Mutated CCDC51 Coding for a Mitochondrial Protein, MITOK Is a Candidate Gene Defect for Autosomal Recessive Rod-Cone Dystrophy
Abstract
:1. Introduction
2. Results
2.1. A Sporadic Case with Mild arRCD
2.2. Identification of a Novel Candidate Gene, CCDC51 Underlying Mild arRCD
2.3. Function and Pathogenic Mechanism of the Mitochondrial Protein MITOK Encoded by CCDC51
2.4. Expression Results of CCDC51, a Novel Candidate Gene Defect Underlying Mild arRCD
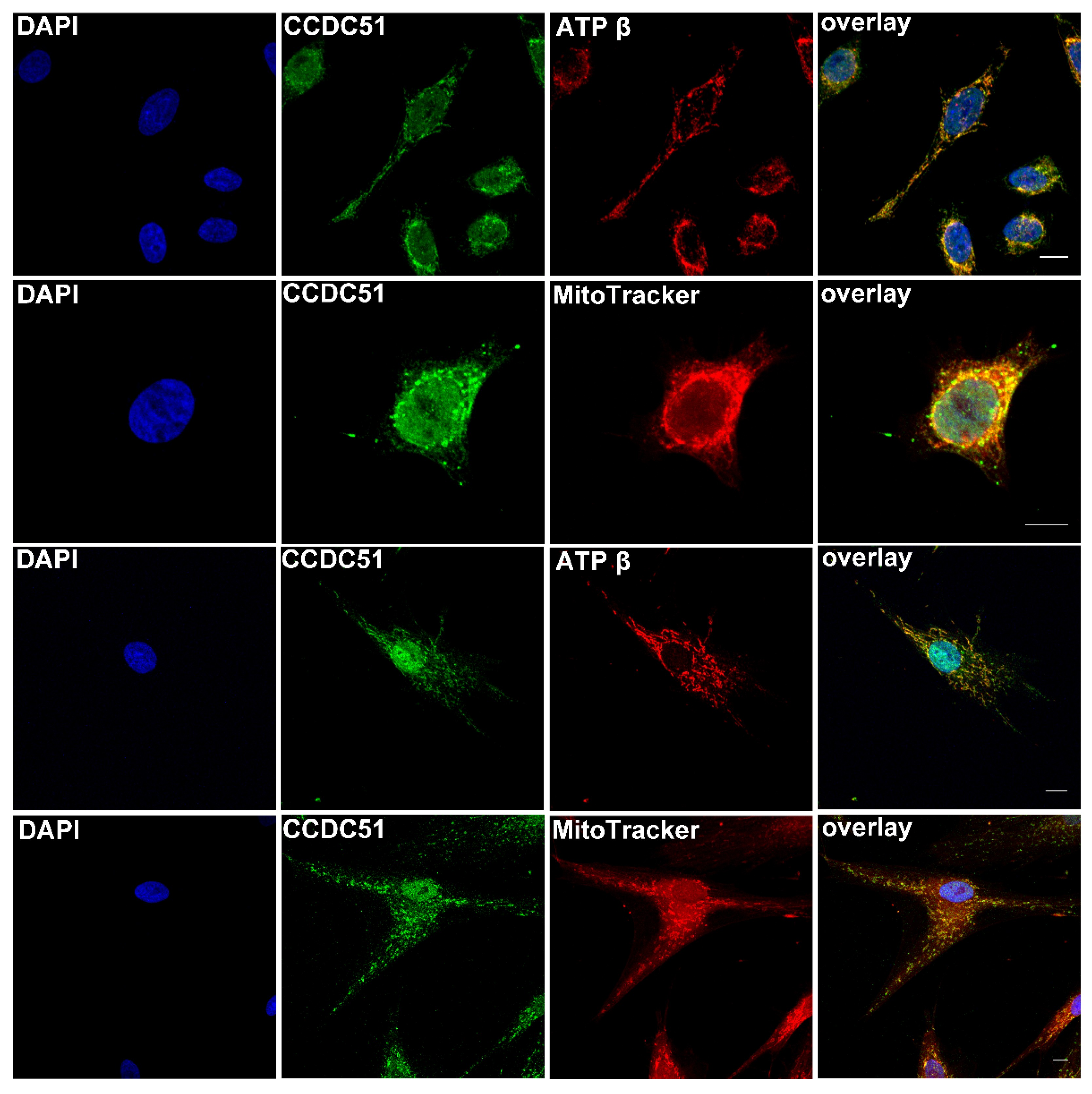
2.5. Detection of Endogenous CCDC51 in Human Cell Lines
2.6. Detection of CCDC51/MITOK in Non-Human Primate, Human, and Mouse Retina
3. Discussion
4. Materials and Methods
4.1. Clinical Examination
4.2. Genetic Analyses
4.3. Gene Expression Analyses
4.4. Protein Localization Studies
4.5. Specificity of Antibody against CCDC51
4.6. Immunolocalization Studies in Human Cells and Retinal Tissue Using Fluorescence Imaging
4.7. Immunohistochemistry in Human Retinal Tissue Using Horseradish Peroxidase
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis pigmentosa. Lancet 2006, 368, 1795–1809. [Google Scholar] [CrossRef]
- Berger, W.; Kloeckener-Gruissem, B.; Neidhardt, J. The molecular basis of human retinal and vitreoretinal diseases. Prog. Retin. Eye Res. 2010, 29, 335–375. [Google Scholar] [CrossRef] [Green Version]
- Carss, K.J.; Arno, G.; Erwood, M.; Stephens, J.; Sanchis-Juan, A.; Hull, S.; Megy, K.; Grozeva, D.; Dewhurst, E.; Malka, S.; et al. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am. J. Hum. Genet. 2017, 100, 75–90. [Google Scholar] [CrossRef] [Green Version]
- Glockle, N.; Kohl, S.; Mohr, J.; Scheurenbrand, T.; Sprecher, A.; Weisschuh, N.; Bernd, A.; Rudolph, G.; Schubach, M.; Poloschek, C.; et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur. J. Hum. Genet. 2014, 22, 99–104. [Google Scholar] [CrossRef]
- Dockery, A.; Stephenson, K.; Keegan, D.; Wynne, N.; Silvestri, G.; Humphries, P.; Kenna, P.F.; Carrigan, M.; Farrar, G.J. Target 5000: Target Capture Sequencing for Inherited Retinal Degenerations. Genes 2017, 8, 304. [Google Scholar] [CrossRef] [Green Version]
- Ellingford, J.; Barton, S.; Bhaskar, S.; O’Sullivan, J.; Williams, S.; Lamb, J.; Panda, B.; Sergouniotis, P.I.; Gillespie, R.; Daiger, S.P.; et al. Molecular findings from 537 individuals with inherited retinal disease. J. Med. Genet. 2016, 53, 761–767. [Google Scholar] [CrossRef] [Green Version]
- Haer-Wigman, L.; Van Zelst-Stams, W.A.G.; Pfundt, R.; Born, L.I.V.D.; Klaver, C.; Verheij, J.B.G.M.; Hoyng, C.B.; Breuning, M.H.; Boon, C.; Kievit, A.J.; et al. Diagnostic exome sequencing in 266 Dutch patients with visual impairment. Eur. J. Hum. Genet. 2017, 25, 591–599. [Google Scholar] [CrossRef]
