An Overview of Physical, Microbiological and Immune Barriers of Oral Mucosa
Abstract
:1. Introduction
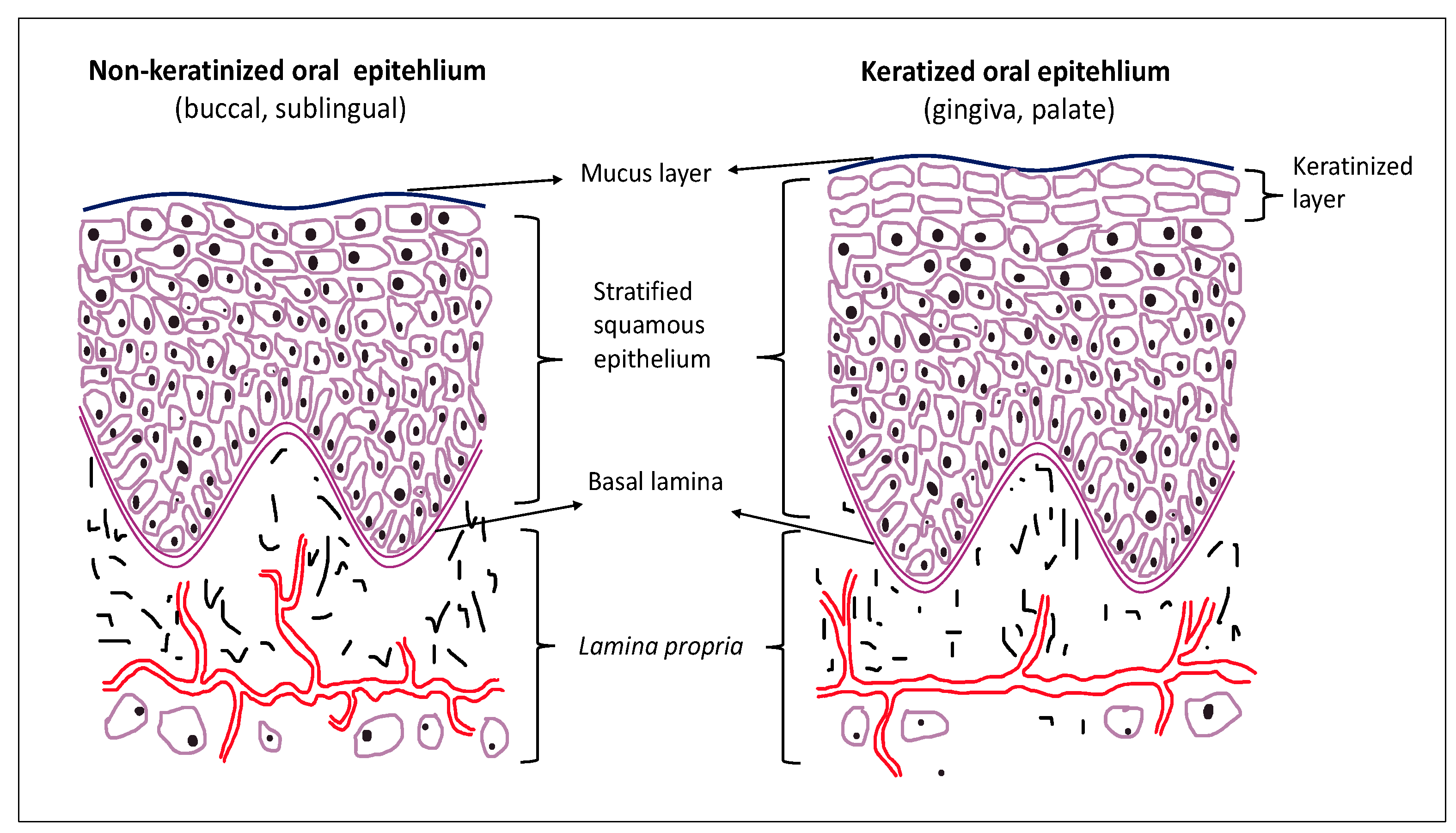
2. Oral Epithelium and Cell Junctions
3. Permeation Barrier of Oral Epithelium to Drugs
4. Oral Microbiota
5. Antimicrobial Peptides
6. Oral Immune System
7. Crosstalk between Immune System and Microbiota
8. Personalized Medicine
9. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Senel, S.; Kremer, M.; Katalin, N.; Squier, C. Delivery of Bioactive Peptides and Proteins across Oral (Buccal) Mucosa. Curr. Pharm. Biotechnol. 2001, 2, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Gadmor, H.; Brown, L. Cell-Cell Interactions in the Oral Mucosa: Tight Junctions and Gap Junctions. In Oral Mucosa in Health and Disease; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2018; pp. 19–30. [Google Scholar]
- Samiei, M.; Ahmadian, E.; Eftekhari, A.; Eghbal, M.A.; Rezaie, F.; Vinken, M. Cell junctions and oral health. EXCLI J. 2019, 18, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Groeger, S.; Meyle, J. Oral Mucosal Epithelial Cells. Front. Immunol. 2019, 10, 208. [Google Scholar] [CrossRef] [Green Version]
- Chiba, H.; Osanai, M.; Murata, M.; Kojima, T.; Sawada, N. Transmembrane proteins of tight junctions. Biochim. Biophys. Acta (BBA) Biomembr. 2008, 1778, 588–600. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.-Y.; Yang, W.-X.; Hu, Y.-J. The role of epithelial tight junctions involved in pathogen infections. Mol. Biol. Rep. 2014, 41, 6591–6610. [Google Scholar] [CrossRef]
- Zihni, C.; Mills, C.; Matter, K.; Balda, M. Tight junctions: From simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016, 17, 564–580. [Google Scholar] [CrossRef] [PubMed]
- Giri, S.; Paudel, D. Tight junction proteins, claudin and occludin in pathological conditions and aging of skin and oral mucosa: A review. Dent. J. Health Sci. Univ. Hokkaido 2019, 38, 17–27. [Google Scholar]
- Leonardo, T.R.; Shi, J.; Chen, D.; Trivedi, H.M.; Chen, L. Differential Expression and Function of Bicellular Tight Junctions in Skin and Oral Wound Healing. Int. J. Mol. Sci. 2020, 21, 2966. [Google Scholar] [CrossRef] [Green Version]
- Beeman, N.E.; Baumgartner, H.K.; Webb, P.G.; Schaack, J.B.; Neville, M.C. Disruption of occludin function in polarized epithelial cells activates the extrinsic pathway of apoptosis leading to cell extrusion without loss of transepithelial resistance. BMC Cell Biol. 2009, 10, 85. [Google Scholar] [CrossRef] [Green Version]
- Bhat, A.A.; Uppada, S.; Achkar, I.; Hashem, S.; Yadav, S.K.; Shanmugakonar, M.; Al-Naemi, H.A.; Haris, M.; Uddin, S. Tight Junction Proteins and Signaling Pathways in Cancer and Inflammation: A Functional Crosstalk. Front. Physiol. 2019, 9, 1942. [Google Scholar] [CrossRef] [Green Version]
