Transcriptome Profile Analysis of Triple-Negative Breast Cancer Cells in Response to a Novel Cytostatic Tetrahydroisoquinoline Compared to Paclitaxel
Abstract
1. Introduction
2. Results
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemistry
5.2. Cell Culture
5.3. Proliferation and Cell Viability Studies
5.4. 3D Tumor Studies
5.5. Imaging
5.6. Microarray WT 2.1 Human Datasets
5.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sturtz, L.A.; Melley, J.; Mamula, K.; Shriver, C.D.; Ellsworth, R.E. Outcome disparities in African American women with triple-negative breast cancer: A comparison of epidemiological and molecular factors between African American and Caucasian women with triple-negative breast cancer. BMC Cancer 2014, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Vidal, G.; Bursac, Z.; Miranda-Carboni, G.; White-Means, S.; Starlard-Davenport, A. Racial disparities in survival outcomes by breast tumor subtype among African American women in Memphis, Tennessee. Cancer Med. 2017, 6, 1776–1786. [Google Scholar] [CrossRef]
- Sakhuja, S.; Deveaux, A.; Wilson, L.E.; Vin-Raviv, N.; Zhang, D.; Braithwaite, D.; Altekruse, S.; Akinyemiju, T. Patterns of de-novo metastasis and breast cancer-specific mortality by race and molecular subtype in the SEER population-based dataset. Breast Cancer Res. Treat. 2020, 186, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Provencher, L.; Dent, R. Emerging trends in the treatment of triple-negative breast cancer in Canada: A survey. Curr. Oncol. 2011, 18, 180–190. [Google Scholar] [CrossRef][Green Version]
- Lorusso, V.; Latorre, A.; Giotta, F. Chemotherapy Options beyond the First Line in HER-Negative Metastatic Breast Cancer. J. Oncol. 2020, 2020, 9645294. [Google Scholar] [CrossRef]
- Ingvarsson, S. Molecular genetics of breast cancer progression. Semin. Cancer Biol. 1999, 9, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Cline, M.J.; Battifora, H.; Yokota, J. Proto-oncogene abnormalities in human breast cancer: Correlations with anatomic features and clinical course of the disease. J. Clin. Oncol. 1987, 5, 999–1006. [Google Scholar] [CrossRef]
- Ping, Z.; Xia, Y.; Shen, T.; Parekh, V.; Siegal, G.P.; Eltoum, I.E.; He, J.; Chen, D.; Deng, M.; Xi, R.; et al. A microscopic landscape of the invasive breast cancer genome. Sci. Rep. 2016, 6, 27545. [Google Scholar] [CrossRef]
- Yang, D.C.; Elliott, R.L.; Head, J.F. Gene targets of antisense therapies in breast cancer. Expert Opin. Ther. Targets 2002, 6, 375–385. [Google Scholar] [CrossRef]
- Wang, S.; Li, X.; Zhang, W.; Gao, Y.; Zhang, K.; Hao, Q.; Li, W.; Wang, Z.; Li, M.; Zhang, W.; et al. Genome-Wide Investigation of Genes Regulated by ERalpha in Breast Cancer Cells. Molecules 2018, 23, 2543. [Google Scholar] [CrossRef]
- Shabnaz, S.; Ahmed, M.U.; Islam, M.S.; Islam, M.R.; Al-Mamun, M.M.; Islam, M.S.; Hasnat, A. Breast cancer risk in relation to TP53 codon 72 and CDH1 gene polymorphisms in the Bangladeshi women. Tumour Biol. 2016, 37, 7229–7237. [Google Scholar] [CrossRef] [PubMed]
- Bareche, Y.; Venet, D.; Ignatiadis, M.; Aftimos, P.; Piccart, M.; Rothe, F.; Sotiriou, C. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann. Oncol. 2018, 29, 895–902. [Google Scholar] [CrossRef]
- Muendlein, A.; Rohde, B.H.; Gasser, K.; Haid, A.; Rauch, S.; Kinz, E.; Drexel, H.; Hofmann, W.; Schindler, V.; Kapoor, R.; et al. Evaluation of BRCA1/2 mutational status among German and Austrian women with triple-negative breast cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 2005–2012. [Google Scholar] [CrossRef] [PubMed]
- Palomba, G.; Budroni, M.; Olmeo, N.; Atzori, F.; Ionta, M.T.; Pisano, M.; Tanda, F.; Cossu, A.; Palmieri, G. Triple-negative breast cancer frequency and type of BRCA mutation: Clues from Sardinia. Oncol. Lett. 2014, 7, 948–952. [Google Scholar] [CrossRef][Green Version]