- Astuti, G.D.N.; Van Den Born, L.I.; Khan, M.I.; Hamel, C.P.; Bocquet, B.; Manès, G.; Quinodoz, M.; Ali, M.; Toomes, C.; McKibbin, M.; et al. Identification of Inherited Retinal Disease-Associated Genetic Variants in 11 Candidate Genes. Genes 2018, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Cremers, F.P.M.; Boon, C.J.F.; Bujakowska, K.; Zeitz, C. Special Issue Introduction: Inherited Retinal Disease: Novel Candidate Genes, Genotype–Phenotype Correlations, and Inheritance Models. Genes 2018, 9, 215. [Google Scholar] [CrossRef] [Green Version]
- Solaguren-Beascoa, M.; Bujakowska, K.M.; Mejecase, C.; Emmenegger, L.; Orhan, E.; Neuille, M.; Mohand-Said, S.; Condroyer, C.; Lancelot, M.E.; Michiels, C.; et al. WDR34, a candidate gene for non-syndromic rodcone dystrophy. Clin. Genet. 2021, 99, 298–302. [Google Scholar] [CrossRef]
- Audo, I.; Bujakowska, K.M.; Léveillard, T.; Mohand-Saïd, S.; Lancelot, M.-E.; Germain, A.; Antonio, A.; Michiels, C.; Saraiva, J.-P.; Letexier, M.; et al. Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J. Rare Dis. 2012, 7, 8. [Google Scholar] [CrossRef] [Green Version]
- Boulanger-Scemama, E.; El Shamieh, S.; Démontant, V.; Condroyer, C.; Antonio, A.; Michiels, C.; Boyard, F.; Saraiva, J.-P.; Letexier, M.; Souied, E.H.; et al. Next-generation sequencing applied to a large French cone and cone-rod dystrophy cohort: Mutation spectrum and new genotype-phenotype correlation. Orphanet J. Rare Dis. 2015, 10, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Audo, I.; Bujakowska, K.; Orhan, E.; Poloschek, C.M.; Defoort-Dhellemmes, S.; Drumare, I.; Kohl, S.; Luu, T.D.; Lecompte, O.; Zrenner, E.; et al. Whole-Exome Sequencing Identifies Mutations in GPR179 Leading to Autosomal-Recessive Complete Congenital Stationary Night Blindness. Am. J. Hum. Genet. 2012, 90, 321–330. [Google Scholar] [CrossRef] [Green Version]
- Zeitz, C.; Jacobson, S.; Hamel, C.P.; Bujakowska, K.; Neuillé, M.; Orhan, E.; Zanlonghi, X.; Lancelot, M.-E.; Michiels, C.; Schwartz, S.B.; et al. Whole-Exome Sequencing Identifies LRIT3 Mutations as a Cause of Autosomal-Recessive Complete Congenital Stationary Night Blindness. Am. J. Hum. Genet. 2013, 92, 67–75. [Google Scholar] [CrossRef] [Green Version]
- El Shamieh, S.; Neuillé, M.; Terray, A.; Orhan, E.; Condroyer, C.; Démontant, V.; Michiels, C.; Antonio, A.; Boyard, F.; Lancelot, M.-E.; et al. Whole-Exome Sequencing Identifies KIZ as a Ciliary Gene Associated with Autosomal-Recessive Rod-Cone Dystrophy. Am. J. Hum. Genet. 2014, 94, 625–633. [Google Scholar] [CrossRef] [Green Version]
- Audo, I.; Bujakowska, K.; Orhan, E.; El Shamieh, S.; Sennlaub, F.; Guillonneau, X.; Antonio, A.; Michiels, C.; Lancelot, M.-E.; Letexier, M.; et al. The familial dementia gene revisited: A missense mutation revealed by whole-exome sequencing identifies ITM2B as a candidate gene underlying a novel autosomal dominant retinal dystrophy in a large family. Hum. Mol. Genet. 2014, 23, 491–501. [Google Scholar] [CrossRef]