- Conacci-Sorrell, M.; Zhurinsky, J.; Ben-Ze’Ev, A. The cadherin-catenin adhesion system in signaling and cancer. J. Clin. Investig. 2002, 109, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Tariq, H.; Bella, J.; Jowitt, T.; Holmes, D.F.; Rouhi, M.; Nie, Z.; Baldock, C.; Garrod, D.; Tabernero, L. Cadherin flexibility provides a key difference between desmosomes and adherens junctions. Proc. Natl. Acad. Sci. USA 2015, 112, 5395–5400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rübsam, M.; Broussard, J.A.; Wickström, S.; Nekrasova, O.; Green, K.J.; Niessen, C.M. Adherens Junctions and Desmosomes Coordinate Mechanics and Signaling to Orchestrate Tissue Morphogenesis and Function: An Evolutionary Perspective. Cold Spring Harb. Perspect. Biol. 2018, 10, a029207. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Cui, Y.; Wei, J.; Sun, J.; Zheng, L.; Xie, J. Gap junction-mediated cell-to-cell communication in oral development and oral diseases: A concise review of research progress. Int. J. Oral Sci. 2020, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pather, S.I.; Rathbone, M.J.; Şenel, S. Current status and the future of buccal drug delivery systems. Expert Opin. Drug Deliv. 2008, 5, 531–542. [Google Scholar] [CrossRef]
- Şenel, S.; Hıncal, A. Drug permeation enhancement via buccal route: Possibilities and limitations. J. Control. Release 2001, 72, 133–144. [Google Scholar] [CrossRef]
- Şenel, S.; Rathbone, M.J.; Cansız, M.; Pather, I. Recent developments in buccal and sublingual delivery systems. Expert Opin. Drug Deliv. 2012, 9, 615–628. [Google Scholar] [CrossRef]
- Rathbone, M.J.; Pather, I.; Şenel, S. Overview of Oral Mucosal Delivery. In Advances in Delivery Science and Technology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2015; pp. 17–29. [Google Scholar]
- Senel, S.; Hoogstraate, A.; Spies, F.; Verhoef, J.; Geest, A.B.-V.; Junginger, H.; Boddé, H. Enhancement of in vitro permeability of porcine buccal mucosa by bile salts: Kinetic and histological studies. J. Control. Release 1994, 32, 45–56. [Google Scholar] [CrossRef]
- Hoogstraate, A.; Senel, S.; Cullander, C.; Verhoef, J.; Junginger, H.; Boddé, H. Effects of bile salts on transport rates and routes of FITC-labelled compounds across porcine buccal epithelium in vitro. J. Control. Release 1996, 40, 211–221. [Google Scholar] [CrossRef]
- Şenel, S.; Çapan, Y.; Sargon, M.; Ikinci, G.; Şolpan, D.; Güven, O.; Boddé, H.; Hincal, A. Enhancement of transbuccal permeation of morphine sulfate by sodium glycodeoxycholate in vitro. J. Control. Release 1997, 45, 153–162. [Google Scholar] [CrossRef]
