The Role of microRNAs in Cholangiocarcinoma
Abstract
:1. Introduction
2. Epidemiology
3. Risk Factors
4. The Role of miRNAs in CCA
4.1. miRNAs Associated with CCA Risk Factors
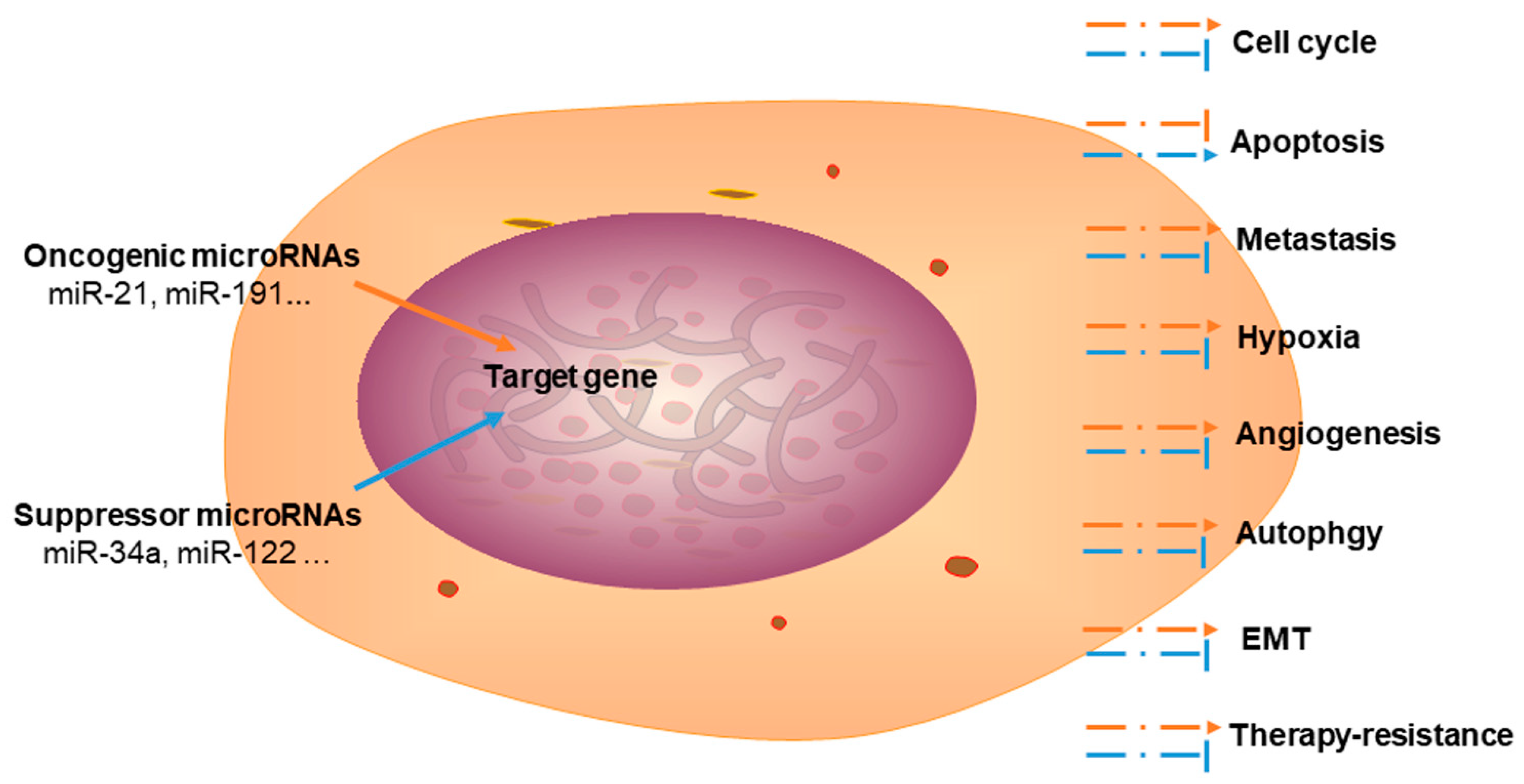
4.2. Dysregulation of miRNAs in CCA
4.2.1. Oncogenic miRNAs in CCA
4.2.2. Tumor Suppressor miRNAs in CCA
4.3. miRNAs as Biomarkers for CCA
4.4. miRNAs in CCA Therapy Resistance
4.5. miRNA-Based Therapies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [CrossRef]
- Rizvi, S.; Khan, S.A.; Hallemeier, C.L.; Kelley, R.K.; Gores, G.J. Cholangiocarcinoma—Evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018, 15, 95–111. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Moeini, A.; Sia, D.; Zhang, Z.; Campreciós, G.; Stueck, A.; Dong, H.; Montal, R.; Torrens, L.; Martinez-Quetglas, I.; Fiel, M.I.; et al. Mixed hepatocellular cholangiocarcinoma tumors: Cholangiolocellular carcinoma is a distinct molecular entity. J. Hepatol. 2017, 66, 952–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esnaola, N.F.; Meyer, J.E.; Karachristos, A.; Maranki, J.L.; Camp, E.R.; Denlinger, C.S. Evaluation and management of intrahepatic and extrahepatic cholangiocarcinoma. Cancer 2016, 122, 1349–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spolverato, G.; Kim, Y.; Alexandrescu, S.; Marques, H.P.; Lamelas, J.; Aldrighetti, L.; Gamblin, T.C.; Maithel, S.K.; Pulitano, C.; Bauer, T.W.; et al. Management and Outcomes of Patients with Recurrent Intrahepatic Cholangiocarcinoma Following Previous Curative-Intent Surgical Resection. Ann. Surg. Oncol. 2016, 23, 235–243. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in Cancer. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 199–227. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syeda, Z.A.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef] [Green Version]
- De Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Wang, F.; Yang, G.-H.; Wang, F.-L.; Ma, Y.-N.; Du, Z.-W.; Zhang, J.-W. Human microRNA clusters: Genomic organization and expression profile in leukemia cell lines. Biochem. Biophys. Res. Commun. 2006, 349, 59–68. [Google Scholar] [CrossRef]
- Cozar, J.; Robles-Fernandez, I.; Rodriguez-Martinez, A.; Puche-Sanz, I.; Vazquez-Alonso, F.; Lorente, J.; Martinez-Gonzalez, L.; Alvarez-Cubero, M. The role of miRNAs as biomarkers in prostate cancer. Mutat. Res. Mutat. Res. 2019, 781, 165–174. [Google Scholar] [CrossRef]
- Bertuccio, P.; Malvezzi, M.; Carioli, G.; Hashim, D.; Boffetta, P.; El-Serag, H.B.; La Vecchia, C.; Negri, E. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J. Hepatol. 2019, 71, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.J.; Jabbour, S.; Parekh, N.; Lin, Y.; Moss, R.A. Increasing mortality in the United States from cholangiocarcinoma: An analysis of the National Center for Health Statistics Database. BMC Gastroenterol. 2016, 16, 117. [Google Scholar] [CrossRef] [Green Version]
- Blechacz, B. Cholangiocarcinoma: Current Knowledge and New Developments. Gut Liver 2017, 11, 13–26. [Google Scholar] [CrossRef]
- Witjes, C.D.M.; Karim-Kos, H.E.; Visser, O.; De Vries, E.; Ijzermans, J.N.M.; De Man, R.A.; Coebergh, J.W.W.; Verhoef, C. Intrahepatic cholangiocarcinoma in a low endemic area: Rising incidence and improved survival. HPB 2012, 14, 777–781. [Google Scholar] [CrossRef] [Green Version]
- Tyson, G.L.; Ilyas, J.A.; Duan, Z.; Green, L.K.; Younes, M.; El-Serag, H.B.; Davila, J.A. Secular Trends in the Incidence of Cholangiocarcinoma in the USA and the Impact of Misclassification. Dig. Dis. Sci. 2014, 59, 3103–3110. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, R.; Sato, Y.; Kobayashi, Y. Cholangiocarcinoma Prognosis Varies over Time Depending on Tumor Site and Pathology. J. Gastrointest. Liver Dis. 2018, 27, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Cai, W.-K.; Sima, H.; Chen, B.-D.; Yang, G.-S. Risk factors for hilar cholangiocarcinoma: A case-control study in China. World J. Gastroenterol. 2011, 17, 249–253. [Google Scholar] [CrossRef]
- Arbelaiz, A.; Azkargorta, M.; Krawczyk, M.; Santos-Laso, A.; Lapitz, A.; Perugorria, M.J.; Erice, O.; Gonzalez, E.; Jimenez-Agüero, R.; La Casta, A.; et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017, 66, 1125–1143. [Google Scholar] [CrossRef]
- Fabris, L.; Fiorotto, R.; Spirli, C.; Cadamuro, M.; Mariotti, V.; Perugorria, M.J.; Banales, J.M.; Strazzabosco, M. Pathobiology of inherited biliary diseases: A roadmap to understand acquired liver diseases. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Prueksapanich, P.; Piyachaturawat, P.; Aumpansub, P.; Ridtitid, W.; Chaiteerakij, R.; Rerknimitr, R. Liver Fluke-Associated Biliary Tract Cancer. Gut Liver 2018, 12, 236–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaib, Y.H.; El-Serag, H.B.; Davila, J.A.; Morgan, R.; McGlynn, K.A. Risk factors of intrahepatic cholangiocarcinoma in the United States: A case-control study. Gastroenterology 2005, 128, 620–626. [Google Scholar] [CrossRef]
- Tan, J.-H.; Zhou, W.-Y.; Zhou, L.; Cao, R.-C.; Zhang, G.-W. Viral hepatitis B and C infections increase the risks of intrahepatic and extrahepatic cholangiocarcinoma: Evidence from a systematic review and meta-analysis. Turk. J. Gastroenterol. 2020, 31, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Petrick, J.L.; Yang, B.; Altekruse, S.F.; Van Dyke, A.L.; Koshiol, J.; Graubard, B.I.; McGlynn, K.A. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma in the United States: A population-based study in SEER-Medicare. PLoS ONE 2017, 12, e0186643. [Google Scholar] [CrossRef] [Green Version]