- Bujakowska, K.; Zhang, Q.; Siemiatkowska, A.M.; Liu, Q.; Place, E.; Falk, M.; Consugar, M.; Lancelot, M.-E.; Antonio, A.; Lonjou, C.; et al. Mutations in IFT172 cause isolated retinal degeneration and Bardet–Biedl syndrome. Hum. Mol. Genet. 2015, 24, 230–242. [Google Scholar] [CrossRef]
- Littink, K.W.; Koenekoop, R.K.; Born, L.I.V.D.; Collin, R.W.J.; Moruz, L.; Veltman, J.; Roosing, S.; Zonneveld, M.N.; Omar, A.; Darvish, M.; et al. Homozygosity Mapping in Patients with Cone–Rod Dystrophy: Novel Mutations and Clinical Characterizations. Investig. Opthalmol. Vis. Sci. 2010, 51, 5943–5951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benayoun, L.; Spiegel, R.; Auslender, N.; Abbasi, A.; Rizel, L.; Hujeirat, Y.; Salama, I.; Garzozi, H.J.; Allon-Shalev, S.; Ben-Yosef, T. Genetic heterogeneity in two consanguineous families segregating early onset retinal degeneration: The pitfalls of homozygosity mapping. Am. J. Med. Genet. Part A 2009, 149A, 650–656. [Google Scholar] [CrossRef]
- Farkas, M.H.; Grant, G.R.; White, J.A.; Sousa, M.E.; Consugar, M.B.; Pierce, E.A. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genom. 2013, 14, 486. [Google Scholar] [CrossRef] [Green Version]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Kleinberger, J.; Maloney, K.; Pollin, T.I.; Jeng, L.J.B. An openly available online tool for implementing the ACMG/AMP standards and guidelines for the interpretation of sequence variants. Genet. Med. 2016, 18, 1165. [Google Scholar] [CrossRef] [Green Version]
- Hartong, D.T.; Dange, M.; McGee, T.L.; Berson, E.L.; Dryja, T.P.; Colman, R.F. Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nat. Genet. 2008, 40, 1230–1234. [Google Scholar] [CrossRef] [Green Version]
- Zenteno, J.C.; García-Montaño, L.A.; Cruz-Aguilar, M.; Ronquillo, J.; Rodas-Serrano, A.; Aguilar-Castul, L.; Matsui, R.; Vencedor-Meraz, C.I.; Arce-González, R.; Graue-Wiechers, F.; et al. Extensive genic and allelic heterogeneity underlying inherited retinal dystrophies in Mexican patients molecularly analyzed by next-generation sequencing. Mol. Genet. Genom. Med. 2019, 8. [Google Scholar] [CrossRef]
- Jespersgaard, C.; Fang, M.; Bertelsen, M.; Dang, X.; Jensen, H.; Chen, Y.; Bech, N.; Dai, L.; Rosenberg, T.; Zhang, J.; et al. Molecular genetic analysis using targeted NGS analysis of 677 individuals with retinal dystrophy. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.; Robinson, A.J. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 2016, 44, D1258–D1261. [Google Scholar] [CrossRef] [Green Version]
- Paggio, A.; Checchetto, V.; Campo, A.; Menabò, R.; Di Marco, G.; Di Lisa, F.; Szabo, I.; Rizzuto, R.; De Stefani, D. Identification of an ATP-sensitive potassium channel in mitochondria. Nature 2019, 572, 609–613. [Google Scholar] [CrossRef]
