Microbial Synthesis and Evaluation of Fungistatic Activity of 3-Butyl-3-hydroxyphthalide, the Mammalian Metabolite of 3-n-Butylidenephthalide
Abstract
:1. Introduction
2. Results and Discussion
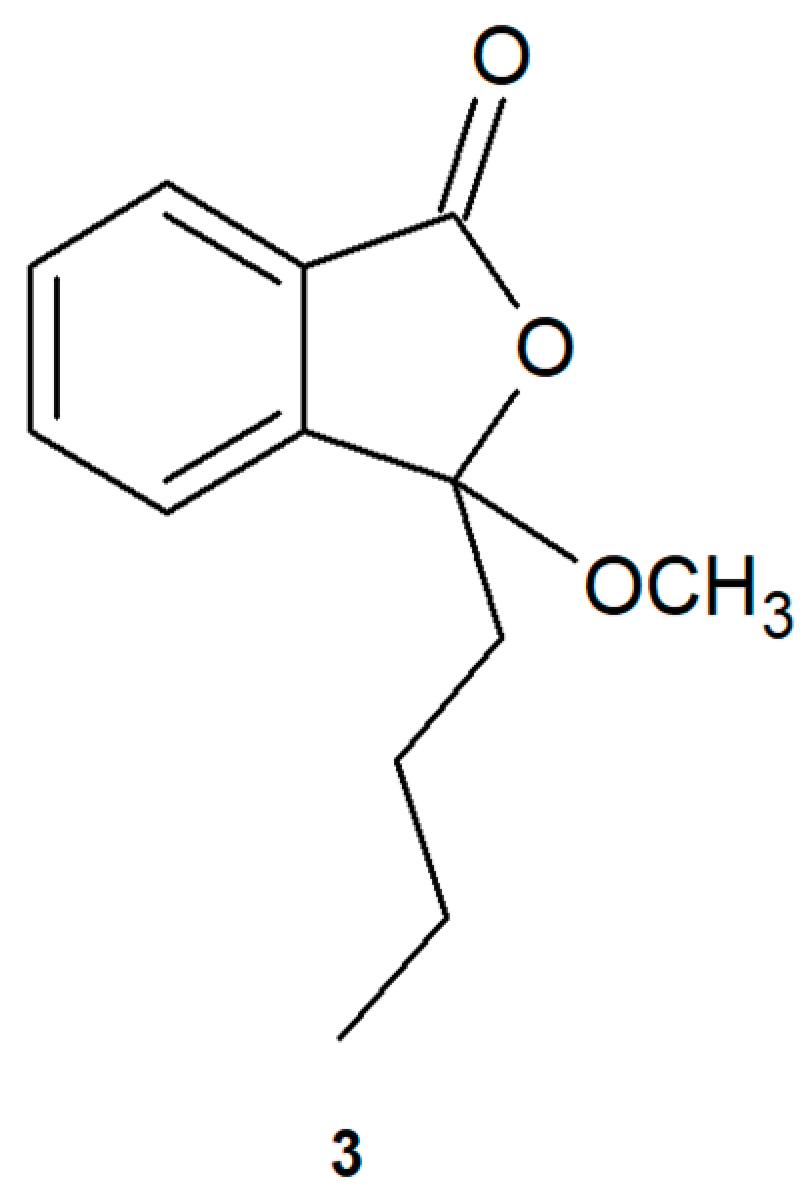
2.1. Biotransformations of 3-n-Butylidenephthalide (1)
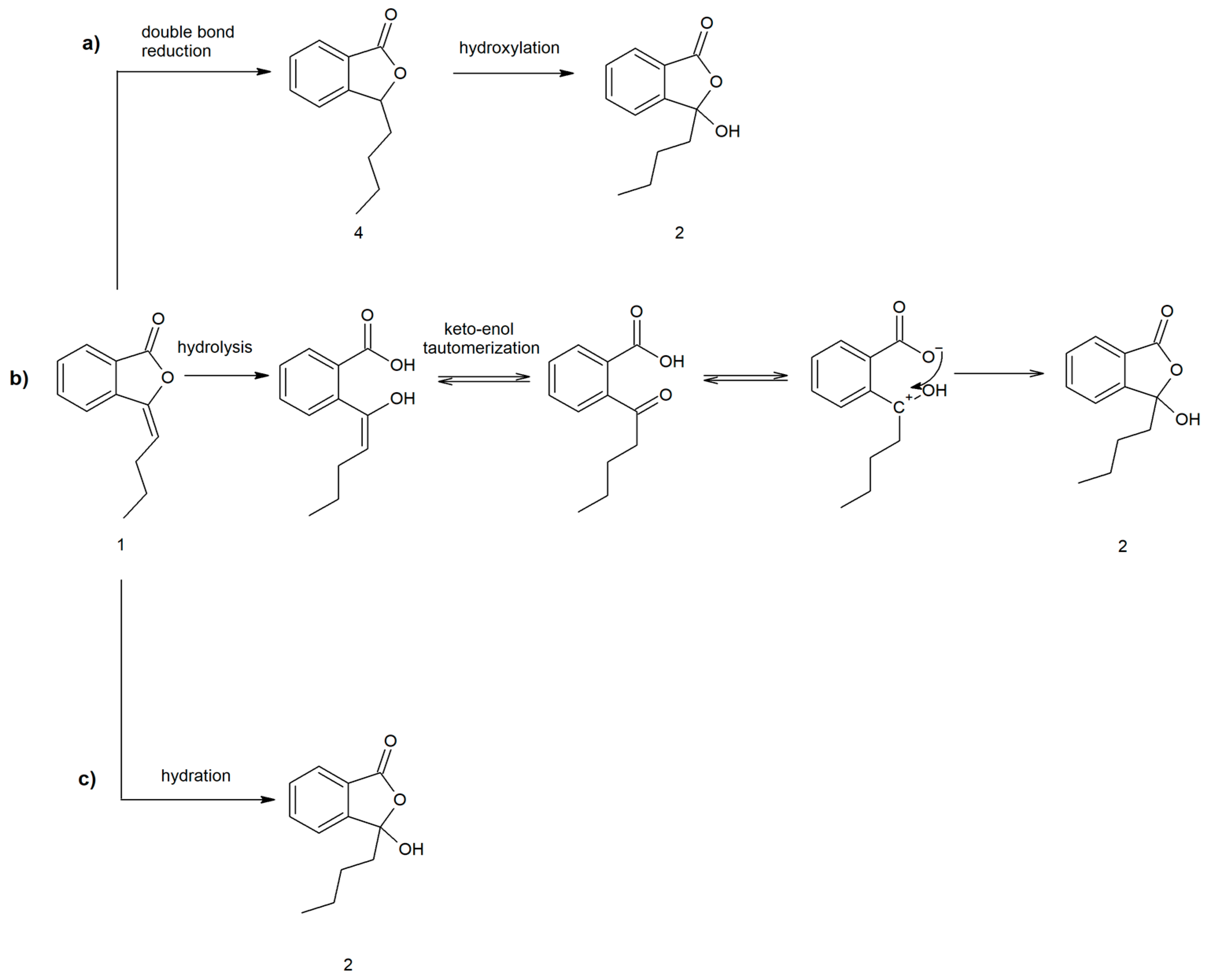
2.2. Proposed Pathways for 3-Butyl-3-hydroxyphthalide (2) Formation
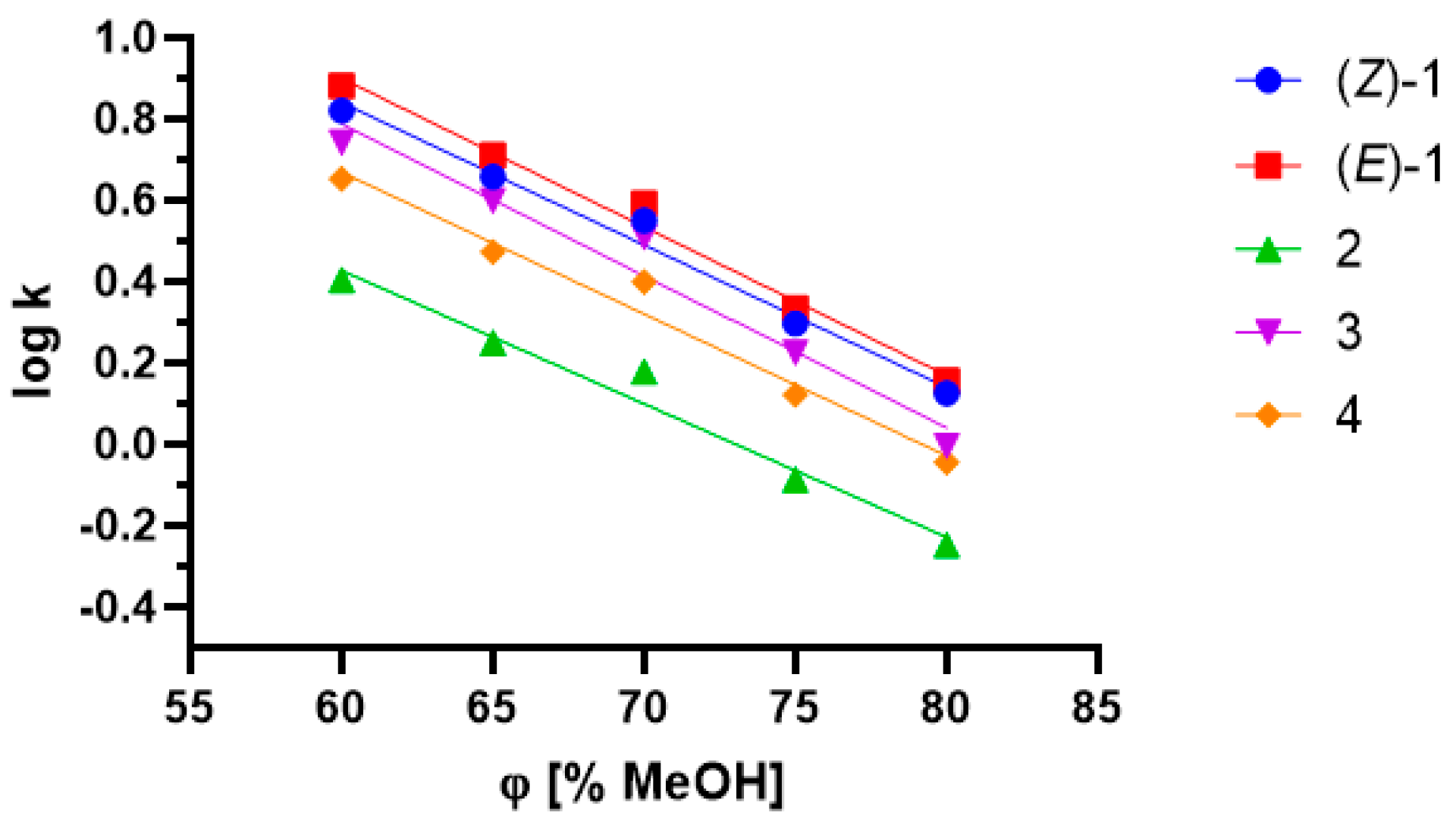
2.3. Influence of Lipophilicity and Fungistatic Activity
3. Materials and Methods
3.1. Compounds
3.2. Microorganisms
3.3. Biotransformations
3.4. Analysis
3.5. Lipophilicity
3.6. Fungistatic Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, H.-C.; Tsai, Y.-J.; Lin, L.-Y.; Wu, C.-S.; Tai, S.-P.; Chen, Y.-C.; Chiang, H.-M. Volatile Compounds from Roots, Stems and Leaves of Angelica Acutiloba Growing in Taiwan. Nat. Prod. Commun. 2014, 9, 583–586. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-R.; Yu, Y.; Zulfajri, M.; Lin, P.-C.; Wang, C.C. Phthalide Derivatives from Angelica Sinensis Decrease Hemoglobin Oxygen Affinity: A New Allosteric-Modulating Mechanism and Potential Use as 2,3-BPG Functional Substitutes. Sci. Rep. 2017, 7, 5504. [Google Scholar] [CrossRef]
- Baananou, S.; Piras, A.; Marongiu, B.; Dessì, M.A.; Falconieri, D.; Porcedda, S.; Rosa, A.; Boughattas, N.A. Antiulcerogenic Activity of Apium Graveolens Seeds Oils Isolated by Supercritical CO2. Afr. J. Pharm. Pharmacol. 2012, 6, 752–762. [Google Scholar] [CrossRef]
- Chae, S.-H.; Kim, S.-I.; Yeon, S.-H.; Lee, S.-W.; Ahn, Y.-J. Adulticidal Activity of Phthalides Identified in Cnidium Officinale Rhizome to B- and Q-Biotypes of Bemisia Tabaci. J. Agric. Food Chem. 2011, 59, 8193–8198. [Google Scholar] [CrossRef] [PubMed]
- Spréa, R.M.; Fernandes, Â.; Finimundy, T.C.; Pereira, C.; Alves, M.J.; Calhelha, R.C.; Canan, C.; Barros, L.; Amaral, J.S.; Ferreira, I.C.F.R. Lovage (Levisticum Officinale W.D.J. Koch) Roots: A Source of Bioactive Compounds towards a Circular Economy. Resources 2020, 9, 81. [Google Scholar] [CrossRef]