- Şenel, S.; Duchêne, D.; Hıncal, A.; Çapan, Y.; Ponchel, G. In vitro studies on enhancing effect of sodium glycocholate on transbuccal permeation of morphine hydrochloride. J. Control. Release 1998, 51, 107–113. [Google Scholar] [CrossRef]
- Raimondi, F.; Santoro, P.; Barone, M.V.; Pappacoda, S.; Barretta, M.L.; Nanayakkara, M.; Apicella, C.; Capasso, L.; Paludetto, R. Bile acids modulate tight junction structure and barrier function of Caco-2 monolayers via EGFR activation. Am. J. Physiol. Liver Physiol. 2008, 294, G906–G913. [Google Scholar] [CrossRef]
- Chen, X.; Oshima, T.; Tomita, T.; Fukui, H.; Watari, J.; Matsumoto, T.; Miwa, H. Acidic bile salts modulate the squamous epithelial barrier function by modulating tight junction proteins. Am. J. Physiol. Liver Physiol. 2011, 301, G203–G209. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Wang, Z.; Li, P.; Yang, Q. The effect of various absorption enhancers on tight junction in the human intestinal Caco-2 cell line. Drug Dev. Ind. Pharm. 2012, 39, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Şenel, S.; Kremer, M.; Kaş, S.; Wertz, P.; Hıncal, A.; Squier, C. Enhancing effect of chitosan on peptide drug delivery across buccal mucosa. Biomaterials 2000, 21, 2067–2071. [Google Scholar] [CrossRef]
- Senel, S. Potential applications of chitosan in oral mucosal delivery. J. Drug Deliv. Sci. Technol. 2010, 20, 23–32. [Google Scholar] [CrossRef]
- Yeh, T.-H.; Hsu, L.-W.; Tseng, M.T.; Lee, P.-L.; Sonjae, K.; Ho, Y.-C.; Sung, H.-W. Mechanism and consequence of chitosan-mediated reversible epithelial tight junction opening. Biomaterials 2011, 32, 6164–6173. [Google Scholar] [CrossRef]
- Smith, J.; Wood, E.; Dornish, M. Effect of Chitosan on Epithelial Cell Tight Junctions. Pharm. Res. 2004, 21, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Sonaje, K.; Chuang, E.-Y.; Lin, K.-J.; Yen, T.-C.; Su, F.-Y.; Tseng, M.T.; Sung, H.-W. Opening of Epithelial Tight Junctions and Enhancement of Paracellular Permeation by Chitosan: Microscopic, Ultrastructural, and Computed-Tomographic Observations. Mol. Pharm. 2012, 9, 1271–1279. [Google Scholar] [CrossRef]
- Tscheik, C.; Blasig, I.E.; Winkler, L. Trends in drug delivery through tissue barriers containing tight junctions. Tissue Barriers 2013, 1, e24565. [Google Scholar] [CrossRef] [Green Version]
- Günzel, D.; Yu, A. Claudins and the Modulation of Tight Junction Permeability. Physiol. Rev. 2013, 93, 525–569. [Google Scholar] [CrossRef] [Green Version]
- Brunner, J.; Ragupathy, S.; Borchard, G. Target specific tight junction modulators. Adv. Drug Deliv. Rev. 2021, 171, 266–288. [Google Scholar] [CrossRef]