- Zhuo, M.-L.; Huang, Y.; Liu, D.-P.; Liang, C.-C. KATP channel: Relation with cell metabolism and role in the cardiovascular system. Int. J. Biochem. Cell Biol. 2005, 37, 751–764. [Google Scholar] [CrossRef]
- Ashcroft, F.M.; Rorsman, P. K(ATP) channels and islet hormone secretion: New insights and controversies. Nat. Rev. Endocrinol. 2013, 9, 660–669. [Google Scholar] [CrossRef] [Green Version]
- Nichols, C.G. KATP channels as molecular sensors of cellular metabolism. Nature 2006, 440, 470–476. [Google Scholar] [CrossRef]
- Xu, M.; Wang, Y.; Ayub, A.; Ashraf, M. Mitochondrial K(ATP) channel activation reduces anoxic injury by restoring mitochondrial membrane potential. Am. J. Physiol. Heart Circ. Physiol. 2001, 281, H1295–H1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvo, S.E.; Clauser, K.; Mootha, V.K. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016, 44, D1251–D1257. [Google Scholar] [CrossRef] [Green Version]
- Consortium, G.T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Jin, X.; Tsueng, G.; Afrasiabi, C.; Su, A.I. BioGPS: Building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016, 44, D313–D316. [Google Scholar] [CrossRef] [Green Version]
- Siegert, S.; Scherf, B.G.; Del Punta, K.; Didkovsky, N.; Heintz, N.; Roska, B. Genetic address book for retinal cell types. Nat. Neurosci. 2009, 12, 1197–1204. [Google Scholar] [CrossRef]
- Siegert, S.; Cabuy, E.; Scherf, B.G.; Kohler, H.; Panda, S.; Le, Y.Z.; Fehling, H.J.; Gaidatzis, D.; Stadler, M.B.; Roska, B. Transcriptional code and disease map for adult retinal cell types. Nat. Neurosci. 2012, 15, 487–495. [Google Scholar] [CrossRef]
- Kalathur, R.K.R.; Gagnière, N.; Berthommier, G.; Poidevin, L.; Raffelsberger, W.; Ripp, R.; Leveillard, T.; Poch, O. Retinobase: A web database, data mining and analysis platform for gene expression data on retina. BMC Genom. 2008, 9, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniele, L.L.; Lillo, C.; Lyubarsky, A.L.; Nikonov, S.S.; Philp, N.; Mears, A.J.; Swaroop, A.; Williams, D.S.; Pugh, E.N. Cone-like Morphological, Molecular, and Electrophysiological Features of the Photoreceptors of the NrlKnockout Mouse. Investig. Opthalmol. Vis. Sci. 2005, 46, 2156–2167. [Google Scholar] [CrossRef] [Green Version]
- Mears, A.J.; Kondo, M.; Swain, P.K.; Takada, Y.; Bush, R.A.; Saunders, T.; Sieving, P.A.; Swaroop, A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001, 29, 447–452. [Google Scholar] [CrossRef]
- Nikonov, S.S.; Daniele, L.L.; Zhu, X.; Craft, C.M.; Swaroop, A.; Pugh, E.N., Jr. Photoreceptors of Nrl−/− mice coexpress functional S- and M-cone opsins having distinct inactivation mechanisms. J. Gen. Physiol. 2005, 125, 287–304. [Google Scholar] [CrossRef] [Green Version]
- Brooks, M.J.; Rajasimha, H.K.; Roger, J.E.; Swaroop, A. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl−/− retinal transcriptomes. Mol. Vis. 2011, 17, 3034–3054. [Google Scholar]