- León, A.; Toscano, R.A.; Tortoriello, J.; Delgado, G. Phthalides and Other Constituents from Ligusticum Porteri; Sedative and Spasmolytic Activities of Some Natural Products and Derivatives. Nat. Prod. Res. 2011, 25, 1234–1242. [Google Scholar] [CrossRef]
- Jia, J.; Wei, C.; Liang, J.; Zhou, A.; Zuo, X.; Song, H.; Wu, L.; Chen, X.; Chen, S.; Zhang, J.; et al. The Effects of DL-3-n-Butylphthalide in Patients with Vascular Cognitive Impairment without Dementia Caused by Subcortical Ischemic Small Vessel Disease: A Multicentre, Randomized, Double-Blind, Placebo-Controlled Trial. Alzheimer’s Dement. 2016, 12, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Yeh, J.-C.; Cindrova-Davies, T.; Belleri, M.; Morbidelli, L.; Miller, N.; Cho, C.-W.C.; Chan, K.; Wang, Y.-T.; Luo, G.-A.; Ziche, M.; et al. The Natural Compound N-Butylidenephthalide Derived from the Volatile Oil of Radix Angelica Sinensis Inhibits Angiogenesis in Vitro and in Vivo. Angiogenesis 2011, 14, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.-M.; Zhang, J.-J.; Li, S.; Chen, S.; Le, W.-D. N-Butylidenephthalide Treatment Prolongs Life Span and Attenuates Motor Neuron Loss in SOD1 G93A Mouse Model of Amyotrophic Lateral Sclerosis. CNS Neurosci. Ther. 2017, 23, 375–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, K.N.; Kim, K.-P.; Cho, K.-H.; Jung, W.-S.; Park, J.-M.; Cho, S.-Y.; Park, S.-K.; Park, T.-H.; Kim, Y.-S.; Lee, E.H. Prevention of Inflammation-Mediated Neurotoxicity by Butylidenephthalide and Its Role in Microglial Activation: BP Inhibits Microglial Inflammatory Response. Cell Biochem. Funct. 2013, 31, 707–712. [Google Scholar] [CrossRef]
- Brindis, F.; Rodríguez, R.; Bye, R.; González-Andrade, M.; Mata, R. (Z)-3-Butylidenephthalide from Ligusticum porteri, an α-Glucosidase Inhibitor. J. Nat. Prod. 2011, 74, 314–320. [Google Scholar] [CrossRef]