- Zhao, X.; Zeng, H.; Lei, L.; Tong, X.; Yang, L.; Yang, Y.; Li, S.; Zhou, Y.; Luo, L.; Huang, J.; et al. Tight junctions and their regulation by non-coding RNAs. Int. J. Biol. Sci. 2021, 17, 712–727. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Tachibana, K.; Krug, S.M.; Kunisawa, J.; Fromm, M.; Kondoh, M. Potential for Tight Junction Protein–Directed Drug Development Using Claudin Binders and Angubindin-1. Int. J. Mol. Sci. 2019, 20, 4016. [Google Scholar] [CrossRef] [Green Version]
- Rathbone, M.J.; Şenel, S.; Pather, I. Design and Development of Systemic Oral Mucosal Drug Delivery Systems. In Advances in Delivery Science and Technology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2015; pp. 149–167. [Google Scholar]
- NuGenerex Family of Companies. Available online: https://generex.com/nugenerex-family-companies/#dx (accessed on 14 July 2021).
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the Normal Bacterial Flora of the Oral Cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef] [Green Version]
- Spratt, D.; Pratten, J. Biofilms and the Oral Cavity. Rev. Environ. Sci. Bio/Technol. 2003, 2, 109–120. [Google Scholar] [CrossRef]
- Guttman, J.A.; Finlay, B.B. Tight junctions as targets of infectious agents. Biochim. Biophys. Acta (BBA) Biomembr. 2009, 1788, 832–841. [Google Scholar] [CrossRef] [Green Version]
- Paradis, T.; Bègue, H.; Basmaciyan, L.; Dalle, F.; Bon, F. Tight Junctions as a Key for Pathogens Invasion in Intestinal Epithelial Cells. Int. J. Mol. Sci. 2021, 22, 2506. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Sulijaya, B.; Yamada-Hara, M.; Tsuzuno, T.; Tabeta, K.; Yamazaki, K. Gingival epithelial barrier: Regulation by beneficial and harmful microbes. Tissue Barriers 2019, 7, e1651158. [Google Scholar] [CrossRef] [PubMed]
- Lederberg, J.; McCray, A.J.T.S. Ome Sweet Omics—A Genealogical Treasury of Words. Scientist 2001, 15, 8. [Google Scholar]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 1–22. [Google Scholar] [CrossRef]
- Baquero, F.; Nombela, C. The microbiome as a human organ. Clin. Microbiol. Infect. 2012, 18, 2–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NIH Human Microbiome Project. Available online: https://hmpdacc.org/hmp/ (accessed on 29 June 2021).
- Lloyd-Price, J.; Mahurkar, A.; Rahnavard, G.; Crabtree, J.; Orvis, J.; Hall, A.B.; Brady, A.; Creasy, H.H.; McCracken, C.; Giglio, M.G.; et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017, 550, 61–66. [Google Scholar] [CrossRef]
- The Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef] [Green Version]
- Expanded Human Oral Microbiome Database (eHOMD). Available online: http://www.homd.org/index.php (accessed on 29 June 2021).