- Wongjarupong, N.; Assavapongpaiboon, B.; Susantitaphong, P.; Cheungpasitporn, W.; Treeprasertsuk, S.; Rerknimitr, R.; Chaiteerakij, R. Non-alcoholic fatty liver disease as a risk factor for cholangiocarcinoma: A systematic review and meta-analysis. BMC Gastroenterol. 2017, 17, 149. [Google Scholar] [CrossRef] [Green Version]
- Petrick, J.L.; Campbell, P.T.; Koshiol, J.; Thistle, J.E.; Andreotti, G.; Beane-Freeman, L.E.; Buring, J.E.; Chan, A.T.; Chong, D.Q.; Doody, M.M.; et al. Tobacco, alcohol use and risk of hepatocellular carcinoma and intrahepatic cholangiocarcinoma: The Liver Cancer Pooling Project. Br. J. Cancer 2018, 118, 1005–1012. [Google Scholar] [CrossRef] [PubMed]
- Labib, P.L.; Goodchild, G.; Pereira, S.P. Molecular Pathogenesis of Cholangiocarcinoma. BMC Cancer 2019, 19, 185. [Google Scholar] [CrossRef]
- Suzuki, Y.; Mori, T.; Abe, N.; Sugiyama, M.; Atomi, Y. Predictive factors for cholangiocarcinoma associated with hepatolithiasis determined on the basis of Japanese Multicenter study. Hepatol. Res. 2012, 42, 166–170. [Google Scholar] [CrossRef]
- Karlsen, T.H.; Folseraas, T.; Thorburn, D.; Vesterhus, M. Primary sclerosing cholangitis—A comprehensive review. J. Hepatol. 2017, 67, 1298–1323. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yang, T.; Wu, M.; Shen, F. Intrahepatic cholangiocarcinoma: Epidemiology, risk factors, diagnosis and surgical management. Cancer Lett. 2016, 379, 198–205. [Google Scholar] [CrossRef]
- Goeppert, B.; Folseraas, T.; Roessler, S.; Kloor, M.; Volckmar, A.; Endris, V.; Buchhalter, I.; Stenzinger, A.; Grzyb, K.; Grimsrud, M.M.; et al. Genomic Characterization of Cholangiocarcinoma in Primary Sclerosing Cholangitis Reveals Therapeutic Opportunities. Hepatology 2020, 72, 1253–1266. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.J.; Crothers, H.; Mytton, J.; Bosch, S.; Iqbal, T.; Ferguson, J.; Hirschfield, G.M. Effects of Primary Sclerosing Cholangitis on Risks of Cancer and Death in People With Inflammatory Bowel Disease, Based on Sex, Race, and Age. Gastroenterology 2020, 159, 915–928. [Google Scholar] [CrossRef]
- Sripa, B.; Kaewkes, S.; Sithithaworn, P.; Mairiang, E.; Laha, T.; Smout, M.; Pairojkul, C.; Bhudhisawasdi, V.; Tesana, S.; Thinkamrop, B.; et al. Liver Fluke Induces Cholangiocarcinoma. PLoS Med. 2007, 4, e201. [Google Scholar] [CrossRef]
- Alsaleh, M.; Leftley, Z.; Barbera, T.A.; Sithithaworn, P.; Khuntikeo, N.; Loilome, W.; Yongvanit, P.; Cox, I.J.; Chamodol, N.; Syms, R.R.; et al. Cholangiocarcinoma: A guide for the nonspecialist. Int. J. Gen. Med. 2018, 12, 13–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Na, B.-K.; Pak, J.H.; Hong, S.-J. Clonorchis sinensis and clonorchiasis. Acta Trop. 2020, 203, 105309. [Google Scholar] [CrossRef]
- Kurokawa, T.; Sato, T.; Andoh, H.; Yasui, O. Cholangiocarcinoma coincident with schistosomiasis japonica. J. Gastroenterol. 2004, 39, 64–68. [Google Scholar] [CrossRef]
- Easterbrook, P.J.; Roberts, T.; Sands, A.; Peeling, R. Diagnosis of viral hepatitis. Curr. Opin. HIV AIDS 2017, 12, 302–314. [Google Scholar] [CrossRef]
- Li, M.; Li, J.; Li, P.; Li, H.; Su, T.; Zhu, R.; Gong, J. Hepatitis B virus infection increases the risk of cholangiocarcinoma: A meta-analysis and systematic review. J. Gastroenterol. Hepatol. 2012, 27, 1561–1568. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, B.; Zhang, H.; Liang, J.; Zeng, W. HBV Infection Status and the Risk of Cholangiocarcinoma in Asia: A Meta-Analysis. BioMed Res. Int. 2016, 2016, 3417976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, W.C.; Patel, T. Are common factors involved in the pathogenesis of primary liver cancers? A meta-analysis of risk factors for intrahepatic cholangiocarcinoma. J. Hepatol. 2012, 57, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Welzel, T.M.; Graubard, B.I.; El–Serag, H.B.; Shaib, Y.H.; Hsing, A.W.; Davila, J.A.; McGlynn, K.A. Risk Factors for Intrahepatic and Extrahepatic Cholangiocarcinoma in the United States: A Population-Based Case-Control Study. Clin. Gastroenterol. Hepatol. 2007, 5, 1221–1228. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Liu, B.; Bi, P.; Wu, T.; Wang, Q.; Zhang, J. An integrated analysis of differential miRNA and mRNA expressions in human gallstones. Mol. BioSyst. 2015, 11, 1004–1011. [Google Scholar] [CrossRef]
- Wu, X.; Yao, C.; Kong, J.; Tian, Y.; Fan, Y.; Zhang, Z.; Han, J.; Wu, S. Molecular mechanism underlying miR-130b-Sp1 transcriptional regulation in LPS-induced upregulation of MUC5AC in the bile duct epithelium. Mol. Med. Rep. 2021, 23, 106. [Google Scholar] [CrossRef]
- Jiang, W.; Deng, X.; Zhu, T.; Wei, Y.; Lei, Z.; Guo, M.; Yang, J. Identification of Cholangiocarcinoma Associated with Hepatolithiasis via the Combination of miRNA and Ultrasound. Cancer Manag. Res. 2020, 12, 1845–1853. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Meng, F.; Zhou, T.; Han, Y.; Kennedy, L.; Venter, J.; Francis, H.; DeMorrow, S.; Onori, P.; Invernizzi, P.; et al. Prolonged darkness reduces liver fibrosis in a mouse model of primary sclerosing cholangitis by miR-200b down-regulation. FASEB J. 2017, 31, 4305–4324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Zhang, J.; Zheng, T.; Chen, H.; Nie, H.; Zheng, B.; Gong, Q. The role of microRNAs in the pathogenesis, grading and treatment of hepatic fibrosis in schistosomiasis. Parasites Vectors 2019, 12, 611. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Lyu, L.; Shen, P.; Wang, J.; Chen, J.; Sun, X.; Chen, L.; Zhang, L.; Zhou, Q.; Duan, Y. rSjP40 protein promotes PPARγ expression in LX-2 cells through microRNA-27b. FASEB J. 2018, 32, 4798–4803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Hu, J.; Wang, X.; Zhao, X.; Li, Z.; Niu, J.; Steer, C.J.; Zheng, G.; Song, G. MicroRNA-378 promotes hepatic inflammation and fibrosis via modulation of the NF-κB-TNFα pathway. J. Hepatol. 2019, 70, 87–96. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Y.; Li, S.; Liu, Y.; Zhou, S.; Cao, S.; Yin, J.; Li, G. MicroRNA-30a ameliorates hepatic fibrosis by inhibiting Beclin1-mediated autophagy. J. Cell. Mol. Med. 2017, 21, 3679–3692. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yang, X.; Wei, R.; Ye, T.; Zhou, J.; Wen, M.; Men, R.; Li, P.; Dong, B.; Liu, L.; et al. MicroRNA-214 promotes hepatic stellate cell activation and liver fibrosis by suppressing Sufu expression. Cell Death Dis. 2018, 9, 718. [Google Scholar] [CrossRef]
- Liang, T.J. Hepatitis B: The virus and disease. Hepatology 2009, 49, S13–S21. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Dong, F.; Xu, Z.; Sharma, S.; Hu, X.; Chen, D.; Zhang, L.; Zhang, J.; Dong, Q. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer 2017, 17, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chen, J.; Liu, Y.; Zeng, X.; Wei, M.; Wu, S.; Xiong, Q.; Song, F.; Yuan, X.; Xiao, Y.; et al. Hepatitis B Virus Induces Autophagy to Promote its Replication by the Axis of miR-192-3p-XIAP Through NF kappa B Signaling. Hepatology 2019, 69, 974–992. [Google Scholar] [CrossRef]
- Gao, K.; Liu, F.; Guo, H.; Li, J.; Zhang, Y.; Mo, Z. miR-224 suppresses HBV replication posttranscriptionally through inhibiting SIRT1-mediated autophagy. Int. J. Clin. Exp. Pathol. 2018, 11, 189–198. [Google Scholar]
- Kohno, T.; Tsuge, M.; Murakami, E.; Hiraga, N.; Abe, H.; Miki, D.; Imamura, M.; Ochi, H.; Hayes, C.N.; Chayama, K. Human microRNA hsa-miR-1231 suppresses hepatitis B virus replication by targeting core mRNA. J. Viral Hepat. 2014, 21, e89–e97. [Google Scholar] [CrossRef]
- Lin, Y.; Deng, W.; Pang, J.; Kemper, T.; Hu, J.; Yin, J.; Zhang, J.; Lu, M. The microRNA-99 family modulates hepatitis B virus replication by promoting IGF-1R/PI3K/Akt/mTOR/ULK1 signaling-induced autophagy. Cell. Microbiol. 2017, 19, e12709. [Google Scholar] [CrossRef] [Green Version]
- Roudot-Thoraval, F. Epidemiology of hepatitis C virus infection. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101596. [Google Scholar] [CrossRef]
- Tian, H.; He, Z. miR-215 Enhances HCV Replication by Targeting TRIM22 and Inactivating NF-κB Signaling. Yonsei Med. J. 2018, 59, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Clément, S.; Sobolewski, C.; Gomes, D.; Rojas, A.; Goossens, N.; Conzelmann, S.; Calo, N.; Negro, F.; Foti, M. Activation of the oncogenic miR-21-5p promotes HCV replication and steatosis induced by the viral core 3a protein. Liver Int. 2019, 39, 1226–1236. [Google Scholar] [CrossRef]
- Kunden, R.D.; Khan, J.Q.; Ghezelbash, S.; Wilson, J.A. The Role of the Liver-Specific microRNA, miRNA-122 in the HCV Replication Cycle. Int. J. Mol. Sci. 2020, 21, 5677. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Aly, H.H.; Tajima, A.; Inoue, I.; Shimotohno, K. Regulation of the hepatitis C virus genome replication by miR-199a. J. Hepatol. 2009, 50, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Liu, X.; Li, W.; Holmes, J.A.; Kruger, A.J.; Yang, C.; Li, Y.; Xu, M.; Ye, H.; Li, S.; et al. Microrna-130a Downregulates HCV Replication through an atg5-Dependent Autophagy Pathway. Cells 2019, 8, 338. [Google Scholar] [CrossRef] [Green Version]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef]
- He, C.; Shi, Y.; Wu, R.; Sun, M.; Fang, L.; Wu, W.; Liu, C.; Tang, M.; Li, Z.; Wang, P.; et al. miR-301a promotes intestinal mucosal inflammation through induction of IL-17A and TNF-α in IBD. Gut 2016, 65, 1938–1950. [Google Scholar] [CrossRef]
- Tian, Y.; Xu, J.; Li, Y.; Zhao, R.; Du, S.; Lv, C.; Wu, W.; Liu, R.; Sheng, X.; Song, Y.; et al. MicroRNA-31 Reduces Inflammatory Signaling and Promotes Regeneration in Colon Epithelium, and Delivery of Mimics in Microspheres Reduces Colitis in Mice. Gastroenterology 2019, 156, 2281–2296.e6. [Google Scholar] [CrossRef]
- Ji, T.; Feng, W.; Zhang, X.; Zang, K.; Zhu, X.; Shang, F. HDAC inhibitors promote pancreatic stellate cell apoptosis and relieve pancreatic fibrosis by upregulating miR-15/16 in chronic pancreatitis. Hum. Cell 2020, 33, 1006–1016. [Google Scholar] [CrossRef]
- Feng, J.; Xing, W.; Xie, L. Regulatory Roles of MicroRNAs in Diabetes. Int. J. Mol. Sci. 2016, 17, 1729. [Google Scholar] [CrossRef]
- Singh, R.; Ha, S.E.; Wei, L.; Jin, B.; Zogg, H.; Poudrier, S.M.; Jorgensen, B.G.; Park, C.; Ronkon, C.F.; Bartlett, A.; et al. miR-10b-5p Rescues Diabetes and Gastrointestinal Dysmotility. Gastroenterology 2021, 160, 1662–1678.e18. [Google Scholar] [CrossRef]
- Ying, W.; Gao, H.; Dos Reis, F.C.G.; Bandyopadhyay, G.; Ofrecio, J.M.; Luo, Z.; Ji, Y.; Jin, Z.; Ly, C.; Olefsky, J.M. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 2021, 33, 781–790.e5. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Kumar, V.; Sud, N.; Mahato, R.I. MicroRNAs in the pathogenesis and treatment of progressive liver injury in NAFLD and liver fibrosis. Adv. Drug Deliv. Rev. 2018, 129, 54–63. [Google Scholar] [CrossRef]