- Frost, A.R.; Böhm, S.V.; Sewduth, R.N.; Josifova, D.; Ogilvie, C.M.; Izatt, L.; Roberts, R.G. Heterozygous deletion of a 2-Mb region including the dystroglycan gene in a patient with mild myopathy, facial hypotonia, oral-motor dyspraxia and white matter abnormalities. Eur. J. Hum. Genet. 2010, 18, 852–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.-H.; Yang, J.-W.; Juranville, J.-F.; Fountoulakis, M.; Lubec, G. Evidence for existence of thirty hypothetical proteins in rat brain. Proteome Sci. 2004, 2. [Google Scholar] [CrossRef] [Green Version]
- Lefevere, E.; Toft-Kehler, A.K.; Vohra, R.; Kolko, M.; Moons, L.; Van Hove, I. Mitochondrial dysfunction underlying outer retinal diseases. Mitochondrion 2017, 36, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.A.; Biousse, V.; Newman, N.J. The neuro-ophthalmology of mitochondrial disease. Surv. Ophthalmol. 2010, 55, 299–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrier, S.A.; Falk, M.J. Mitochondrial disorders and the eye. Curr. Opin. Ophthalmol. 2011, 22, 325–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falk, M.J. Neurodevelopmental Manifestations of Mitochondrial Disease. J. Dev. Behav. Pediatr. 2010, 31, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Thompson, D.; Khan, N.W.; Othman, M.I.; Chang, B.; Jia, L.; Grahek, G.; Wu, Z.; Hiriyanna, S.; Nellissery, J.; Li, T.; et al. Rd9 Is a Naturally Occurring Mouse Model of a Common Form of Retinitis Pigmentosa Caused by Mutations in RPGR-ORF15. PLoS ONE 2012, 7, e35865. [Google Scholar] [CrossRef]
- Chang, B.; Hawes, N.; Hurd, R.; Davisson, M.; Nusinowitz, S.; Heckenlively, J. Retinal degeneration mutants in the mouse. Vis. Res. 2002, 42, 517–525. [Google Scholar] [CrossRef] [Green Version]
- van de Pavert, S.; Kantardzhieva, A.; Malysheva, A.; Meuleman, J.; Versteeg, I.; Levelt, C.; Klooster, J.; Geiger, S.; Seeliger, M.W.; Rashbass, P.; et al. Crumbs homologue 1 is required for maintenance of photoreceptor cell polarization and adhesion during light exposure. J. Cell Sci. 2004, 117, 4169–4177. [Google Scholar] [CrossRef] [Green Version]
- Bujakowska, K.; Maubaret, C.; Chakarova, C.F.; Tanimoto, N.; Beck, S.C.; Fahl, E.; Humphries, M.M.; Kenna, P.F.; Makarov, E.; Makarova, O.; et al. Study of Gene-Targeted Mouse Models of Splicing Factor GenePrpf31Implicated in Human Autosomal Dominant Retinitis Pigmentosa (RP). Investig. Opthalmol. Vis. Sci. 2009, 50, 5927–5933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebeling, M.C.; Geng, Z.; Kapphahn, R.J.; Roehrich, H.; Montezuma, S.R.; Dutton, J.R.; Ferrington, D.A. Impaired Mitochondrial Function in iPSC-Retinal Pigment Epithelium with the Complement Factor H Polymorphism for Age-Related Macular Degeneration. Cells 2021, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- Brantova, O.; Tesařová, M.; Hansíková, H.; Elleder, M.; Zeman, J.; Sladkova, J. Ultrastructural Changes of Mitochondria in the Cultivated Skin Fibroblasts of Patients with Point Mutations in Mitochondrial DNA. Ultrastruct. Pathol. 2006, 30, 239–245. [Google Scholar] [CrossRef]