- Kilian, M. The oral microbiome—Friend or foe? Eur. J. Oral Sci. 2018, 126, 5–12. [Google Scholar] [CrossRef]
- Lu, M.; Xuan, S.; Wang, Z. Oral microbiota: A new view of body health. Food Sci. Hum. Wellness 2019, 8, 8–15. [Google Scholar] [CrossRef]
- Moutsopoulos, N.M.; Konkel, J.E. Tissue-Specific Immunity at the Oral Mucosal Barrier. Trends Immunol. 2018, 39, 276–287. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-S.; Kao, C.-Y. Current understanding of the gut microbiota shaping mechanisms. J. Biomed. Sci. 2019, 26, 1–11. [Google Scholar] [CrossRef]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Genet. 2018, 16, 745–759. [Google Scholar] [CrossRef]
- Poveda-Roda, R.; Jiménez, Y.; Carbonell, E.; Gavaldá, C.; Margaix-Muñoz, M.M.; Sarrión-Pérez, G. Bacteremia originating in the oral cavity. A review. Medicina Oral Patología Oral Cirugia Bucal 2008, 13, 355–362. [Google Scholar]
- Bascones-Martinez, M.M.-C.A.J.H.M.A.; Muñoz-Corcuera, M.; Meurman, J. Odontogenic Infections in the Etiology of Infective Endocarditis. Cardiovasc. Hematol. Disord. Targets 2009, 9, 231–235. [Google Scholar] [CrossRef]
- Horliana, A.C.R.T.; Chambrone, L.; Foz, A.M.; Artese, H.; Rabelo, M.D.S.; Pannuti, C.M.; Romito, G.A. Dissemination of Periodontal Pathogens in the Bloodstream after Periodontal Procedures: A Systematic Review. PLoS ONE 2014, 9, e98271. [Google Scholar] [CrossRef] [Green Version]
- Wilson, W.R.; Taubert, K.A.; Gewitz, M.H.; Lockhart, P.B.; Baddour, L.M.; E Levison, M.; Bolger, A.F.; Cabell, C.H.; Takahashi, M.; Baltimore, R.S.; et al. Prevention of Infective Endocarditis. Circulation 2007, 116, 1736–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Rosa, G.R.M.; Gattuso, G.; Pedullà, E.; Rapisarda, E.; Nicolosi, D.; Salmeri, M. Association of oral dysbiosis with oral cancer development (Review). Oncol. Lett. 2020, 19, 3045–3058. [Google Scholar] [CrossRef] [Green Version]
- Chakraborti, S.; Das, R.K. Dysbiosis of Oral Microbiota and Its Effect on Epithelial-Mesenchymal Transition: A Review. SN Compr. Clin. Med. 2020, 2, 2324–2335. [Google Scholar] [CrossRef]
- Sugita, K.; Soyka, M.B.; Wawrzyniak, P.; Rinaldi, A.O.; Mitamura, Y.; Akdis, M.; Akdis, C.A. Outside-in hypothesis revisited. Ann. Allergy, Asthma Immunol. 2020, 125, 517–527. [Google Scholar] [CrossRef]
- Sözener, Z.C.; Cevhertas, L.; Nadeau, K.; Akdis, M.; Akdis, C.A. Environmental factors in epithelial barrier dysfunction. J. Allergy Clin. Immunol. 2020, 145, 1517–1528. [Google Scholar] [CrossRef]
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012, 32, 143–171. [Google Scholar] [CrossRef] [Green Version]
- Gorr, S.-U.; Abdolhosseini, M. Antimicrobial peptides and periodontal disease. J. Clin. Periodontol. 2011, 38, 126–141. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Khurshid, Z.; Naseem, M.; Sheikh, Z.; Najeeb, S.; Shahab, S.; Zafar, M.S. Oral antimicrobial peptides: Types and role in the oral cavity. Saudi Pharm. J. 2016, 24, 515–524. [Google Scholar] [CrossRef] [Green Version]
- A Dale, B.; Fredericks, L.P. Antimicrobial peptides in the oral environment: Expression and function in health and disease. Curr. Issues Mol. Biol. 2005, 7, 119–133. [Google Scholar]