- Hanin, G.; Yayon, N.; Tzur, Y.; Haviv, R.; Bennett, E.R.; Udi, S.; Krishnamoorthy, Y.R.; Kotsiliti, E.; Zangen, R.; Efron, B.; et al. miRNA-132 induces hepatic steatosis and hyperlipidaemia by synergistic multitarget suppression. Gut 2018, 67, 1124–1134. [Google Scholar] [CrossRef] [Green Version]
- Heo, M.J.; Kim, T.H.; You, J.S.; Blaya, D.; Sancho-Bru, P.; Kim, S.G. Alcohol dysregulates miR-148a in hepatocytes through FoxO1, facilitating pyroptosis via TXNIP overexpression. Gut 2019, 68, 708–720. [Google Scholar] [CrossRef] [Green Version]
- Bala, S.; Csak, T.; Saha, B.; Zatsiorsky, J.; Kodys, K.; Catalano, D.; Satishchandran, A.; Szabo, G. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J. Hepatol. 2016, 64, 1378–1387. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Bai, R.; Li, M.; Ye, H.; Wu, C.; Wang, C.; Li, S.; Tan, L.; Mai, D.; Li, G.; et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 2019, 10, 1858. [Google Scholar] [CrossRef]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018, 9, 5395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, P.; Chi, X.; Du, Q.; Luo, J.; Cui, X.; Dong, K.; Bing, Y.; Heres, C.; Geller, D.A. miR-383 promotes cholangiocarcinoma cell proliferation, migration, and invasion through targeting IRF. J. Cell. Biochem. 2018, 119, 9720–9729. [Google Scholar] [CrossRef]
- Li, J.; Yao, L.; Li, G.; Ma, D.; Sun, C.; Gao, S.; Zhang, P.; Gao, F. miR-221 Promotes Epithelial-Mesenchymal Transition through Targeting PTEN and Forms a Positive Feedback Loop with β-catenin/c-Jun Signaling Pathway in Extra-Hepatic Cholangiocarcinoma. PLoS ONE 2015, 10, e0141168. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Huang, F.; Deng, G.; Nie, W.; Huang, W.; Zeng, X. miR-31 promotes oncogenesis in intrahepatic cholangiocarcinoma cells via the direct suppression of RASA1. Exp. Ther. Med. 2013, 6, 1265–1270. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.-X. Long non-coding RNA MEG3 represses cholangiocarcinoma by regulating miR-361-5p/TRAF3 axis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7356–7368. [Google Scholar] [CrossRef]
- Zhang, J.W.; Wang, X.; Li, G.C.; Wang, D.; Han, S.; Zhang, Y.D.; Luo, C.H.; Wang, H.W.; Jiang, W.J.; Li, C.X.; et al. MiR-30a-5p promotes cholangiocarcinoma cell proliferation through targeting SOCS3. J. Cancer 2020, 11, 3604–3614. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Byrnes, K.; Han, C.; Wang, Y.; Wu, T. miR-21 Targets 15-PGDH and Promotes Cholangiocarcinoma Growth. Mol. Cancer Res. 2014, 12, 890–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Zhao, J.; Guo, X.; Feng, Y.; Zhang, B.; Tian, L. LncRNA MT1JP plays a protective role in intrahepatic cholangiocarcinoma by regulating miR-18a-5p/FBP1 axis. BMC Cancer 2021, 21, 142. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.-Y.; Yu, J.-H.; Zhang, W.-G.; Wang, Z.-D.; Dong, Q.; Tai, S.; Cui, Y.-F.; Li, H. MicroRNA-421 functions as an oncogenic miRNA in biliary tract cancer through down-regulating farnesoid X receptor expression. Gene 2012, 493, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, Z.-Q.; Yang, Z.-R.; Tong, D.-N.; Guan, J.; Shi, B.-J.; Nie, J.; Ding, X.-T.; Li, B.; Zhou, G.-W.; et al. MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology 2017, 66, 136–151. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xie, C.; Pan, S.; Liang, Y.; Han, J.; Lan, Y.; Sun, J.; Li, K.; Sun, B.; Yang, G.; et al. N-myc downstream-regulated gene 2 inhibits human cholangiocarcinoma progression and is regulated by leukemia inhibitory factor/MicroRNA-181c negative feedback pathway. Hepatology 2016, 64, 1606–1622. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Yin, J.; Cong, J. Downregulation of microRNA-193-3p inhibits the progression of intrahepatic cholangiocarcinoma cells by upregulating TGFBR3. Exp. Ther. Med. 2018, 15, 4508–4514. [Google Scholar] [CrossRef] [Green Version]
- Ursu, S.; Majid, S.; Garger, C.; De Semir, D.; Bezrookove, V.; Desprez, P.-Y.; McAllister, S.; Soroceanu, L.; Nosrati, M.; Yimam, K.; et al. Novel tumor suppressor role of miRNA-876 in cholangiocarcinoma. Oncogenesis 2019, 8, 42. [Google Scholar] [CrossRef] [Green Version]
- Loeffler, M.A.; Hu, J.; Kirchner, M.; Wei, X.; Xiao, Y.; Albrecht, T.; De La Torre, C.; Sticht, C.; Banales, J.M.; Vogel, M.N.; et al. miRNA profiling of biliary intraepithelial neoplasia reveals stepwise tumorigenesis in distal cholangiocarcinoma via the miR-451a/ATF2 axis. J. Pathol. 2020, 252, 239–251. [Google Scholar] [CrossRef]
- Fu, W.; Yu, G.; Liang, J.; Fan, P.; Dong, K.; Zhang, B.; Chen, X.; Zhu, H.; Chu, L. miR-144-5p and miR-451a Inhibit the Growth of Cholangiocarcinoma Cells Through Decreasing the Expression of ST8SIA4. Front. Oncol. 2021, 10, 563486. [Google Scholar] [CrossRef]
- Kwon, H.; Song, K.; Han, C.; Zhang, J.; Lu, L.; Chen, W.; Wu, T. Epigenetic Silencing of miRNA-34a in Human Cholangiocarcinoma via EZH2 and DNA Methylation: Impact on Regulation of Notch Pathway. Am. J. Pathol. 2017, 187, 2288–2299. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Shi, B.; Zhang, K. miR-186 Suppresses the Progression of Cholangiocarcinoma Cells through Inhibition of Twist1. Oncol. Res. 2019, 27, 1061–1068. [Google Scholar] [CrossRef]
- Utaijaratrasmi, P.; Vaeteewoottacharn, K.; Tsunematsu, T.; Jamjantra, P.; Wongkham, S.; Pairojkul, C.; Khuntikeo, N.; Ishimaru, N.; Sirivatanauksorn, Y.; Pongpaibul, A.; et al. The microRNA-15a-PAI-2 axis in cholangiocarcinoma-associated fibroblasts promotes migration of cancer cells. Mol. Cancer 2018, 17, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, A.; Zhao, L.; Kang, Q.; Li, J.; Chen, K.; Fu, H. Transcription factor HIF1α promotes proliferation, migration, and invasion of cholangiocarcinoma via long noncoding RNA H19/microRNA-612/Bcl-2 axis. Transl. Res. 2020, 224, 26–39. [Google Scholar] [CrossRef]