- Zhou, Z.R.; Wang, X.Y.; Yu, X.L.; Mei, X.; Chen, X.X.; Hu, Q.C.; Yang, Z.Z.; Guo, X.M. Building radiation-resistant model in triple-negative breast cancer to screening radioresistance-related molecular markers. Ann. Transl. Med. 2020, 8, 108. [Google Scholar] [CrossRef]
- Li, M.; Li, H.; Liu, F.; Bi, R.; Tu, X.; Chen, L.; Ye, S.; Cheng, X. Characterization of ovarian clear cell carcinoma using target drug-based molecular biomarkers: Implications for personalized cancer therapy. J. Ovarian Res. 2017, 10, 9. [Google Scholar] [CrossRef]
- Reddy, T.P.; Rosato, R.R.; Li, X.; Moulder, S.; Piwnica-Worms, H.; Chang, J.C. A comprehensive overview of metaplastic breast cancer: Clinical features and molecular aberrations. Breast Cancer Res. 2020, 22, 121. [Google Scholar] [CrossRef]
- Chen, J.; Liu, C.; Cen, J.; Liang, T.; Xue, J.; Zeng, H.; Zhang, Z.; Xu, G.; Yu, C.; Lu, Z.; et al. KEGG-expressed genes and pathways in triple-negative breast cancer: Protocol for a systematic review and data mining. Medicine 2020, 99, e19986. [Google Scholar] [CrossRef]
- Shome, R.; Ghosh, S.S. Tweaking EMT, and MDR dynamics to constrain triple-negative breast cancer invasiveness by EGFR and Wnt/beta-catenin signaling regulation. Cell Oncol. 2021, 44, 405–422. [Google Scholar] [CrossRef]
- Ring, A.; Kaur, P.; Lang, J.E. EP300 knockdown reduces cancer stem cell phenotype, tumor growth and metastasis in triple-negative breast cancer. BMC Cancer 2020, 20, 1076. [Google Scholar] [CrossRef] [PubMed]
- Jurj, A.; Pop, L.A.; Zanoaga, O.; Ciocan-Cartita, C.A.; Cojocneanu, R.; Moldovan, C.; Raduly, L.; Pop-Bica, C.; Trif, M.; Irimie, A.; et al. New Insights in Gene Expression Alteration as Effect of Paclitaxel Drug Resistance in Triple-Negative Breast Cancer Cells. Cell Physiol. Biochem. 2020, 54, 648–664. [Google Scholar] [PubMed]
- Zhu, C.; Ge, C.; He, J.; Zhang, X.; Feng, G.; Fan, S. Identification of Key Genes and Pathways Associated with Irradiation in Breast Cancer Tissue and Breast Cancer Cell Lines. Dose Response 2020, 18. [Google Scholar] [CrossRef] [PubMed]
- Marczyk, M.; Patwardhan, G.A.; Zhao, J.; Qu, R.; Li, X.; Wali, V.B.; Gupta, A.K.; Pillai, M.M.; Kluger, Y.; Yan, Q.; et al. Multi-Omics Investigation of Innate Navitoclax Resistance in Triple-Negative Breast Cancer Cells. Cancers 2020, 12, 2551. [Google Scholar] [CrossRef]
- Li, W.; You, Y.; Zhang, X.; Song, Y.; Xiang, H.; Peng, X.; Qin, J.; Tan, G. Amplification of chromosome 8q21-qter associated with the acquired paclitaxel resistance of nasopharyngeal carcinoma cells. Int. J. Clin. Exp. Pathol. 2015, 8, 12346–12356. [Google Scholar] [PubMed]
- Loftus, P.G.; Watson, L.; Deedigan, L.M.; Camarillo-Retamosa, E.; Dwyer, R.M.; O’Flynn, L.; Alagesan, S.; Griffin, M.; O’Brien, T.; Kerin, M.J.; et al. Targeting stromal cell Syndecan-2 reduces breast tumour growth, metastasis and limits immune evasion. Int. J. Cancer 2021, 148, 1245–1259. [Google Scholar] [CrossRef]
- Lian, B.; Pei, Y.C.; Jiang, Y.Z.; Xue, M.Z.; Li, D.Q.; Li, X.G.; Zheng, Y.Z.; Liu, X.Y.; Qiao, F.; Sun, W.L.; et al. Truncated HDAC9 identified by the integrated genome-wide screen as the key modulator for paclitaxel resistance in triple-negative breast cancer. Theranostics 2020, 10, 11092–11109. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Jiang, M.; Zhang, F.; Ma, Y.; Wang, H.; Xu, Y. ZGRF1 Is Associated with Poor Prognosis in Triple-Negative Breast Cancer and Promotes Cancer Stemness Based on Bioinformatics. Oncol. Targets Ther. 2020, 13, 2843–2854. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, J.; Zhang, W.; Liu, G.; Yin, D.; Li, J.; Zhang, S.; Li, H. Establishment of paclitaxel-resistant cell line and the underlying mechanism on drug resistance. Int. J. Gynecol. Cancer 2012, 22, 1450–1456. [Google Scholar] [CrossRef]