- Ouhara, K.; Komatsuzawa, H.; Yamada, S.; Shiba, H.; Fujiwara, T.; Ohara, M.; Sayama, K.; Hashimoto, K.; Kurihara, H.; Sugai, M. Susceptibilities of periodontopathogenic and cariogenic bacteria to antibacterial peptides, β-defensins and LL37, produced by human epithelial cells. J. Antimicrob. Chemother. 2005, 55, 888–896. [Google Scholar] [CrossRef]
- Ji, S.; Choi, Y. Innate immune response to oral bacteria and the immune evasive characteristics of periodontal pathogens. J. Periodontal Implant. Sci. 2013, 43, 3–11. [Google Scholar] [CrossRef]
- Hans, M.; Hans, V.M. Epithelial Antimicrobial Peptides: Guardian of the Oral Cavity. Int. J. Pept. 2014, 2014, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salem, A.; Almahmoudi, R.; Hagström, J.; Stark, H.; Nordström, D.; Salo, T.; Eklund, K.K. Human β-Defensin 2 Expression in Oral Epithelium: Potential Therapeutic Targets in Oral Lichen Planus. Int. J. Mol. Sci. 2019, 20, 1780. [Google Scholar] [CrossRef] [Green Version]
- Muniz, L.R.; Knosp, C.; Yeretssian, G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front. Immunol. 2012, 3, 310. [Google Scholar] [CrossRef] [Green Version]
- Zong, X.; Fu, J.; Xu, B.; Wang, Y.; Jin, M. Interplay between gut microbiota and antimicrobial peptides. Anim. Nutr. 2020, 6, 389–396. [Google Scholar] [CrossRef]
- Wu, R.-Q.; Zhang, D.; Tu, E.; Chen, Q.-M.; Chen, W. The mucosal immune system in the oral cavity—An orchestra of T cell diversity. Int. J. Oral Sci. 2014, 6, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, C.A.T.N.Z.M.L.E. The microbiome and innate immunity. Nat. Cell Biol. 2016, 535, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [Green Version]
- Cua, D.J.; Tato, C.M. Innate IL-17-producing cells: The sentinels of the immune system. Nat. Rev. Immunol. 2010, 10, 479–489. [Google Scholar] [CrossRef]
- Günther, J.; Seyfert, H.-M. The first line of defence: Insights into mechanisms and relevance of phagocytosis in epithelial cells. Semin. Immunopathol. 2018, 40, 555–565. [Google Scholar] [CrossRef] [Green Version]
- Iwakura, Y.; Ishigame, H.; Saijo, S.; Nakae, S. Functional Specialization of Interleukin-17 Family Members. Immunity 2011, 34, 149–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abusleme, L.; Moutsopoulos, N.M. IL-17: Overview and role in oral immunity and microbiome. Oral Dis. 2017, 23, 854–865. [Google Scholar] [CrossRef]
- Amatya, N.; Garg, A.V.; Gaffen, S.L. IL-17 Signaling: The Yin and the Yang. Trends Immunol. 2017, 38, 310–322. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.-S.; Tang, Y.-L.; Pang, X.; Zheng, M.; Liang, X.-H. The maintenance of an oral epithelial barrier. Life Sci. 2019, 227, 129–136. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Dutzan, N.; Kajikawa, T.; Abusleme, L.; Greenwell-Wild, T.; Zuazo, C.E.; Ikeuchi, T.; Brenchley, L.; Abe, T.; Hurabielle, C.; Martin, D.; et al. A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci. Transl. Med. 2018, 10, eaat0797. [Google Scholar] [CrossRef] [Green Version]
- Gaffen, S.L.; Moutsopoulos, N.M. Regulation of host-microbe interactions at oral mucosal barriers by type 17 immunity. Sci. Immunol. 2020, 5, eaau4594. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, L.; Ren, W.; Li, S.; Zheng, J.; Li, S.; Jiang, C.; Yang, S.; Zhi, K. The Role of the Immune Response in the Development of Medication-Related Osteonecrosis of the Jaw. Front. Immunol. 2021, 12, 606043. [Google Scholar] [CrossRef]