- Sim, Y.; Shin, S. Combinatorial Anti-Trichophyton Effects of Ligusticum Chuanxiong Essential Oil Components with Antibiotics. Arch. Pharm. Res. 2008, 31, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Liu, W.; Huang, X.; Hao, L.; Li, Y.; Sun, S. Antifungal Activity and Potential Mechanism of N-Butylphthalide Alone and in Combination With Fluconazole Against Candida Albicans. Front. Microbiol. 2019, 10, 1461. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, F.L.L.D.; Santos, N.C.; de Paula Cavalcante, C.S.; Andreu, D.; Baptista, G.R.; Gonçalves, S. Antibiofilm Activity on Candida Albicans and Mechanism of Action on Biomembrane Models of the Antimicrobial Peptide Ctn [15–34]. Int. J. Mol. Sci. 2020, 21, 8339. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Verma, R.; Murari, A.; Agrawal, A. Oral Candidiasis: An Overview. J. Oral Maxillofac. Pathol. 2014, 18, 81. [Google Scholar] [CrossRef]
- Spettel, K.; Barousch, W.; Makristathis, A.; Zeller, I.; Nehr, M.; Selitsch, B.; Lackner, M.; Rath, P.-M.; Steinmann, J.; Willinger, B. Analysis of Antifungal Resistance Genes in Candida Albicans and Candida Glabrata Using next Generation Sequencing. PLoS ONE 2019, 14, e0210397. [Google Scholar] [CrossRef]
- Yassin, M.T.; Mostafa, A.A.; Al-Askar, A.A.; Bdeer, R. In Vitro Antifungal Resistance Profile of Candida Strains Isolated from Saudi Women Suffering from Vulvovaginitis. Eur. J. Med. Res. 2020, 25, 1. [Google Scholar] [CrossRef] [PubMed]
- Bühler, T.; Medinger, M.; Bouitbir, J.; Krähenbühl, S.; Leuppi-Taegtmeyer, A. Hepatotoxicity Due to Azole Antimycotic Agents in a HLA B * 35:02-Positive Patient. Front. Pharmacol. 2019, 10, 645. [Google Scholar] [CrossRef]
- Van Daele, R.; Spriet, I.; Wauters, J.; Maertens, J.; Mercier, T.; Van Hecke, S.; Brüggemann, R. Antifungal Drugs: What Brings the Future? Med Mycol. 2019, 57, S328–S343. [Google Scholar] [CrossRef] [Green Version]