- Wu, J.; Yang, B.; Zhang, Y.; Feng, X.; He, B.; Xie, H.; Zhou, L.; Wu, J.; Zheng, S. miR-424-5p represses the metastasis and invasion of intrahepatic cholangiocarcinoma by targeting ARK5. Int. J. Biol. Sci. 2019, 15, 1591–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.; Wei, C.; Lin, J.; Dong, S.; Gao, D.; Chen, J.; Zhao, Y.; Liu, B. UHRF1 is regulated by miR-124-3p and promotes cell proliferation in intrahepatic cholangiocarcinoma. J. Cell. Physiol. 2019, 234, 19875–19885. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Wang, Y.; Li, W.; Shi, L.; Geng, Z. Micro RNA-551b-3p inhibits tumour growth of human cholangiocarcinoma by targeting Cyclin D1. J. Cell. Mol. Med. 2019, 23, 4945–4954. [Google Scholar] [CrossRef] [Green Version]
- Liao, G.; Liu, X.; Wu, D.; Duan, F.; Xie, X.; Wen, S.; Li, Y.; Li, S. MORC2 promotes cell growth and metastasis in human cholangiocarcinoma and is negatively regulated by miR-186-5p. Aging 2019, 11, 3639–3649. [Google Scholar] [CrossRef]
- Chen, T.; Lei, S.; Zeng, Z.; Pan, S.; Zhang, J.; Xue, Y.; Sun, Y.; Lan, J.; Xu, S.; Mao, D.; et al. MicroRNA-137 suppresses the proliferation, migration and invasion of cholangiocarcinoma cells by targeting WNT2B. Int. J. Mol. Med. 2020, 45, 886–896. [Google Scholar] [CrossRef]
- Leng, K.; Xu, Y.; Kang, P.; Qin, W.; Cai, H.; Wang, H.; Ji, D.; Jiang, X.; Li, J.; Li, Z.; et al. Akirin2 is modulated by miR-490-3p and facilitates angiogenesis in cholangiocarcinoma through the IL-6/STAT3/VEGFA signaling pathway. Cell Death Dis. 2019, 10, 262. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.; Liu, Y.; Wu, Y.; Zhu, S.; Cai, R.; Zhou, L.; Yin, X. microRNA-329 suppresses epithelial-to-mesenchymal transition and lymph node metastasis in bile duct cancer by inhibiting laminin subunit beta. J. Cell. Physiol. 2019, 234, 17786–17799. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Sun, Z.; Li, O.; Guo, C.; Yi, W.; Tan, Z.; Jiang, B. Leptin stimulates the epithelial-mesenchymal transition and pro-angiogenic capability of cholangiocarcinoma cells through the miR-122/PKM2 axis. Int. J. Oncol. 2019, 55, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Liu, G.; Zhang, M.; Zhang, Z.; Jia, Y.; Peng, L.; Zhu, Y.; Hu, J.; Huang, R.; Sun, X. miR-122-5p Inhibits the Proliferation, Invasion and Growth of Bile Duct Carcinoma Cells by Targeting ALDOA. Cell. Physiol. Biochem. 2018, 48, 2596–2606. [Google Scholar] [CrossRef]
- Palumbo, T.; Poultsides, G.A.; Kouraklis, G.; Liakakos, T.; Drakaki, A.; Peros, G.; Hatziapostolou, M.; Iliopoulos, D. A functional microRNA library screen reveals miR-410 as a novel anti-apoptotic regulator of cholangiocarcinoma. BMC Cancer 2016, 16, 353. [Google Scholar] [CrossRef] [Green Version]
- Mansini, A.P.; Pisarello, M.J.L.; Thelen, K.M.; Cruz-Reyes, M.; Peixoto, E.; Jin, S.; Howard, B.N.; Trussoni, C.E.; Gajdos, G.B.; LaRusso, N.F.; et al. MicroRNA (miR)-433 and miR-22 dysregulations induce histone-deacetylase-6 overexpression and ciliary loss in cholangiocarcinoma. Hepatology 2018, 68, 561–573. [Google Scholar] [CrossRef]
- Yang, R.; Chen, Y.; Tang, C.; Li, H.; Wang, B.; Yan, Q.; Hu, J.; Zou, S. MicroRNA-144 suppresses cholangiocarcinoma cell proliferation and invasion through targeting platelet activating factor acetylhydrolase isoform 1b. BMC Cancer 2014, 14, 917. [Google Scholar] [CrossRef] [Green Version]
- Zu, C.; Liu, S.; Cao, W.; Liu, Z.; Qiang, H.; Li, Y.; Cheng, C.; Ji, L.; Li, J.; Li, J. MiR-590-3p suppresses epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma by inhibiting SIP1 expression. Oncotarget 2017, 8, 34698–34708. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Han, C.; Zhu, H.; Song, K.; Wu, T. miR-101 Inhibits Cholangiocarcinoma Angiogenesis through Targeting Vascular Endothelial Growth Factor (VEGF). Am. J. Pathol. 2013, 182, 1629–1639. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, L.; Tang, L.; Luo, J.; Ji, H.; Zhang, W.; Zhou, J.; Li, Q.; Miao, L. Long noncoding RNA SNHG6 promotes proliferation and angiogenesis of cholangiocarcinoma cells through sponging miR-101-3p and activation of E2F8. J. Cancer 2020, 11, 3002–3012. [Google Scholar] [CrossRef] [Green Version]
- Fan, F.; Lu, J.; Yu, W.; Zhang, Y.; Xu, S.; Pang, L.; Zhu, B. MicroRNA-26b-5p regulates cell proliferation, invasion and metastasis in human intrahepatic cholangiocarcinoma by targeting S100A7. Oncol. Lett. 2018, 15, 386–392. [Google Scholar] [CrossRef]
- Pan, X.; Wang, G.; Wang, B. MicroRNA-1182 and let-7a exert synergistic inhibition on invasion, migration and autophagy of cholangiocarcinoma cells through down-regulation of NUAK1. Cancer Cell Int. 2021, 21, 161. [Google Scholar] [CrossRef]
- Pan, X.; Wang, G.; Wang, B. Ectopic expression of microRNA-874 represses epithelial mesenchymal transition through the NF-κB pathway via CCNE1 in cholangiocarcinoma. Cell. Signal. 2021, 82, 109927. [Google Scholar] [CrossRef]
- Lixin, S.; Wei, S.; Haibin, S.; Qingfu, L.; Tiemin, P. miR-885-5p inhibits proliferation and metastasis by targeting IGF2BP1 and GALNT3 in human intrahepatic cholangiocarcinoma. Mol. Carcinog. 2020, 59, 1371–1381. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-F.; Li, Z.-R.; Cheng, Z.-Q.; Yin, X.-M.; Wu, J.-S. Decrease of miR-622 expression promoted the proliferation, migration and invasion of cholangiocarcinoma cells by targeting regulation of c-Myc. Biomed. Pharmacother. 2017, 96, 7–13. [Google Scholar] [CrossRef]