- Papi, P.; Rosella, D.; Giardino, R.; Cicalini, E.; Piccoli, L.; Pompa, G. Medication-related osteonecrosis of the jaw: Clinical and practical guidelines. J. Int. Soc. Prev. Community Dent. 2016, 6, 97–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bullock, G.; Miller, C.; McKechnie, A.; Hearnden, V. Synthetic Hydroxyapatite Inhibits Bisphosphonate Toxicity to the Oral Mucosa In Vitro. Materials 2020, 13, 2086. [Google Scholar] [CrossRef] [PubMed]
- Gare, J.; Kanoute, A.; Meda, N.; Viennot, S.; Bourgeois, D.; Carrouel, F. Periodontal Conditions and Pathogens Associated with Pre-Eclampsia: A Scoping Review. Int. J. Environ. Res. Public Health 2021, 18, 7194. [Google Scholar] [CrossRef] [PubMed]
- Daalderop, L.; Wieland, B.; Tomsin, K.; Reyes, L.; Kramer, B.; Vanterpool, S.; Been, J. Periodontal Disease and Pregnancy Outcomes: Overview of Systematic Reviews. JDR Clin. Transl. Res. 2017, 3, 10–27. [Google Scholar] [CrossRef]
- Novak, N.; Haberstok, J.; Bieber, T.; Allam, J.-P. The immune privilege of the oral mucosa. Trends Mol. Med. 2008, 14, 191–198. [Google Scholar] [CrossRef]
- Naafs, M. Oral Mucosal Immune Suppression, Tolerance and Silencing: A Mini-Review. Mod. Approaches Dent. Oral Heal. Care 2018, 1, 001–012. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.C.; Khodadadi, H.; Baban, B. Innate immunity and oral microbiome: A personalized, predictive, and preventive approach to the management of oral diseases. EPMA J. 2019, 10, 43–50. [Google Scholar] [CrossRef]
- Shapiro, H.; A Thaiss, C.; Levy, M.; Elinav, E. The cross talk between microbiota and the immune system: Metabolites take center stage. Curr. Opin. Immunol. 2014, 30, 54–62. [Google Scholar] [CrossRef]
- Golubnitschaja, O.; Kinkorova, J.; Costigliola, V. Predictive, Preventive and Personalised Medicine as the hardcore of ‘Horizon 2020’: EPMA position paper. EPMA J. 2014, 5, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belibasakis, G.N.; Bostanci, N.; Marsh, P.D.; Zaura, E. Applications of the oral microbiome in personalized dentistry. Arch. Oral Biol. 2019, 104, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Garcia, I.; Kuska, R.; Somerman, M. Expanding the Foundation for Personalized Medicine. J. Dent. Res. 2013, 92, S3–S10. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.S.; Shetty, S.; Vannala, V. Embracing Personalized Medicine in Dentistry. J. Pharm. Bioallied Sci. 2019, 11, S92–S96. [Google Scholar] [CrossRef] [PubMed]
- Polverini, P.J. Personalized medicine and the future of dental practice. Pers. Med. 2018, 15, 449–451. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Şenel, S. An Overview of Physical, Microbiological and Immune Barriers of Oral Mucosa. Int. J. Mol. Sci. 2021, 22, 7821. https://doi.org/10.3390/ijms22157821
Şenel S. An Overview of Physical, Microbiological and Immune Barriers of Oral Mucosa. International Journal of Molecular Sciences. 2021; 22(15):7821. https://doi.org/10.3390/ijms22157821
Chicago/Turabian StyleŞenel, Sevda. 2021. "An Overview of Physical, Microbiological and Immune Barriers of Oral Mucosa" International Journal of Molecular Sciences 22, no. 15: 7821. https://doi.org/10.3390/ijms22157821
APA StyleŞenel, S. (2021). An Overview of Physical, Microbiological and Immune Barriers of Oral Mucosa. International Journal of Molecular Sciences, 22(15), 7821. https://doi.org/10.3390/ijms22157821