- Kebamo, S.; Tesema, S. The Role of Biotransformation in Drug Discovery and Development. J. Drug Metab. Toxicol. 2015, 6. [Google Scholar] [CrossRef]
- Duan, F.; Xu, W.; Liu, J.; Jia, Z.; Chen, K.; Chen, Y.; Wang, M.; Ma, K.; Dong, J.; Chen, L.; et al. Preparing the Key Metabolite of Z -Ligustilide in Vivo by a Specific Electrochemical Reaction. J. Sep. Sci. 2018, 41, 2799–2807. [Google Scholar] [CrossRef] [PubMed]
- Shanu-Wilson, J.; Evans, L.; Wrigley, S.; Steele, J.; Atherton, J.; Boer, J. Biotransformation: Impact and Application of Metabolism in Drug Discovery. ACS Med. Chem. Lett. 2020, 11, 2087–2107. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.L.; Kelly, D.E. Microbial Cytochromes P450: Biodiversity and Biotechnology. Where Do Cytochromes P450 Come from, What Do They Do and What Can They Do for Us? Philos. Trans. R. Soc. B: Biol. Sci. 2013, 368, 20120476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, W.; Feng, J.; Chen, X.; Bao, Y.-J.; Wang, Y.; Wu, Q.; Ma, Y.; Zhu, D. Distinct Regioselectivity of Fungal P450 Enzymes for Steroidal Hydroxylation. Appl. Environ. Microbiol. 2019, 85, e01182-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hüttel, W.; Hoffmeister, D. Fungal Biotransformations in Pharmaceutical Sciences. In Industrial Applications; Hofrichter, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 293–317. [Google Scholar]
- Diao, X.; Deng, P.; Xie, C.; Li, X.; Zhong, D.; Zhang, Y.; Chen, X. Metabolism and Pharmacokinetics of 3-n-Butylphthalide (NBP) in Humans: The Role of Cytochrome P450s and Alcohol Dehydrogenase in Biotransformation. Drug Metab. Dispos. 2013, 41, 430–444. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.; Ko, N.L.; Ma, B.; Tam, Y.K.; Lin, G. Metabolic Conversion from Co-Existing Ingredient Leading to Significant Systemic Exposure of Z-Butylidenephthalide, a Minor Ingredient in Chuanxiong Rhizoma in Rats. Curr. Drug Metab. 2012, 13, 524–534. [Google Scholar] [CrossRef]