- Zhu, H.; Jiang, X.; Zhou, X.; Dong, X.; Xie, K.; Yang, C.; Jiang, H.; Sun, X.; Lu, J. Neuropilin-1 regulated by miR-320 contributes to the growth and metastasis of cholangiocarcinoma cells. Liver Int. 2018, 38, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Jiang, J.; Yu, Y.; Tian, R.; Guo, X.; Li, X.; Shen, M.; Xu, M.; Zhu, F.; Shi, C.; et al. Direct targeting of SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma tumourigenesis and metastasis. Br. J. Cancer 2013, 109, 3092–3104. [Google Scholar] [CrossRef] [Green Version]
- Wagenaar, T.R.; Zabludoff, S.; Ahn, S.-M.; Allerson, C.; Arlt, H.; Baffa, R.; Cao, H.; Davis, S.; Garcia-Echeverria, C.; Gaur, R.; et al. Anti–miR-21 Suppresses Hepatocellular Carcinoma Growth via Broad Transcriptional Network Deregulation. Mol. Cancer Res. 2015, 13, 1009–1021. [Google Scholar] [CrossRef] [Green Version]
- Najjary, S.; Mohammadzadeh, R.; Mokhtarzadeh, A.; Mohammadi, A.; Kojabad, A.B.; Baradaran, B. Role of miR-21 as an authentic oncogene in mediating drug resistance in breast cancer. Gene 2020, 738, 144453. [Google Scholar] [CrossRef]
- Selaru, F.M.; Olaru, A.V.; Kan, T.; David, S.; Cheng, Y.; Mori, Y.; Yang, J.; Paun, B.; Jin, Z.; Agarwal, R.; et al. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology 2009, 49, 1595–1601. [Google Scholar] [CrossRef]
- Wang, L.-J.; He, C.-C.; Sui, X.; Cai, M.-J.; Zhou, C.-Y.; Ma, J.-L.; Wu, L.; Wang, H.; Han, S.-X.; Zhu, Q. MiR-21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTEN. Oncotarget 2015, 6, 5932–5946. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Zhu, J.; Wu, B.; Chen, J.; Zhu, Z.; Cai, P.; Guo, W.; Gu, Z.; Wang, J.; Huang, S. Diagnostic and prognostic value of microRNAs in cholangiocarcinoma: A systematic review and meta-analysis. Cancer Manag. Res. 2018, 10, 2125–2139. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Wang, Y.-N.; Song, D.-J.; Tan, J.-P.; Cao, Y.; Fan, J.; Wang, Z.; Zhou, J. A High-Accuracy Model Based on Plasma miRNAs Diagnoses Intrahepatic Cholangiocarcinoma: A Single Center with 1001 Samples. Diagnostics 2021, 11, 610. [Google Scholar] [CrossRef]
- Slabáková, E.; Culig, Z.; Remšík, J.; Souček, K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017, 8, e3100. [Google Scholar] [CrossRef]
- Kong, L.; Wu, Q.; Zhao, L.; Ye, J.; Li, N.; Yang, H. Upregulated lncRNA-UCA1 contributes to metastasis of bile duct carcinoma through regulation of miR-122/CLIC1and activation of the ERK/MAPK signaling pathway. Cell Cycle 2019, 18, 1212–1228. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Y.; Mackowiak, B.; Gao, B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut 2021, 70, 784–795. [Google Scholar] [CrossRef]
- Mishra, N.K.; Niu, M.; Southekal, S.; Bajpai, P.; Elkholy, A.; Manne, U.; Guda, C. Identification of Prognostic Markers in Cholangiocarcinoma Using Altered DNA Methylation and Gene Expression Profiles. Front. Genet. 2020, 11, 522125. [Google Scholar] [CrossRef]
- Xu, Y.; Yao, Y.; Jiang, X.; Zhong, X.; Wang, Z.; Li, C.; Kang, P.; Leng, K.; Ji, D.; Li, Z.; et al. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 81. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.-J.; Zhang, K.-L.; Zhang, N.; Ma, X.-W.; Yan, S.-W.; Cao, D.-H.; Shi, S.-J. Serum miR-26a as a diagnostic and prognostic biomarker in cholangiocarcinoma. Oncotarget 2015, 6, 18631–18640. [Google Scholar] [CrossRef] [Green Version]
- Salem, P.E.S.; Ghazala, R.A.; El Gendi, A.M.; Emara, D.M.; Ahmed, N.M. The association between circulating MicroRNA-150 level and cholangiocarcinoma. J. Clin. Lab. Anal. 2020, 34, e23397. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chen, Y. Increased Expression of miR-29a and Its Prognostic Significance in Patients with Cholangiocarcinoma. Oncol. Res. Treat. 2017, 40, 128–132. [Google Scholar] [CrossRef]
- Silakit, R.; Loilome, W.; Yongvanit, P.; Chusorn, P.; Techasen, A.; Boonmars, T.; Khuntikeo, N.; Chamadol, N.; Pairojkul, C.; Namwat, N. Circulating miR-192 in liver fluke-associated cholangiocarcinoma patients: A prospective prognostic indicator. J. Hepato Biliary Pancreat. Sci. 2014, 21, 864–872. [Google Scholar] [CrossRef]
- McNally, M.E.; Collins, A.; Wojcik, S.E.; Liu, J.; Henry, J.C.; Jiang, J.; Schmittgen, T.; Bloomston, M. Concomitant dysregulation of microRNAs miR-151-3p and miR-126 correlates with improved survival in resected cholangiocarcinoma. HPB 2013, 15, 260–264. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Q.; Feng, F.; Zhu, L.; Zheng, Y.; Luo, X.; Liu, C.; Yi, B.; Jiang, X. Circulating miR-106a is a Novel Prognostic and Lymph Node Metastasis Indicator for Cholangiocarcinoma. Sci. Rep. 2015, 5, 16103. [Google Scholar] [CrossRef]
- Zhang, R.-X.; Zheng, Z.; Li, K.; Wu, X.-H.; Zhu, L. Both plasma and tumor tissue miR-146a high expression correlates with prolonged overall survival of surgical patients with intrahepatic cholangiocarcinoma. Medicine 2017, 96, e8267. [Google Scholar] [CrossRef]
- Ishigami, K.; Nosho, K.; Kanno, S.; Mitsuhashi, K.; Igarashi, H.; Shitani, M.; Motoya, M.; Kimura, Y.; Hasegawa, T.; Kaneto, H.; et al. MicroRNA-31 reflects IL-6 expression in cancer tissue and is related with poor prognosis in bile duct cancer. Carcinogenesis 2018, 39, 1127–1134. [Google Scholar] [CrossRef]
- Li, J.; Gao, B.; Huang, Z.; Duan, T.; Li, D.; Zhang, S.; Zhao, Y.; Liu, L.; Wang, Q.; Chen, Z.; et al. Prognostic significance of microRNA-203 in cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 9512–9516. [Google Scholar]