- Peng, X.; Tan, G. Differential expression of taxol resistance and taxol resistance reversal related genes in nasopharyngeal carcinoma by cDNA microarray. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2012, 37, 48–52. [Google Scholar]
- Lai, D.; Ho, K.C.; Hao, Y.; Yang, X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011, 71, 2728–2738. [Google Scholar] [CrossRef]
- Li, W.; Tan, G.L.; Ma, Y.H.; Peng, X.W.; He, G.X. Role of folate receptor 1 in paclitaxel-resistance of nasopharyngeal carcinoma cells]. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 2010, 45, 1035–1040. [Google Scholar]
- Karam, J.A.; Huang, S.; Fan, J.; Stanfield, J.; Schultz, R.A.; Pong, R.C.; Sun, X.; Mason, R.P.; Xie, X.J.; Niu, G.; et al. Upregulation of TRAG3 gene in urothelial carcinoma of the bladder. Int. J. Cancer 2011, 128, 2823–2832. [Google Scholar] [CrossRef] [PubMed]
- Moelans, C.B.; Verschuur-Maes, A.H.; van Diest, P.J. Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. J. Pathol. 2011, 225, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, W.; Li, H.; Ma, Y.; He, G.; Tan, G. Genomic methylation profiling combined with gene expression microarray reveals the aberrant methylation mechanism involved in nasopharyngeal carcinoma taxol resistance. Anticancer Drugs 2012, 23, 856–864. [Google Scholar] [CrossRef]
- Peng, X.; Cao, P.; He, D.; Han, S.; Zhou, J.; Tan, G.; Li, W.; Yu, F.; Yu, J.; Li, Z.; et al. MiR-634 sensitizes nasopharyngeal carcinoma cells to paclitaxel and inhibits cell growth both in vitro and in vivo. Int. J. Clin. Exp. Pathol. 2014, 7, 6784–6791. [Google Scholar]
- Schettini, F.; Giuliano, M.; De Placido, S.; Arpino, G. Nab-paclitaxel for the treatment of triple-negative breast cancer: Rationale, clinical data, and future perspectives. Cancer Treat. Rev. 2016, 50, 129–141. [Google Scholar] [CrossRef]
- Gangapuram, M.; Jean, R.; Mazzio, E.; Badisa, R.; Eyunni, S.; Goodman, C.B.; Redda, K.K.; Soliman, K.F. Substituted Tetrahydroisoquinolines as Microtubule-destabilizing Agents in Triple-Negative Human Breast Cancer Cells. Anticancer Res. 2016, 36, 5043–5052. [Google Scholar] [CrossRef]
- Hajek, R. Paclitaxel (Taxol). Cas. Lek. Ces. 1996, 135, 393–396. [Google Scholar]
- Paclitaxel and docetaxel in breast and ovarian cancer. Drug Ther. Bull. 1997, 35, 43–46. [CrossRef]
- Geney, R.; Sun, L.; Pera, P.; Bernacki, R.J.; Xia, S.; Horwitz, S.B.; Simmerling, C.L.; Ojima, I. Use of the tubulin bound paclitaxel conformation for structure-based rational drug design. Chem. Biol. 2005, 12, 339–348. [Google Scholar] [CrossRef]
- El-Sahli, S.; Hua, K.; Sulaiman, A.; Chambers, J.; Li, L.; Farah, E.; McGarry, S.; Liu, D.; Zheng, P.; Lee, S.H.; et al. A triple-drug nanotherapy to target breast cancer cells, cancer stem cells, and tumor vasculature. Cell Death Dis. 2021, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Castle, B.T.; McCubbin, S.; Prahl, L.S.; Bernens, J.N.; Sept, D.; Odde, D.J. Mechanisms of kinetic stabilization by the drugs paclitaxel and vinblastine. Mol. Biol. Cell 2017, 28, 1238–1257. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sun, X. Regulation of paclitaxel activity by microtubule-associated proteins in cancer chemotherapy. Cancer Chemother. Pharmacol. 2017, 80, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Naaz, F.; Haider, M.R.; Shafi, S.; Yar, M.S. Anti-tubulin agents of natural origin: Targeting taxol, vinca, and colchicine binding domains. Eur. J. Med. Chem. 2019, 171, 310–331. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.C.; Cassimeris, L. Reorganization of paclitaxel-stabilized microtubule arrays at mitotic entry: Roles of depolymerizing kinesins and severing proteins. Cancer Biol. Ther. 2019, 20, 1337–1347. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Liu, X.; Guan, Q.; Ding, N.; Wei, Q.; Tong, B.; Zhao, M.; Zhang, W.; Ma, L. 5-(3,4,5-trimethoxybenzoyl)-4-methyl-2-(p-tolyl) imidazole (BZML) targets tubulin and DNA to induce anticancer activity and overcome multidrug resistance in colorectal cancer cells. Chem. Biol. Interact. 2020, 315, 108886. [Google Scholar] [CrossRef]