- Nycz-Empel, A.; Bober, K.; Wyszomirski, M.; Kisiel, E.; Zięba, A. The Application of CA and PCA to the Evaluation of Lipophilicity and Physicochemical Properties of Tetracyclic Diazaphenothiazine Derivatives. J. Anal. Methods Chem. 2019, 2019, 8131235. [Google Scholar] [CrossRef]
- Ciura, K.; Fedorowicz, J.; Andrić, F.; Greber, K.E.; Gurgielewicz, A.; Sawicki, W.; Sączewski, J. Lipophilicity Determination of Quaternary (Fluoro)Quinolones by Chromatographic and Theoretical Approaches. Int. J. Mol. Sci. 2019, 20, 5288. [Google Scholar] [CrossRef] [Green Version]
- Klose, M.; Theiner, S.; Varbanov, H.; Hoefer, D.; Pichler, V.; Galanski, M.; Meier-Menches, S.; Keppler, B. Development and Validation of Liquid Chromatography-Based Methods to Assess the Lipophilicity of Cytotoxic Platinum (IV) Complexes. Inorganics 2018, 6, 130. [Google Scholar] [CrossRef] [Green Version]
- Chmiel, T.; Mieszkowska, A.; Kempińska-Kupczyk, D.; Kot-Wasik, A.; Namieśnik, J.; Mazerska, Z. The Impact of Lipophilicity on Environmental Processes, Drug Delivery and Bioavailability of Food Components. Microchem. J. 2019, 146, 393–406. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and Drug-like Compounds: The Rule-of-Five Revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Fernandes, T.R.; Segorbe, D.; Prusky, D.; Di Pietro, A. How Alkalinization Drives Fungal Pathogenicity. PLoS Pathog. 2017, 13, e1006621. [Google Scholar] [CrossRef]
- Zöllner, A.; Buchheit, D.; Meyer, M.R.; Maurer, H.H.; Peters, F.T.; Bureik, M. Production of Human Phase 1 and 2 Metabolites by Whole-Cell Biotransformation with Recombinant Microbes. Bioanalysis 2010, 2, 1277–1290. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-I.; McCarty, R.M.; Liu, H. The Enzymology of Organic Transformations: A Survey of Name Reactions in Biological Systems. Angew. Chem. Int. Ed. 2017, 56, 3446–3489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, R.; Ko, N.L.; Li, S.