- Chen, Q.; Wang, C.; Zhang, H.; Li, Y.; Cao, Y.; Zhang, Y.; Liu, S.; Li, Z.; Xin, X.; Han, X. Expression levels of serum miRNA-195 in different types of patients with cholangiocarcinoma and its value to determine the prognosis thereof. Oncol. Lett. 2018, 15, 5947–5951. [Google Scholar] [CrossRef] [Green Version]
- Meijer, L.L.; Puik, J.R.; Le Large, T.Y.; Heger, M.; Dijk, F.; Funel, N.; Wurdinger, T.; Garajová, I.; Van Grieken, N.C.; Van De Wiel, M.A.; et al. Unravelling the Diagnostic Dilemma: A MicroRNA Panel of Circulating MiR-16 and MiR-877 as A Diagnostic Classifier for Distal Bile Duct Tumors. Cancers 2019, 11, 1181. [Google Scholar] [CrossRef] [Green Version]
- Phelip, J.-M.; Edeline, J.; Blanc, J.-F.; Barbier, E.; Michel, P.; Bourgeois, V.; Neuzillet, C.; Malka, D.; Manfredi, S.; Desrame, J.; et al. Modified FOLFIRINOX versus CisGem first-line chemotherapy for locally advanced non resectable or metastatic biliary tract cancer (AMEBICA)-PRODIGE 38: Study protocol for a randomized controlled multicenter phase II/III study. Dig. Liver Dis. 2019, 51, 318–320. [Google Scholar] [CrossRef]
- Shroff, R.T.; Javle, M.M.; Xiao, L.; Kaseb, A.O.; Varadhachary, G.R.; Wolff, R.A.; Raghav, K.P.S.; Iwasaki, M.; Masci, P.; Ramanathan, R.K.; et al. Gemcitabine, Cisplatin, and nab-Paclitaxel for the Treatment of Advanced Biliary Tract Cancers: A Phase 2 Clinical Trial. JAMA Oncol. 2019, 5, 824–830. [Google Scholar] [CrossRef]
- Silakit, R.; Kitirat, Y.; Thongchot, S.; Loilome, W.; Techasen, A.; Ungarreevittaya, P.; Khuntikeo, N.; Yongvanit, P.; Yang, J.H.; Kim, N.H.; et al. Potential role of HIF-1-responsive microRNA210/HIF3 axis on gemcitabine resistance in cholangiocarcinoma cells. PLoS ONE 2018, 13, e0199827. [Google Scholar] [CrossRef]
- Asukai, K.; Kawamoto, K.; Eguchi, H.; Konno, M.; Asai, A.; Iwagami, Y.; Yamada, D.; Asaoka, T.; Noda, T.; Wada, H.; et al. Micro-RNA-130a-3p Regulates Gemcitabine Resistance via PPARG in Cholangiocarcinoma. Ann. Surg. Oncol. 2017, 24, 2344–2352. [Google Scholar] [CrossRef]
- Li, Q.; Xia, X.; Ji, J.; Ma, J.; Tao, L.; Mo, L.; Chen, W. MiR-199a-3p enhances cisplatin sensitivity of cholangiocarcinoma cells by inhibiting mTOR signaling pathway and expression of MDR1. Oncotarget 2017, 8, 33621–33630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, D.; Yan, Y.; Shui, S.; Wu, G.; Ren, J.; Wang, Y.; Han, X. miR-106b regulates the 5-fluorouracil resistance by targeting Zbtb7a in cholangiocarcinoma. Oncotarget 2017, 8, 52913–52922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morishita, A.; Oura, K.; Tadokoro, T.; Fujita, K.; Tani, J.; Masaki, T. MicroRNAs in the Pathogenesis of Hepatocellular Carcinoma: A Review. Cancers 2021, 13, 514. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]

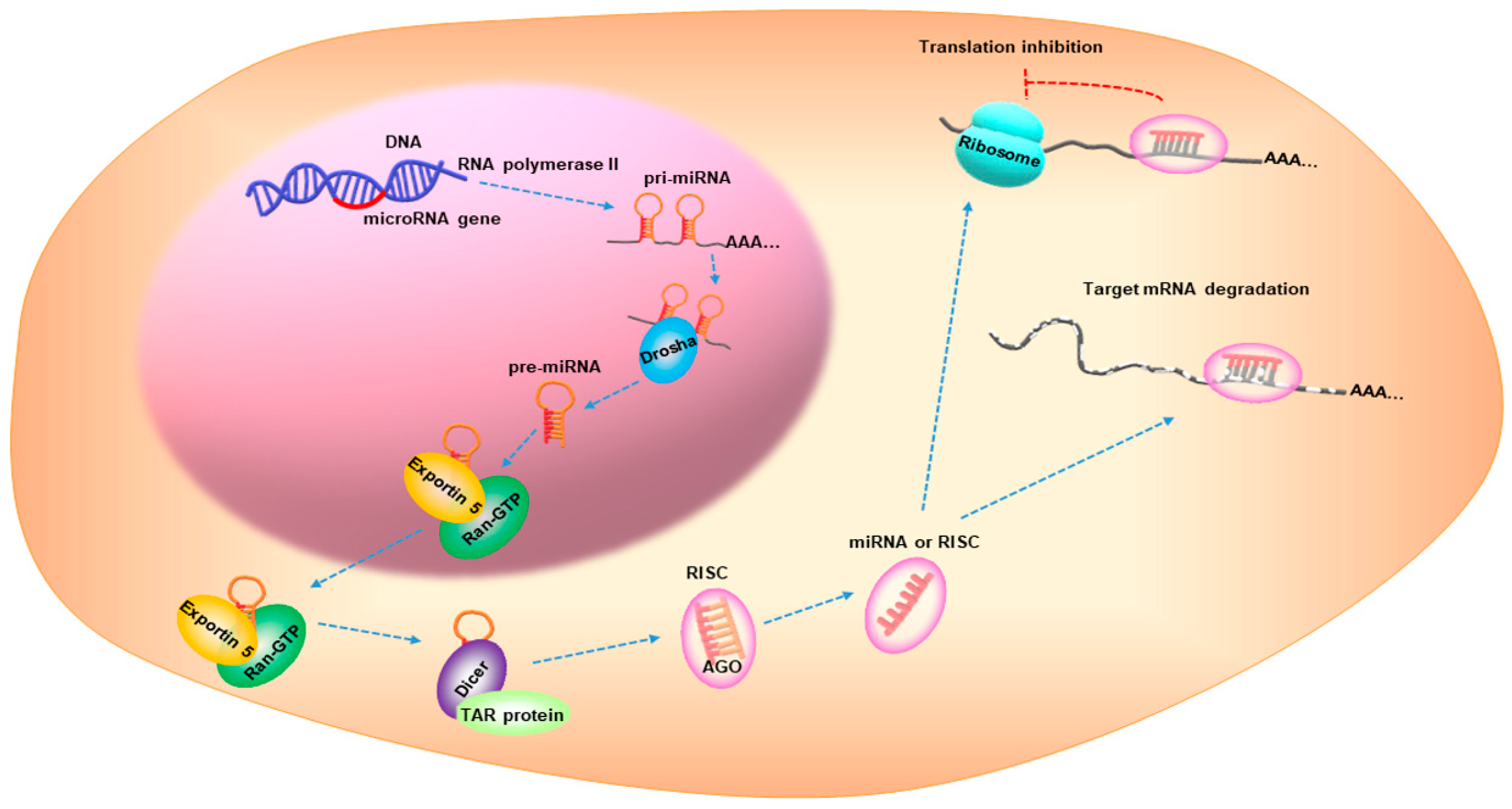
| Risk Factor | Related microRNA | Expression | Functions | References |
|---|---|---|---|---|
| Lithiasis | ||||
| Cholecystolithiasis | miR-210 | upregulated | Targets ATP11A to regulate the ABC transporter pathway | [43] |
| Hepatolithiasis | miR-130b | downregulated | miR-130b-Sp1-MUC5AC signaling pathway | [44] |
| Choledocholithiasis | unknown | unknown | unknown | |
| Cholestatic liver diseases | ||||
| PSC | miR-200b | upregulated | Promotes biliary hyperplasia and liver fibrosis | [46] |
| FPLD | unknown | unknown | unknown | |
| Parasitic infection | ||||
| miR-21 miR-96 miR-351 miR-146a/b miR-27b | unknown | Promotes fibrosis | [47,48] | |
| Cirrhosis | ||||
| miR-378 | upregulated | NF-κB-TNFα axis | [49] | |
| miR-30a | downregulated | Targets Beclin1-mediated autophagy | [50] | |
| miR-214-3p | upregulated | Decreases Sufu and promotes HSC activation | [51] | |
| Infections | ||||
| HBV | miR-192-3p | downregulated | Inhibits autophagy and suppresses HBV replication | [54] |
| miR-224, miR-1231 | unknown | Suppresses HBV replication | [55,56] | |
| miR-99 family | unknown | Promotes HBV replication | [57] | |
| HCV | miR-215 | unknown | Promotes HCV replication via targeting TRIM22 | [59] |
| miR-21-5p | upregulated | Promotes HCV replication | [60] | |
| miR-199a | unknown | Suppresses HCV replication | [62] | |
| miR-130a | unknown | Suppresses HCV replication | [63] | |
| Inflammatory disorders | ||||
| IBD | miR-301a | upregulated | Promotes mucosal inflammation | [65] |
| miR-31 | upregulated | Reduces inflammatory response | [66] | |
| Chronic pancreatitis | miR-15/16 | downregulated | Alleviates apoptosis and fibrosis | [67] |
| Metabolic abnormalities | ||||
| Diabetes | miR-10b-5p | unknown | Regulates diabetes by KLF11-KIT pathway | [69] |
| miR-690 | unknown | Improves insulin sensitivity | [70] | |
| NAFLD | miR-132 | upregulated | Alters serum and hepatic lipid profiles | [72] |
| Lifestyle | ||||
| Alcohol | miR-148a | downregulated | Regulates hepatocyte apoptosis | [73] |
| Tobacco | miR-25-3p | unknown | Promotes the development and progression of cancers | [75,76] |
| miRNA | Target Gene | Mechanism | References |
|---|---|---|---|
| Oncogenic miRNA | |||
| miR-383 | Interferon regulatory factor 1 (IRF1) | Proliferation, migration, and invasion | [77] |
| miR-221 | Phosphatase and tensin homolog (PTEN) | Epithelial–mesenchymal transition (EMT) | [78] |
| miR-31 | RAS p21 GTPase activating protein 1 (RASA1) | Proliferation and apoptosis | [79] |
| miR-361-5p | TNF receptor-associated factor 3 (TRAF3) | Apoptosis | [80] |
| miR-30a-5p | Suppressor of cytokine signaling 3 (SOCS3) | Proliferation and apoptosis | [81] |