- Xie, S.; Ogden, A.; Aneja, R.; Zhou, J. Microtubule-Binding Proteins as Promising Biomarkers of Paclitaxel Sensitivity in Cancer Chemotherapy. Med. Res. Rev. 2016, 36, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.N.; Zheng, L.L.; Wang, D.; Liang, X.X.; Gao, F.; Zhou, X.L. Recent advances in microtubule-stabilizing agents. Eur. J. Med. Chem. 2018, 143, 806–828. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, W.; Tang, K.; Chen, X.; Feng, Z.; Chen, J. Silencing Aurora-A leads to re-sensitization of breast cancer cells to Taxol through downregulation of SRC-mediated ERK and mTOR pathways. Oncol. Rep. 2017, 38, 2011–2022. [Google Scholar] [CrossRef][Green Version]
- Li, Y.; Tang, K.; Zhang, H.; Zhang, Y.; Zhou, W.; Chen, X. Function of Aurora kinase A in Taxol-resistant breast cancer and its correlation with P-gp. Mol. Med. Rep. 2011, 4, 739–746. [Google Scholar]
- Habu, T.; Matsumoto, T. p31(comet) inactivates the chemically induced Mad2-dependent spindle assembly checkpoint and leads to resistance to anti-mitotic drugs. Springerplus 2013, 2, 562. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marcus, A.I.; Peters, U.; Thomas, S.L.; Garrett, S.; Zelnak, A.; Kapoor, T.M.; Giannakakou, P. Mitotic kinesin inhibitors induce mitotic arrest and cell death in Taxol-resistant and -sensitive cancer cells. J. Biol. Chem. 2005, 280, 11569–11577. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Escolano, M.; Montoyo-Pujol, Y.G.; Ortiz-Martinez, F.; Ponce, J.J.; Delgado-Garcia, S.; Martin, T.A.; Ballester, H.; Aranda, F.I.; Castellon-Molla, E.; Sempere-Ortells, J.M.; et al. ID1, and ID4 Are Biomarkers of Tumor Aggressiveness and Poor Outcome in Immunophenotypes of Breast Cancer. Cancers 2021, 13, 492. [Google Scholar] [CrossRef] [PubMed]
- Zekan, D.S.; Dahman, A.; Hajiran, A.J.; Luchey, A.M.; Chahoud, J.; Spiess, P.E. Prognostic predictors of lymph node metastasis in penile cancer: A systematic review. Int. Braz. J. Urol. 2021, 47, 943–956. [Google Scholar]
- Sharma, P.; Patel, D.; Chaudhary, J. Id1 and Id3 expression is associated with increasing grade of prostate cancer: Id3 preferentially regulates CDKN1B. Cancer Med. 2012, 1, 187–197. [Google Scholar] [CrossRef]
- Thankamony, A.P.; Murali, R.; Karthikeyan, N.; Varghese, B.A.; Teo, W.S.; McFarland, A.; Roden, D.L.; Holliday, H.; Konrad, C.V.; Cazet, A.; et al. Targeting the Id1-Kif11 Axis in Triple-Negative Breast Cancer Using Combination Therapy. Biomolecules 2020, 10, 1295. [Google Scholar] [CrossRef]
- Teo, W.S.; Holliday, H.; Karthikeyan, N.; Cazet, A.S.; Roden, D.L.; Harvey, K.; Konrad, C.V.; Murali, R.; Varghese, B.A.; Thankamony, A.P.; et al. Id Proteins Promote a Cancer Stem Cell Phenotype in Mouse Models of Triple-Negative Breast Cancer via Negative Regulation of Robo1. Front. Cell Dev. Biol. 2020, 8, 552. [Google Scholar] [CrossRef]
- Tasdemir, N.; Ding, K.; Savariau, L.; Levine, K.M.; Du, T.; Elangovan, A.; Bossart, E.A.; Lee, A.V.; Davidson, N.E.; Oesterreich, S. Proteomic and transcriptomic profiling identifies mediators of anchorage-independent growth and roles of inhibitor of differentiation proteins in invasive lobular carcinoma. Sci. Rep. 2020, 10, 11487. [Google Scholar] [CrossRef]
- Wahdan-Alaswad, R.; Harrell, J.C.; Fan, Z.; Edgerton, S.M.; Liu, B.; Thor, A.D. Metformin attenuates transforming growth factor-beta (TGF-beta) mediated oncogenesis in mesenchymal stem-like/claudin-low triple-negative breast cancer. Cell Cycle 2016, 15, 1046–1059. [Google Scholar] [CrossRef]