-L.; Tam, Y.K.; Lin, G. Pharmacokinetics and Metabolism of Ligustilide, a Major Bioactive Component in Rhizoma Chuanxiong, in the Rat. Drug Metab. Dispos. 2008, 36, 400–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engleder, M.; Pichler, H. On the Current Role of Hydratases in Biocatalysis. Appl. Microbiol. Biotechnol. 2018, 102, 5841–5858. [Google Scholar] [CrossRef] [Green Version]
- Li, C.-Y.; Qi, L.-W.; Li, P. Correlative Analysis of Metabolite Profiling of Danggui Buxue Tang in Rat Biological Fluids by Rapid Resolution LC–TOF/MS. J. Pharm. Biomed. Anal. 2011, 55, 146–160. [Google Scholar] [CrossRef]
- Zheng, B.; West, L.M. Estimating The Lipophilicity Of Natural Products Using A Polymeric Reversed Phase Hplc Method. J. Liq. Chromatogr. Relat. Technol. 2009, 33, 118–132. [Google Scholar] [CrossRef]
- Sima, I.A.; Kot-Wasik, A.; Wasik, A.; Namieśnik, J.; Sârbu, C. Assessment of Lipophilicity Indices Derived from Retention Behavior of Antioxidant Compounds in RP-HPLC. Molecules 2017, 22, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamaian, R.; Moţ, A.; Silaghi-Dumitrescu, R.; Ionuţ, I.; Stana, A.; Oniga, O.; Nastasă, C.; Benedec, D.; Tiperciuc, B. Study of the Relationships between the Structure, Lipophilicity and Biological Activity of Some Thiazolyl-Carbonyl-Thiosemicarbazides and Thiazolyl-Azoles. Molecules 2015, 20, 22188–22201. [Google Scholar] [CrossRef]
- Ghazal, H.S.; Dyas, A.M.; Ford, J.L.; Hutcheon, G.A. The Impact of Food Components on the Intrinsic Dissolution Rate of Ketoconazole. Drug Dev. Ind. Pharm. 2015, 41, 1647–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsume, Y.; Mudie, D.M.; Langguth, P.; Amidon, G.E.; Amidon, G.L. The Biopharmaceutics Classification System: Subclasses for in Vivo Predictive Dissolution (IPD) Methodology and IVIVC. Eur. J. Pharm. Sci. 2014, 57, 152–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrêa, J.C.R.; Salgado, H.R.N. Review of Fluconazole Properties and Analytical Methods for Its Determination. Crit. Rev. Anal. Chem. 2011, 41, 124–132. [Google Scholar] [CrossRef]
