; Cho, E.-G.; Park, Y.-J. PowerCore: A program applying the advanced M strategy with a heuristic search for establishing core sets. Bioinformation 2007, 23, 2155–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, J.J. Isolation ofplant DNA from fresh tissue. Focus 1990, 12, 39–40. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Picard Toolkit. Broad Institute, GitHub Repository. 2019. Available online: http://Broadinstitute.Github.Io/Picard (accessed on 30 January 2021).
- Van der Auwera, G.A.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2020. [Google Scholar]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef] [Green Version]
- Francis, R.M. Pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [Green Version]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]


| Ecotype | SNP | Insertion | Deletion | Structure Variation | Total Accessions |
|---|---|---|---|---|---|
| Te_Japonica | 0 | 1 | 0 | 0 | 279 |
| Tr_Japonica | 1 | 1 | 0 | 0 | 26 |
| Indica | 23 | 2 | 1 | 0 | 102 |
| Aus | 23 | 2 | 0 | 0 | 9 |
| Aromatic | 0 | 1 | 0 | 0 | 2 |
| Admixture | 15 | 2 | 0 | 0 | 3 |
| Wild | 105 | 38 | 36 | 2 | 54 |
| Trait | Salinity Level (mM) | Mean ± SD | Range | Median | IQR |
|---|---|---|---|---|---|
| GP | 200 | 21.57 ± 7.94 | 0–30 | 24.33 | 29.517 |
| 0 | 28.47 ± 2.72 | 0–30 | 29 | 29.585 | |
| GE | 200 | 3.17 ± 5.37 | 0–27 | 0.33 | 28.0218 |
| 0 | 22.93 ± 10.19 | 0–30 | 28 | 48.131 | |
| GI | 200 | 3.55 ± 1.84 | 0–9.24 | 3.59 | 7.5618 |
| 0 | 8.49 ± 2.28 | 0–14.83 | 9.27 | 13.21 | |
| MGT | 200 | 7.31 ± 1.57 | 3.34–10 | 7.24 | 6.7405 |
| 0 | 3.87 ± 1.83 | 2–10 | 3.14 | 6.1583 | |
| GR | 200 | 0.14 ± 0.03 | 0.1–0.3 | 0.14 | 0.2735 |
| 0 | 0.29 ± 0.07 | 0.1–0.5 | 0.32 | 0.4516 | |
| SL | 200 | 0.36 ± 0.27 | 0.19–2.34 | 0.25 | 2.1924 |
| 0 | 10.13 ± 2.93 | 0.98–19.43 | 9.89 | 15.911 | |
| RL | 200 | 0.87 ± 0.51 | 0.2–8.11 | 0.81 | 6.7159 |
| 0 | 5.92 ± 1.31 | 1.86–11.01 | 5.67 | 7.854 | |
| TDW | 200 | 0.68 ± 0.08 | 0.36–1.05 | 0.68 | 0.9325 |
| 0 | 0.51 ± 0.07 | 0.26–0.78 | 0.5 | 0.695 |
| Parameters (Phenotypes) | Formula |
|---|---|
| Germination percentage (GP) | (number of germinated seeds/total number of seeds) × 100 |
| Germination energy (GE) | (number of germinated seeds on day 4/total number of seeds) × 100 |
| Germination index (GI) | Σ (Nd/d) |
| Mean germination time (MGT) | Σ (d × n)/Σ nd nd: the number of germinated seeds on each day d: number of days after the start of the experiment |
| Germination rate (GR) | Σ N/Σ (N × d) N: the number of seeds that germinated on day d d: the days during the experiment |
| Root length (RL) | Length of root after 10 days |
| Shoot length (SL) | Length of shoot after 10 days |
| Total dry weight TDW) | Total dry weight of shoot and root (80 °C for 24 h) |
| Relative GP | GP in condition/GP in control |
| Relative GE | GE in condition/GE in control |
| Relative GI | GI in condition/GI in control |
| Relative MGT | MGT in condition/MGT in control |
| Relative GR | GR in condition/GR in control |
| Relative RL | RL in condition/RL in control |
| Relative SL | SL in condition/SL in control |
| Relative TDW | TDW in condition/TDW in control |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Min, M.-H.; Maung, T.Z.; Cao, Y.; Phitaktansakul, R.; Lee, G.-S.; Chu, S.-H.; Kim, K.-W.; Park, Y.-J. Haplotype Analysis of BADH1 by Next-Generation Sequencing Reveals Association with Salt Tolerance in Rice during Domestication. Int. J. Mol. Sci. 2021, 22, 7578. https://doi.org/10.3390/ijms22147578
Min M-H, Maung TZ, Cao Y, Phitaktansakul R, Lee G-S, Chu S-H, Kim K-W, Park Y-J. Haplotype Analysis of BADH1 by Next-Generation Sequencing Reveals Association with Salt Tolerance in Rice during Domestication. International Journal of Molecular Sciences. 2021; 22(14):7578. https://doi.org/10.3390/ijms22147578
Chicago/Turabian StyleMin, Myeong-Hyeon, Thant Zin Maung, Yuan Cao, Rungnapa Phitaktansakul, Gang-Seob Lee, Sang-Ho Chu, Kyu-Won Kim, and Yong-Jin Park. 2021. "Haplotype Analysis of BADH1 by Next-Generation Sequencing Reveals Association with Salt Tolerance in Rice during Domestication" International Journal of Molecular Sciences 22, no. 14: 7578. https://doi.org/10.3390/ijms22147578
APA StyleMin, M.-H., Maung, T. Z., Cao, Y., Phitaktansakul, R., Lee, G.-S., Chu, S.-H., Kim, K.-W., & Park, Y.-J. (2021). Haplotype Analysis of BADH1 by Next-Generation Sequencing Reveals Association with Salt Tolerance in Rice during Domestication. International Journal of Molecular Sciences, 22(14), 7578. https://doi.org/10.3390/ijms22147578