- Castanon, E.; Bosch-Barrera, J.; Lopez, I.; Collado, V.; Moreno, M.; Lopez-Picazo, J.M.; Arbea, L.; Lozano, M.D.; Calvo, A.; Gil-Bazo, I. Id1 and Id3 co-expression correlates with clinical outcome in stage III-N2 non-small cell lung cancer patients treated with definitive chemoradiotherapy. J. Transl. Med. 2013, 11, 13. [Google Scholar] [CrossRef]
- Chen, Y.H.; Wu, Z.Q.; Zhao, Y.L.; Si, Y.L.; Guo, M.Z.; Han, W.D. FHL2 inhibits the Id3-promoted proliferation and invasive growth of human MCF-7 breast cancer cells. Chin. Med. J. 2012, 125, 2329–2333. [Google Scholar] [PubMed]
- Lin, S.; Mei, W.; Lai, H.; Li, X.; Weng, H.; Xiong, J.; Lin, X.; Zeng, T.; Zhang, Q.; Liu, X.; et al. Cigarette smoking promotes keratinocyte malignancy via generation of cancer stem-like cells. J. Cancer 2021, 12, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Bo, Z.; Gong, W.; Guo, Y. Inhibitor of Differentiation 1 (Id1) in Cancer and Cancer Therapy. Int. J. Med. Sci. 2020, 17, 995–1005. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Kreso, A.; Ryan, P.; Hermans, K.G.; Gibson, L.; Wang, Y.; Tsatsanis, A.; Gallinger, S.; Dick, J.E. ID1, and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell 2012, 21, 777–792. [Google Scholar] [CrossRef]
- Yang, H.Y.; Liu, H.L.; Liu, G.Y.; Zhu, H.; Meng, Q.W.; Qu, L.D.; Liu, L.X.; Jiang, H.C. Expression and prognostic values of Id-1 and Id-3 in gastric adenocarcinoma. J. Surg. Res. 2011, 167, 258–266. [Google Scholar] [CrossRef]
- Shuno, Y.; Tsuno, N.H.; Okaji, Y.; Tsuchiya, T.; Sakurai, D.; Nishikawa, T.; Yoshikawa, N.; Sasaki, K.; Hongo, K.; Tsurita, G.; et al. Id1/Id3 knockdown inhibits metastatic potential of pancreatic cancer. J. Surg. Res. 2010, 161, 76–82. [Google Scholar] [CrossRef]
- Cheung, H.W.; Ling, M.T.; Tsao, S.W.; Wong, Y.C.; Wang, X. Id-1-induced Raf/MEK pathway activation is essential for its protective role against taxol-induced apoptosis in nasopharyngeal carcinoma cells. Carcinogenesis 2004, 25, 881–887. [Google Scholar] [CrossRef]
- Meng, J.; Liu, K.; Shao, Y.; Feng, X.; Ji, Z.; Chang, B.; Wang, Y.; Xu, L.; Yang, G. ID1 confers cancer cell chemoresistance through STAT3/ATF6-mediated induction of autophagy. Cell Death Dis. 2020, 11, 137. [Google Scholar] [CrossRef]
- Liu, K.; Chen, X.; Wu, L.; Chen, S.; Fang, N.; Cai, L.; Jia, J. ID1 mediates resistance to osimertinib in EGFR T790M-positive non-small cell lung cancer through epithelial-mesenchymal transition. BMC Pulm. Med. 2021, 21, 163. [Google Scholar] [CrossRef]
- He, Y.; Wei, T.; Ye, Z.; Orme, J.J.; Lin, D.; Sheng, H.; Fazli, L.; Jeffrey Karnes, R.; Jimenez, R.; Wang, L.; et al. A noncanonical AR addiction drives enzalutamide resistance in prostate cancer. Nat. Commun. 2021, 12, 1521. [Google Scholar] [CrossRef]
- Kim, H.; Chung, H.; Kim, H.J.; Lee, J.Y.; Oh, M.Y.; Kim, Y.; Kong, G. Id-1 regulates Bcl-2 and Bax expression through p53 and NF-kappaB in MCF-7 breast cancer cells. Breast Cancer Res. Treat. 2008, 112, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, C.; Zhao, X.; Maroni, D.; Band, V.; Naramura, M. Distinct effects of EGFR ligands on human mammary epithelial cell differentiation. PLoS ONE 2013, 8, e75907. [Google Scholar] [CrossRef] [PubMed]
- Olsen, D.A.; Bechmann, T.; Ostergaard, B.; Wamberg, P.A.; Jakobsen, E.H.; Brandslund, I. Increased concentrations of growth factors and activation of the EGFR system in breast cancer. Clin. Chem. Lab. Med. 2012, 50, 1809–1818. [Google Scholar] [CrossRef]
- Peterson, E.A.; Jenkins, E.C.; Lofgren, K.A.; Chandiramani, N.; Liu, H.; Aranda, E.; Barnett, M.; Kenny, P.A. Amphiregulin Is a Critical Downstream Effector of Estrogen Signaling in ERalpha-Positive Breast Cancer. Cancer Res. 2015, 75, 4830–4838. [Google Scholar] [CrossRef]