- Bennion, B.J.; Be, N.A.; McNerney, M.W.; Lao, V.; Carlson, E.M.; Valdez, C.A.; Malfatti, M.A.; Enright, H.A.; Nguyen, T.H.; Lightstone, F.C.; et al. Predicting a Drug’s Membrane Permeability: A Computational Model Validated With in Vitro Permeability Assay Data. J. Phys. Chem. B 2017, 121, 5228–5237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momin, R.A.; Nair, M.G. Mosquitocidal, Nematicidal, and Antifungal Compounds from Apium Graveolens L. Seeds. J. Agric. Food Chem. 2001, 49, 142–145. [Google Scholar] [CrossRef]
- Pannek, J.; Gach, J.; Boratyński, F.; Olejniczak, T. Antimicrobial Activity of Extracts and Phthalides Occurring in Apiaceae Plants: Antimicrobial Activity of Phthalides. Phytother. Res. 2018, 32, 1459–1487. [Google Scholar] [CrossRef]
- Fan, L.; Luo, B.; Luo, Z.; Zhang, L.; Fan, J.; Xue, W.; Tang, L.; Li, Y. Synthesis and Antifungal Activities of 3-Substituted Phthalide Derivatives. Z. Für Nat. B 2019, 74, 811–818. [Google Scholar] [CrossRef]
- Xiao, B.; Yin, J.; Park, M.; Liu, J.; Li, J.L.; Kim, E.L.; Hong, J.; Chung, H.Y.; Jung, J.H. Design and Synthesis of Marine Fungal Phthalide Derivatives as PPAR-γ Agonists. Bioorganic Med. Chem. 2012, 20, 4954–4961. [Google Scholar] [CrossRef]

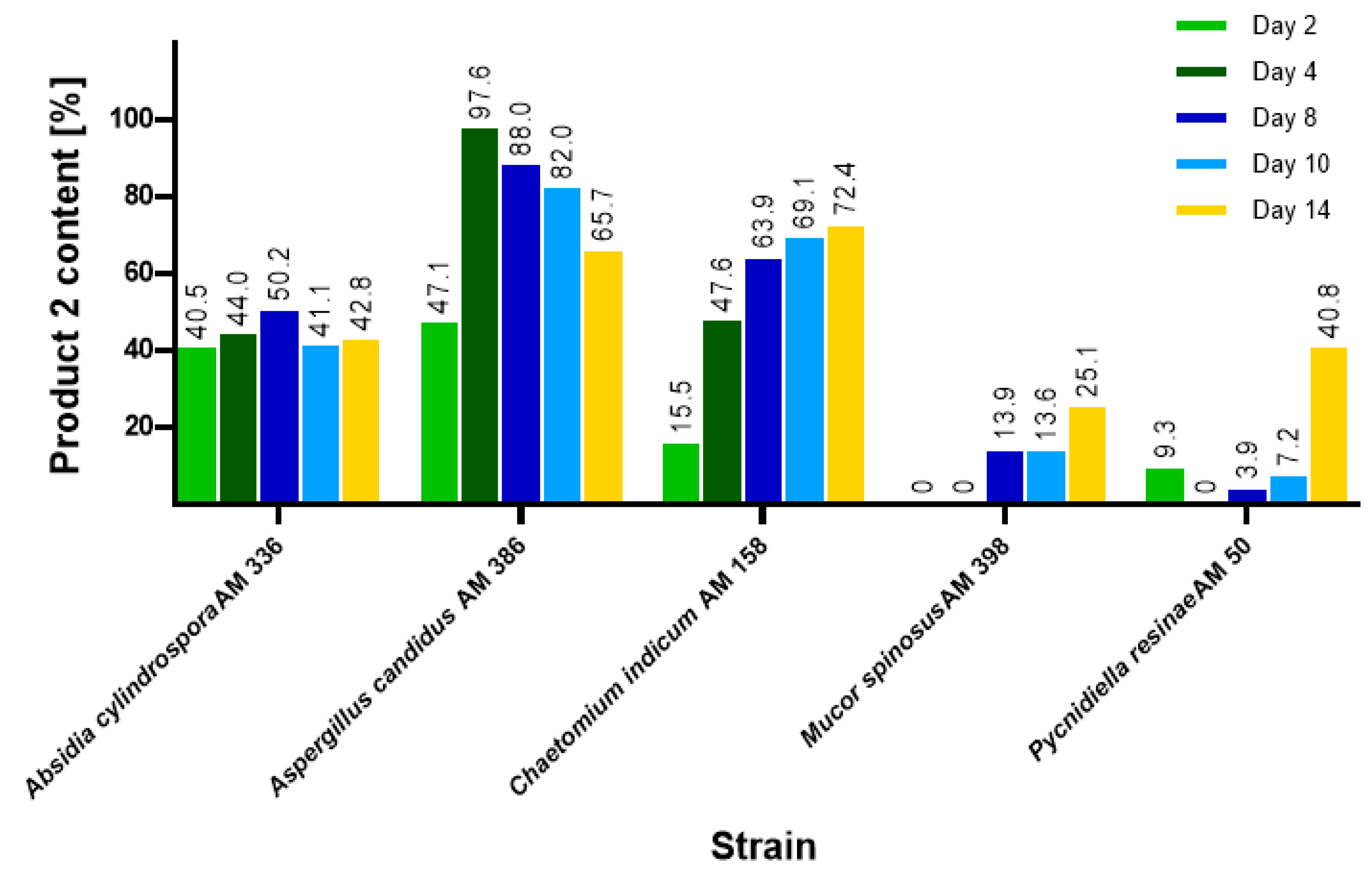



| Strain | Time of the Process [Days] | 3-Butyl-3-hydroxyphthalide (2) | |
|---|---|---|---|
| (Based on HPLC) [%] | (Isolation Yield) [%] | ||
| Chaetomium indicum AM 158 | 14 | 69.8 | 36.6 |
| Absidia cylindospora AM 336 | 14 | 57.0 | 29.2 |
| Aspergillus candidus AM 386 | 8 | 82.2 | 45.4 |
| Compound | Log kw ** | Standard Error for the Slope | Standard Error for the Intercept | Coefficient of Determination | φ0 ** | Log P (Calculated) |