| miR-21 | 15-hydroxyprostaglandin dehydrogenase (15-PGDH/HPGD) | Cell growth | [82] |
| miR-18a-5p | Fructose-bisphosphatase 1 (FBP1) | Proliferation, invasion, and apoptosis | [83] |
| miR-421 | Farnesoid X receptor (FXR) | Proliferation and migration | [84] |
| miR-191 | Ten-eleven translocation 1 (TET1) | Proliferation, invasion, and migration | [85] |
| miR-181c | N-myc downstream-regulated gene 2 (NDRG2) | Proliferation, chemoresistance, and metastasis | [86] |
| miR-193-3p | Transforming growth factor-β receptor type 3 (TGFBR3) | Proliferation, migration, and invasion | [87] |
| Suppressor miRNA | |||
| miR-876 | B-cell lymphoma-extra large (BCL-XL) | Proliferation and apoptosis | [88] |
| miR-451a | Activating transcription factor 2 (ATF2), ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 (ST8SIA4) | Migration, invasion, and proliferation | [89,90] |
| miR-34a | Notch receptor 1 (NOTCH1), Notch receptor 2 (NOTCH2), Jagged canonical Notch ligand 1 (JAG1) | Proliferation | [91] |
| miR-186 | Twist family BHLH transcription factor 1 (TWIST1) | Proliferation, migration, invasion, and EMT | [92] |
| miR-15a | Plasminogen activator inhibitor type-2 (PAI-2) | Migration | [93] |
| miR-612 | BCL-2 | Proliferation, migration, and invasion | [94] |
| miR-424-5p | AMPK-related protein kinase 5 (ARK5) | Metastasis, invasion, migration, and EMT | [95] |
| miR-124-3p | Ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1) | Proliferation and cell cycle arrest | [96] |
| miR-551b-3p | Cyclin D1 (CCND1) | Proliferation and apoptosis | [97] |
| miR-186-5p | Microrchidia family CW-type zinc finger 2 (MORC2) | Cell growth and metastasis | [98] |
| miR-137 | Wnt family member 2B (WNT2B) | Proliferation, migration, and invasion | [99] |
| miR-490-3p | AKIRIN2 | Proliferation, migration, invasion, and angiogenesis | [100] |
| miR-329 | Laminin subunit beta 3 (LAMB3) | Proliferation, migration, and invasion | [101] |
| miR-122-5p | Aldolase, Fructose-Bisphosphate A (ALDOA) | Proliferation, invasion, apoptosis, and EMT | [102,103] |
| miR-410 | X-linked inhibitor of apoptosis protein (XIAP) | Apoptosis | [104] |
| miR-22 | Histone deacetylase 6 (HDAC6) | Proliferation and migration | [105] |
| miR-433 | Histone deacetylase 6 (HDAC6) | Proliferation and migration | [105] |
| miR-144 | Platelet-activating factor acetylhydrolase isoform 1b (LIS1), ST8SIA4 | Proliferation and invasion | [90,106] |
| miR-590-3p | Sphingosine-1-phosphate receptor 1 (SIP1) | Invasion, migration, and EMT | [107] |
| miR-101 | Vascular endothelial growth factor (VEGF), Cyclooxygenase-2 (COX-2), E2F transcription factor 8 (E2F8) | Angiogenesis and proliferation | [108,109] |
| miR-26b-5p | S100 calcium-binding protein A7 (S100A7) | Proliferation, migration, and invasion | [110] |
| miR-1182 | NUAK1 (also known as ARK5) | Invasion, migration, and autophagy | [111] |
| let-7a | NUAK1 | Invasion, migration, and autophagy | [111] |
| miR-874 | Cyclin E1 (CCNE1) | Invasion and EMT | [112] |
| miR-885-5p | Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1), Polypeptide N-acetylgalactosaminyltransferase 3 (GALNT3) | Proliferation and metastasis | [113] |
| miR-622 | C-MYC | Proliferation, migration, and invasion | [114] |
| miR-320 | Neuropilin-1 (NRP-1) | Proliferation, invasion, and angiogenesis | [115] |
| miR-200b/c | SUZ12 Polycomb repressive complex 2 subunit (SUZ12)/Rho associated coiled-coil containing protein kinase 2 (ROCK2) | Tumorigenesis and metastasis | [116] |
| miRNA | Expression | Detectable Location | Tumor Type (Background) | Biomarker Category | References |
|---|---|---|---|---|---|
| miR-21, miR-221 | Upregulated | Plasma | Hepatolithiasis-CCA | Diagnosis/Prognosis | [45,121] |
| miR-26a | Upregulated | Serum | CCA | Diagnosis/Prognosis | [128] |
| miR-150-5p | Downregulated | Serum | CCA | Diagnosis | [129] |
| miR-29a | Upregulated | Tissue | CCA | Prognosis | [130] |
| miR-192 | Upregulated | Serum | CCA | Prognosis | [131] |
| miR-151-3p | Upregulated | Tissue | Resected CCA | Prognosis | [132] |
| miR-126 | Downregulated | Tissue | Resected CCA | Prognosis | [132] |
| miR-106a | Downregulated | Serum | CCA | Prognosis | [133] |
| miR-146a | Upregulated | Tissue | iCCA | Prognosis | [134] |
| miR-31 | Upregulated | Tissue | CCA | Prognosis | [135] |
| miR-203 | Downregulated | Tissue | CCA | Prognosis | [136] |
| miR-191 | Upregulated | Tissue | iCCA | Prognosis | [85] |
| miR-195 | Downregulated | Serum | CCA | Prognosis | [137] |
| miR-16 | Downregulated | Plasma | dCCA | Diagnosis | [138] |
| miR-877 | Upregulated | Plasma | dCCA | Diagnosis | [138] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, T.; Morishita, A.; Kobara, H.; Masaki, T. The Role of microRNAs in Cholangiocarcinoma. Int. J. Mol. Sci. 2021, 22, 7627. https://doi.org/10.3390/ijms22147627
Shi T, Morishita A, Kobara H, Masaki T. The Role of microRNAs in Cholangiocarcinoma. International Journal of Molecular Sciences. 2021; 22(14):7627. https://doi.org/10.3390/ijms22147627
Chicago/Turabian StyleShi, Tingting, Asahiro Morishita, Hideki Kobara, and Tsutomu Masaki. 2021. "The Role of microRNAs in Cholangiocarcinoma" International Journal of Molecular Sciences 22, no. 14: 7627. https://doi.org/10.3390/ijms22147627
APA StyleShi, T., Morishita, A., Kobara, H., & Masaki, T. (2021). The Role of microRNAs in Cholangiocarcinoma. International Journal of Molecular Sciences, 22(14), 7627. https://doi.org/10.3390/ijms22147627