- Schmucker, H.; Blanding, W.M.; Mook, J.M.; Wade, J.F.; Park, J.P.; Kwist, K.; Shah, H.; Booth, B.W. Amphiregulin regulates proliferation and migration of HER2-positive breast cancer cells. Cell Oncol. 2018, 41, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.B.; Soloff, A.C.; Ethier, S.P.; Yeh, E.S. Perspectives on Epidermal Growth Factor Receptor Regulation in Triple-Negative Breast Cancer: Ligand-Mediated Mechanisms of Receptor Regulation and Potential for Clinical Targeting. Adv. Cancer Res. 2015, 127, 253–281. [Google Scholar] [PubMed]
- Kefaloyianni, E.; Keerthi Raja, M.R.; Schumacher, J.; Muthu, M.L.; Krishnadoss, V.; Waikar, S.S.; Herrlich, A. Proximal Tubule-Derived Amphiregulin Amplifies and Integrates Profibrotic EGF Receptor Signals in Kidney Fibrosis. J. Am. Soc. Nephrol. 2019, 30, 2370–2383. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jing, Y.; Ding, L.; Zhang, X.; Song, Y.; Chen, S.; Zhao, X.; Huang, X.; Pu, Y.; Wang, Z.; et al. Epiregulin reprograms cancer-associated fibroblasts and facilitates oral squamous cell carcinoma invasion via JAK2-STAT3 pathway. J. Exp. Clin. Cancer Res. 2019, 38, 274. [Google Scholar] [CrossRef]
- Rajaram, M.; Li, J.; Egeblad, M.; Powers, R.S. System-wide analysis reveals a complex network of tumor-fibroblast interactions involved in tumorigenicity. PLoS Genet. 2013, 9, e1003789. [Google Scholar] [CrossRef]
- Xiang, G.; Liu, F.; Liu, J.; Meng, Q.; Li, N.; Niu, Y. Prognostic role of amphiregulin and the correlation with androgen receptor in invasive breast cancer. Pathol. Res. Pract. 2019, 215, 152414. [Google Scholar] [CrossRef]
- Higginbotham, J.N.; Demory Beckler, M.; Gephart, J.D.; Franklin, J.L.; Bogatcheva, G.; Kremers, G.J.; Piston, D.W.; Ayers, G.D.; McConnell, R.E.; Tyska, M.J.; et al. Amphiregulin exosomes increase cancer cell invasion. Curr. Biol. 2011, 21, 779–786. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef]
- West, N.R. Coordination of Immune-Stroma Crosstalk by IL-6 Family Cytokines. Front. Immunol. 2019, 10, 1093. [Google Scholar] [CrossRef]
- Peng, Z.P.; Jiang, Z.Z.; Guo, H.F.; Zhou, M.M.; Huang, Y.F.; Ning, W.R.; Huang, J.H.; Zheng, L.; Wu, Y. Glycolytic activation of monocytes regulates the accumulation and function of neutrophils in human hepatocellular carcinoma. J. Hepatol. 2020, 73, 906–917. [Google Scholar] [CrossRef]
- Tanaka, H.; Nishioka, Y.; Yokoyama, Y.; Higashiyama, S.; Matsuura, N.; Matsuura, S.; Hieda, M. Nuclear envelope-localized EGF family protein amphiregulin activates breast cancer cell migration in an EGF-like domain-independent manner. Biochem. Biophys. Res. Commun. 2012, 420, 721–726. [Google Scholar] [CrossRef]
- Rego, S.L.; Helms, R.S.; Dreau, D. Tumor necrosis factor-alpha-converting enzyme activities, and tumor-associated macrophages in breast cancer. Immunol. Res. 2014, 58, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Taverna, S.; Pucci, M.; Giallombardo, M.; Di Bella, M.A.; Santarpia, M.; Reclusa, P.; Gil-Bazo, I.; Rolfo, C.; Alessandro, R. Amphiregulin contained in NSCLC-exosomes induces osteoclast differentiation through the activation of EGFR pathway. Sci. Rep. 2017, 7, 3170. [Google Scholar] [CrossRef]
- Kim, J.W.; Kim, D.K.; Min, A.; Lee, K.H.; Nam, H.J.; Kim, J.H.; Kim, J.S.; Kim, T.Y.; Im, S.A.; Park, I.A. Amphiregulin confers trastuzumab resistance via AKT and ERK activation in HER2-positive breast cancer. J. Cancer Res. Clin. Oncol. 2016, 142, 157–165. [Google Scholar] [CrossRef]
- Revillion, F.; Lhotellier, V.; Hornez, L.; Bonneterre, J.; Peyrat, J.P. ErbB/HER ligands in human breast cancer, and relationships with their receptors, the bio-pathological features and prognosis. Ann. Oncol. 2008, 19, 73–80. [Google Scholar] [CrossRef]
- Katoh, Y.; Katoh, M. Canonical WNT signaling pathway and human AREG. Int. J. Mol. Med. 2006, 17, 1163–1166. [Google Scholar] [CrossRef]