|---|---|---|---|---|---|---|
| (Z)-1 | 2.9422 | 0.0025 | 0.1747 | 0.9851 | 84.063 | 3.38 |
| *(E)-1 * | 3.094 | 0.0024 | 0.1673 | 0.9875 | 84.536 | 3.38 |
| 2 | 2.397 | 0.0032 | 0.2256 | 0.9721 | 73.079 | 2.09 |
| 3 | 3.029 | 0.0042 | 0.2958 | 0.9633 | 81.200 | 2.86 |
| 4 | 2.7623 | 0.0032 | 0.2256 | 0.9752 | 79.149 | 3.00 |
| Compound | C. albicans 636/20 | C. albicans 595/20 | C. albicans 38 | C. albicans ATTC 90028 |
|---|---|---|---|---|
| 1 | 88 | <50 | <50 | 110 |
| 2 | 203 | >250 1 | 250 | >250 1 |
| 3 | 244 | 115 | >250 1 | >250 1 |
| 4 | 123 | <50 | 87 | 89 |
| Fluconazole | >250 1 | 0.89 | 0.44 | 4.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gach, J.; Olejniczak, T.; Krężel, P.; Boratyński, F. Microbial Synthesis and Evaluation of Fungistatic Activity of 3-Butyl-3-hydroxyphthalide, the Mammalian Metabolite of 3-n-Butylidenephthalide. Int. J. Mol. Sci. 2021, 22, 7600. https://doi.org/10.3390/ijms22147600
Gach J, Olejniczak T, Krężel P, Boratyński F. Microbial Synthesis and Evaluation of Fungistatic Activity of 3-Butyl-3-hydroxyphthalide, the Mammalian Metabolite of 3-n-Butylidenephthalide. International Journal of Molecular Sciences. 2021; 22(14):7600. https://doi.org/10.3390/ijms22147600
Chicago/Turabian StyleGach, Joanna, Teresa Olejniczak, Piotr Krężel, and Filip Boratyński. 2021. "Microbial Synthesis and Evaluation of Fungistatic Activity of 3-Butyl-3-hydroxyphthalide, the Mammalian Metabolite of 3-n-Butylidenephthalide" International Journal of Molecular Sciences 22, no. 14: 7600. https://doi.org/10.3390/ijms22147600
APA StyleGach, J., Olejniczak, T., Krężel, P., & Boratyński, F. (2021). Microbial Synthesis and Evaluation of Fungistatic Activity of 3-Butyl-3-hydroxyphthalide, the Mammalian Metabolite of 3-n-Butylidenephthalide. International Journal of Molecular Sciences, 22(14), 7600. https://doi.org/10.3390/ijms22147600