- Nickerson, N.K.; Mill, C.P.; Wu, H.J.; Riese, D.J., 2nd; Foley, J. Autocrine-derived epidermal growth factor receptor ligands contribute to the recruitment of tumor-associated macrophage and growth of basal breast cancer cells in vivo. Oncol. Res. 2013, 20, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Riese, D.J., 2nd; Cullum, R.L. Epiregulin: Roles in normal physiology and cancer. Semin. Cell Dev. Biol. 2014, 28, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zhou, Y.; Yong, H.; Wang, X.; Zhao, W.; Ding, G.; Zhu, J.; Li, X.; Feng, Z.; Wang, B. Elevated epiregulin expression predicts poor prognosis in gastric cancer. Pathol. Res. Pract. 2019, 215, 873–879. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Jin, Q.; Chen, C.; Liu, Y.; Ye, X.; Jiang, Y.; Ji, F.; Qian, H.; Gan, D.; Yue, S.; et al. The miR-186-3p/EREG axis orchestrates tamoxifen resistance and aerobic glycolysis in breast cancer cells. Oncogene 2019, 38, 5551–5565. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.; Han, Y.; Xia, W.; Zhang, L.; Xu, S.; Ju, H.; Zhang, X.; Ren, G.; Liu, L.; et al. EREG-driven oncogenesis of Head and Neck Squamous Cell Carcinoma exhibits higher sensitivity to Erlotinib therapy. Theranostics 2020, 10, 10589–10605. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, A.; Pina-Oviedo, S.; Yuan, Y.; Huczynski, A.; Chambers, T.C. Limitations of an ex vivo breast cancer model for studying the mechanism of action of the anticancer drug paclitaxel. Eur. J. Pharmacol. 2021, 891, 173780. [Google Scholar] [CrossRef]
- Bonnet, J.; Rigal, L.; Mondesert, O.; Morin, R.; Corsaut, G.; Vigneau, M.; Ducommun, B.; Lobjois, V. Mitotic arrest affects clustering of tumor cells. Cell Div. 2021, 16, 2. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39 (Suppl. 1), D561–D568. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
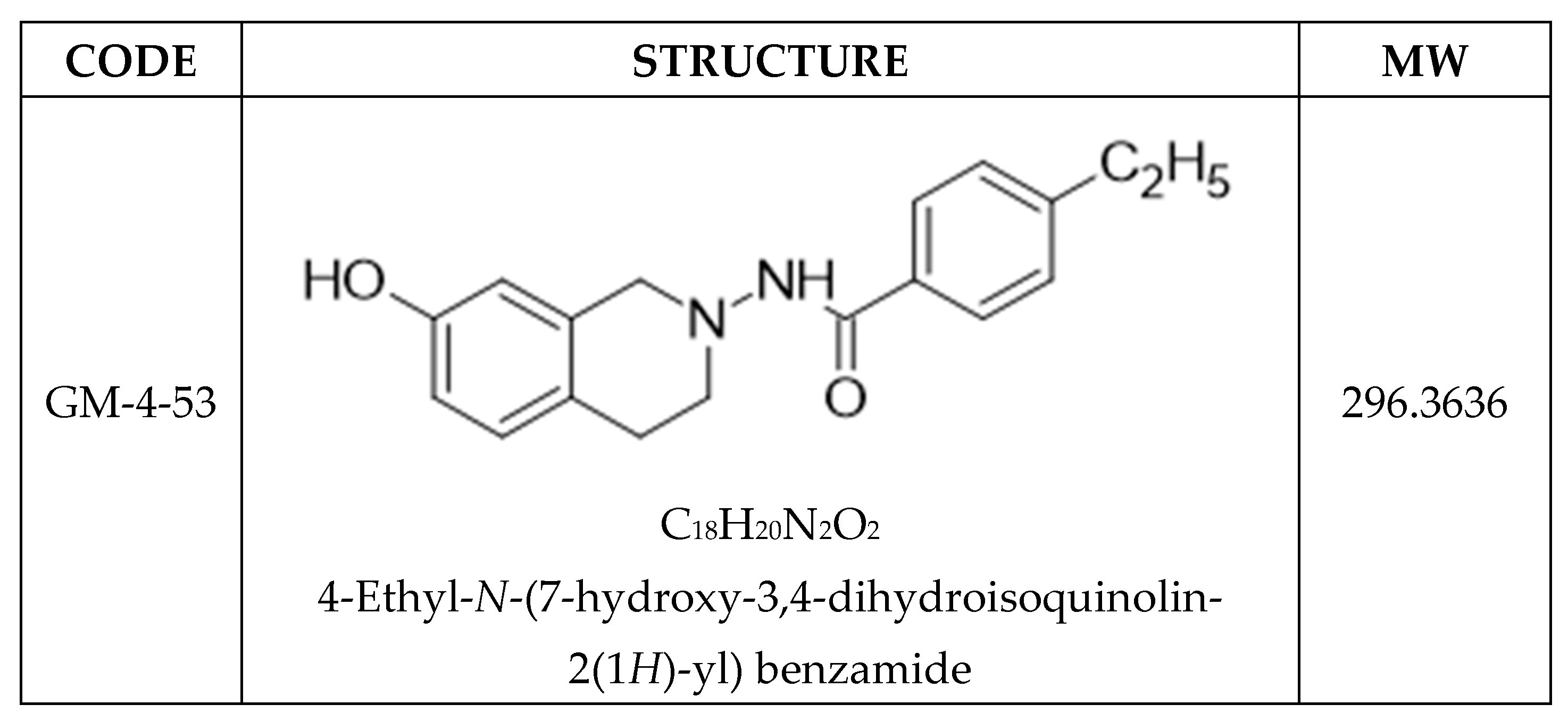





| MCF-7 | Ishikawa | MDA-MB-231 | |
|---|---|---|---|
| Human Breast Cancer (ER+, PR+) | Human Endometrial Cancer | Human Breast Cancer (TNBC) (ER−) (PR−) | |
| IG50 (nM) | IG50 (nM) | IG50 (nM) | |
| GM-4-53 | 674.01 | 269.61 | 261.26 |
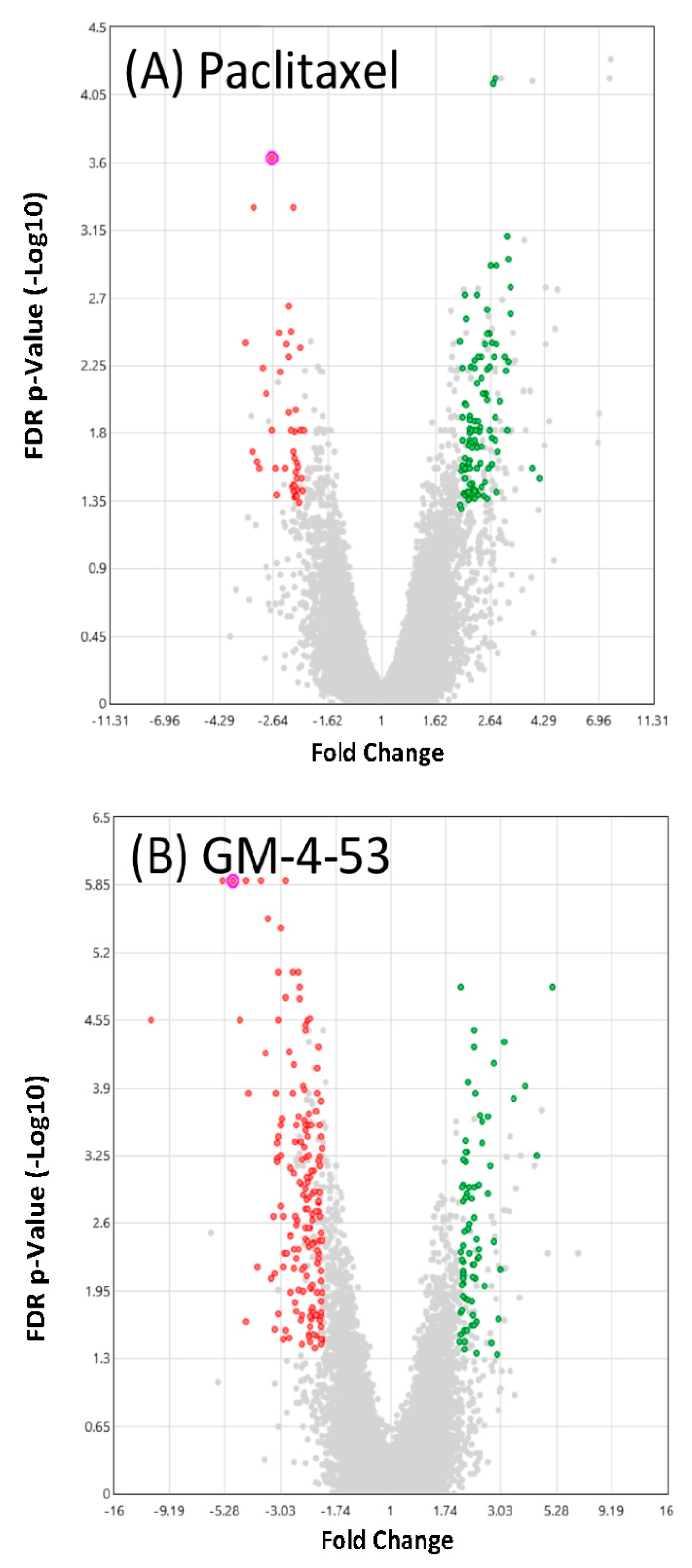
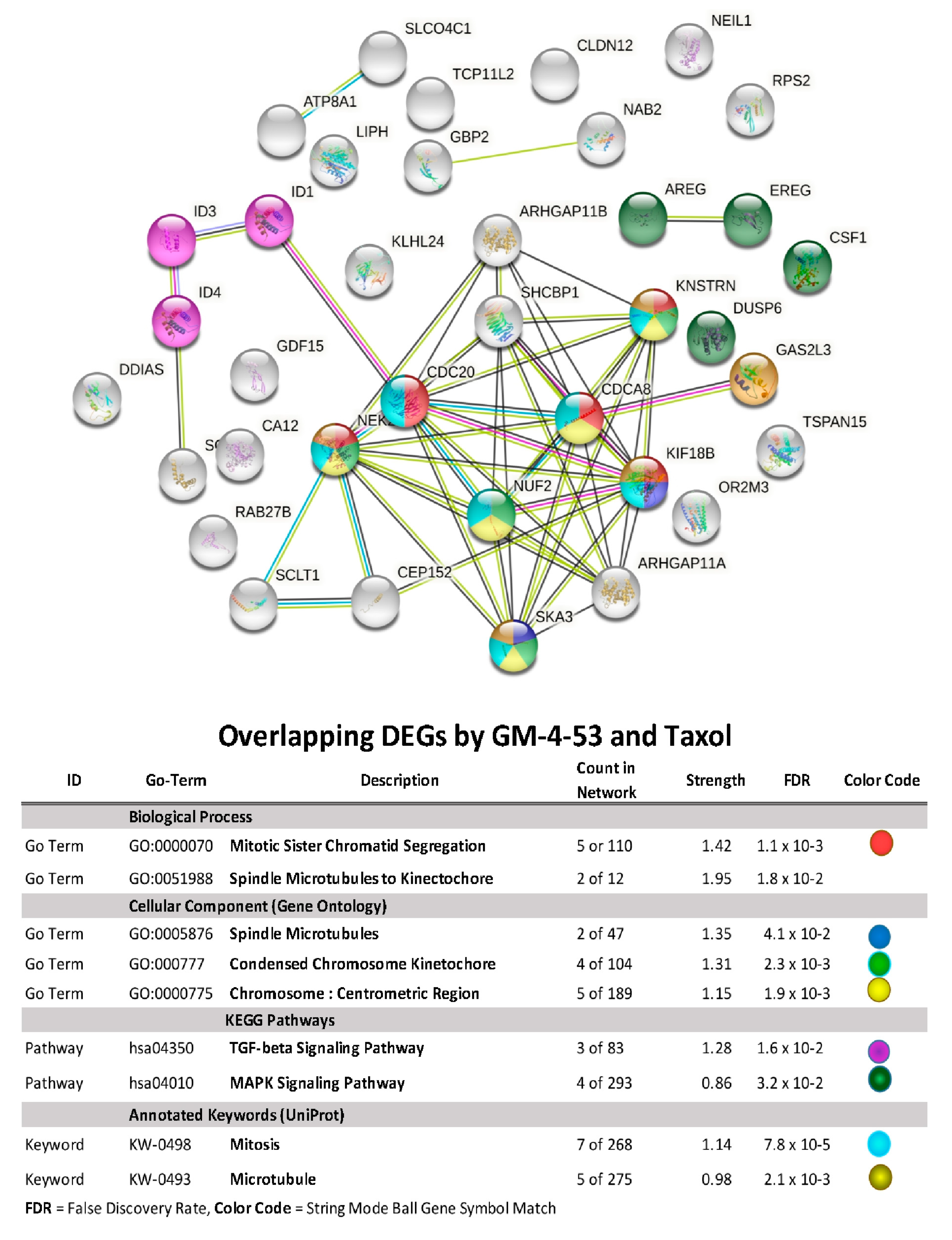
| Paclitaxel | GM-4-53 | ||||||
|---|---|---|---|---|---|---|---|
| Symbol | Description | FC | p-Value | FDR p-Value | FC | p-Value | FDR p-Value |
| AREG | amphiregulin | −3.06 | 1.0 × 10−4 | 2.5 × 10−2 | −2.74 | 1.0 × 10−4 | 1.2 × 10−2 |
| ARHGAP11A | Rho GTPase activating protein 11A | 2.64 | 3.3 × 10−6 | 1.2 × 10−3 | 2.08 | 3.3 × 10−6 | 1.1 × 10−3 |
| ARHGAP11B | Rho GTPase activating protein 11B | 3.10 | 6.8 × 10−6 | 5.3 × 10−3 | 2.38 | 3.9 × 10−5 | 5.5 × 10−3 |
| ATP8A1 | ATPase, (APLT), class I, 8A, m1 | −2.27 | 2.2 × 10−6 | 3.3 × 10−3 | −2.88 | 4.9 × 10−9 | 1.7 × 10−5 |
| CA12 | carbonic anhydrase XII | −3.17 | 8.6 × 10−8 | 5.0 × 10−4 | −4.88 | 3.0 × 10−11 | 1.3 × 10−6 |
| CDC20 | cell division cycle 20 | 2.49 | 3.0 × 10−4 | 4.2 × 10−2 | 2.11 | 7.0 × 10−4 | 3.5 × 10−2 |
| CDCA8 | cell division cycle associated 8 | 2.30 | 6.7 × 10−5 | 1.9 × 10−2 | 2.20 | 3.2 × 10−5 | 4.9 × 10−3 |
| CEP152 | centrosomal protein 152 kDa | 2.25 | 2.0 × 10−4 | 3.4 × 10−2 | 2.41 | 3.6 × 10−5 | 5.2 × 10−3 |
| CLDN12 | claudin 12 | 3.07 | 1.6 × 10−7 | 8.0 × 10−4 | 2.11 | 5.0 × 10−6 | 1.4 × 10−3 |
| CSF1 | colony stimulating factor 1 | −2.27 | 2.1 × 10−6 | 3.3 × 10−3 | −3.70 | 8.8 × 10−11 | 1.3 × 10−6 |
| DDIAS | DNA damage-induced apoptosis suppressor | 2.13 | 1.6 × 10−6 | 2.8 × 10−3 | 2.11 | 7.1 × 10−7 | 4.0 × 10−4 |
| DUSP6 | dual specificity phosphatase 6 | −2.21 | 9.2 × 10−8 | 5.0 × 10−4 | −2.86 | 8.8 × 10−11 | 1.3 × 10−6 |
| EREG | epiregulin | −3.19 | 8.6 × 10−5 | 2.1 × 10−2 | −2.96 | 9.4 × 10−6 | 2.2 × 10−3 |
| GAS2L3 | growth arrest-specific 2 like 3 | 2.61 | 7.9 × 10−6 | 5.8 × 10−3 | 2.08 | 5.9 × 10−5 | 7.3 × 10−3 |
| GBP2 | guanylate binding protein 2, interferon-inducible | −2.81 | 1.7 × 10−5 | 8.6 × 10−3 | −3.00 | 1.2 × 10−6 | 6.0 × 10−4 |
| GDF15 | growth differentiation factor 15 | −2.18 | 4.0 × 10−4 | 4.2 × 10−2 | −2.03 | 6.0 × 10−5 | 7.3 × 10−3 |
| ID1 | inhibitor of DNA binding 1, dom-HLHP | −2.15 | 2.0 × 10−4 | 3.2 × 10−2 | −5.43 | 1.3 × 10−10 | 1.3 × 10−6 |
| ID3 | inhibitor of DNA binding 3, dom-HLHP | −2.21 | 8.5 × 10−4 | 2.1 × 10−2 | −4.32 | 1.4 × 10−10 | 1.3 × 10−6 |
| ID4 | inhibitor of DNA binding 4, dom-HLHP | −2.49 | 1.0 × 10−5 | 6.2 × 10−3 | −3.16 | 1.4 × 10−7 | 1.0 × 10−4 |
| KIF18B | kinesin family member 18B | 2.19 | 2.8 × 10−5 | 1.2 × 10−2 | 2.24 | 5.2 × 10−6 | 1.4 × 10−3 |
| KLHL24 | kelch-like family member 24 | −2.12 | 1.0 × 10−4 | 2.7 × 10−2 | −2.62 | 7.6 × 10−7 | 4.0 × 10−4 |
| KNSTRN | kinetochore-loc. astrin/SPAG5bp | 2.80 | 3.6 × 10−7 | 1.2 × 10−3 | 2.15 | 4.5 × 10−6 | 1.3 × 10−3 |
| LIPH | lipase, member H | −2.51 | 2.5 × 10−6 | 3.4 × 10−3 | −2.38 | 8.8 × 10−7 | 5.0 × 10−4 |
| LPAL2 | lipoprotein, Lp(a)-like 2, pg | −2.11 | 4.0 × 10−4 | 4.6 × 10−2 | −2.58 | 1.3 × 10−5 | 2.6 × 10−3 |
| NAB2 | NGFI-A binding protein 2 (EGR1 binding protein 2) | −2.05 | 3.0 × 10−4 | 3.9 × 10−2 | −2.20 | 8.6 × 10−6 | 2.0 × 10−3 |
| NEIL1 | nei-like DNA glycosylase 1 | −2.01 | 4.7 × 10−5 | 1.5 × 10−2 | −2.31 | 6.5 × 10−7 | 4.0 × 10−4 |
| NEK2 | NIMA-related kinase 2 | 3.05 | 4.7 × 10−5 | 1.5 × 10−2 | 2.00 | 7.0 × 10−4 | 3.5 × 10−2 |
| NUF2 | NUF2, NDC80 kinetochore com c | 2.67 | 5.9 × 10−5 | 1.7 × 10−2 | 2.03 | 5.0 × 10−4 | 2.9 × 10−2 |
| PELO; ITGA1 | pelota hom (Dros); integrin alpha 1 | −2.21 | 2.0 × 10−4 | 3.5 × 10−2 | −2.66 | 2.3 × 10−6 | 8.0 × 10−4 |
| RAB27B | RAB27B, member RAS oncogene family | −2.26 | 4.1 × 10−5 | 1.5 × 10−2 | −2.27 | 1.1 × 10−6 | 6.0 × 10−4 |
| RPS2 | ribosomal protein S2 | 3.17 | 5.4 × 10−7 | 1.7 × 10−3 | 2.41 | 3.3 × 10−6 | 1.1 × 10−3 |
| SCLT1 | sodium channel/clathrin linker 1 | 3.09 | 2.7 × 10−7 | 1.1 × 10−3 | 2.30 | 3.5 × 10−6 | 1.1 × 10−3 |
| SHCBP1 | SHC SH2-domain binding protein 1 | 2.39 | 5.1 × 10−5 | 1.6 × 10−2 | 2.06 | 6.4 × 10−5 | 7.7 × 10−3 |
| SKA3 | Spindle & Kinetochore complex s3 | 2.33 | 1.0 × 10−4 | 2.7 × 10−2 | 2.12 | 2.0 × 10−4 | 1.3 × 10−2 |
| SLCO4C1 | SC organic anion transporter fm4C1 | −3.40 | 3.7 × 10−6 | 4.0 × 10−3 | −4.53 | 1.2 × 10−8 | 2.8 × 10−5 |
| SOX4 | SRY box 4 | −2.31 | 5.4 × 10−6 | 4.9 × 10−3 | −3.08 | 2.0 × 10−9 | 9.6 × 10−6 |
| TSPAN15 | tetraspanin 15 | −2.31 | 1.1 × 10−6 | 2.3 × 10−3 | −2.41 | 9.7 × 10−8 | 1.0 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gangapuram, M.; Mazzio, E.A.; Redda, K.K.; Soliman, K.F.A. Transcriptome Profile Analysis of Triple-Negative Breast Cancer Cells in Response to a Novel Cytostatic Tetrahydroisoquinoline Compared to Paclitaxel. Int. J. Mol. Sci. 2021, 22, 7694. https://doi.org/10.3390/ijms22147694
Gangapuram M, Mazzio EA, Redda KK, Soliman KFA. Transcriptome Profile Analysis of Triple-Negative Breast Cancer Cells in Response to a Novel Cytostatic Tetrahydroisoquinoline Compared to Paclitaxel. International Journal of Molecular Sciences. 2021; 22(14):7694. https://doi.org/10.3390/ijms22147694
Chicago/Turabian StyleGangapuram, Madhavi, Elizabeth A. Mazzio, Kinfe K. Redda, and Karam F. A. Soliman. 2021. "Transcriptome Profile Analysis of Triple-Negative Breast Cancer Cells in Response to a Novel Cytostatic Tetrahydroisoquinoline Compared to Paclitaxel" International Journal of Molecular Sciences 22, no. 14: 7694. https://doi.org/10.3390/ijms22147694
APA StyleGangapuram, M., Mazzio, E. A., Redda, K. K., & Soliman, K. F. A. (2021). Transcriptome Profile Analysis of Triple-Negative Breast Cancer Cells in Response to a Novel Cytostatic Tetrahydroisoquinoline Compared to Paclitaxel. International Journal of Molecular Sciences, 22(14), 7694. https://doi.org/10.3390/ijms22147694