- Audo, I.; Friedrich, A.; Mohand-Saïd, S.; Lancelot, M.-E.; Antonio, A.; Moskova-Doumanova, V.; Poch, O.; Bhattacharya, S.; Sahel, J.-A.; Zeitz, C. An Unusual Retinal Phenotype Associated With a Novel Mutation in RHO. Arch. Ophthalmol. 2010, 128, 1036–1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audo, I.; Lancelot, M.E.; Mohand-Said, S.; Antonio, A.; Germain, A.; Sahel, J.A.; Bhattacharya, S.S.; Zeitz, C. Novel C2orf71 mutations account for approximately 1% of cases in a large French arRP cohort. Hum. Mutat. 2011, 32, E2091–E2103. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, C.; Riahi, Z.; Chantot-Bastaraud, S.; Smagghe, L.; Letexier, M.; Marcaillou, C.; Lefèvre, G.M.; Hardelin, J.-P.; El-Amraoui, A.; Singh-Estivalet, A.; et al. An innovative strategy for the molecular diagnosis of Usher syndrome identifies causal biallelic mutations in 93% of European patients. Eur. J. Hum. Genet. 2016, 24, 1730–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-W.; Yang, H.-J.; Brooks, M.J.; Zelinger, L.; Karakulah, G.; Gotoh, N.; Boleda, A.; Gieser, L.; Giuste, F.; Whitaker, D.; et al. NRL-Regulated Transcriptome Dynamics of Developing Rod Photoreceptors. Cell Rep. 2016, 17, 2460–2473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orhan, E.; Prezeau, L.; El Shamieh, S.; Bujakowska, K.M.; Michiels, C.; Zagar, Y.; Vol, C.; Bhattacharya, S.S.; Sahel, J.-A.; Sennlaub, F.; et al. Further Insights Into GPR179: Expression, Localization, and Associated Pathogenic Mechanisms Leading to Complete Congenital Stationary Night Blindness. Investig. Opthalmol. Vis. Sci. 2013, 54, 8041–8050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Shamieh, S.; Méjécase, C.; Bertelli, M.; Terray, A.; Michiels, C.; Condroyer, C.; Fouquet, S.; Sadoun, M.; Clérin, E.; Liu, B.; et al. Further Insights into the Ciliary Gene and Protein KIZ and Its Murine Ortholog PLK1S1 Mutated in Rod-Cone Dystrophy. Genes 2017, 8, 277. [Google Scholar] [CrossRef]
- Khateb, S.; Nassisi, M.; Bujakowska, K.M.; Méjécase, C.; Condroyer, C.; Antonio, A.; Foussard, M.; Démontant, V.; Mohand-Saïd, S.; Sahel, J.-A.; et al. Longitudinal Clinical Follow-up and Genetic Spectrum of Patients with Rod-Cone Dystrophy Associated With Mutations inPDE6AandPDE6B. JAMA Ophthalmol. 2019, 137, 669–679. [Google Scholar] [CrossRef]
- Khateb, S.; Mohand-Saïd, S.; Nassisi, M.; Bonnet, C.; Roux, A.-F.; Andrieu, C.; Antonio, A.; Condroyer, C.; Zeitz, C.; Devisme, C.; et al. Phenotypic Characteristics of Rod–Cone Dystrophy Associated with Myo7a Mutations in a Large French Cohort. Retina 2020, 40, 1603–1615. [Google Scholar] [CrossRef] [PubMed]
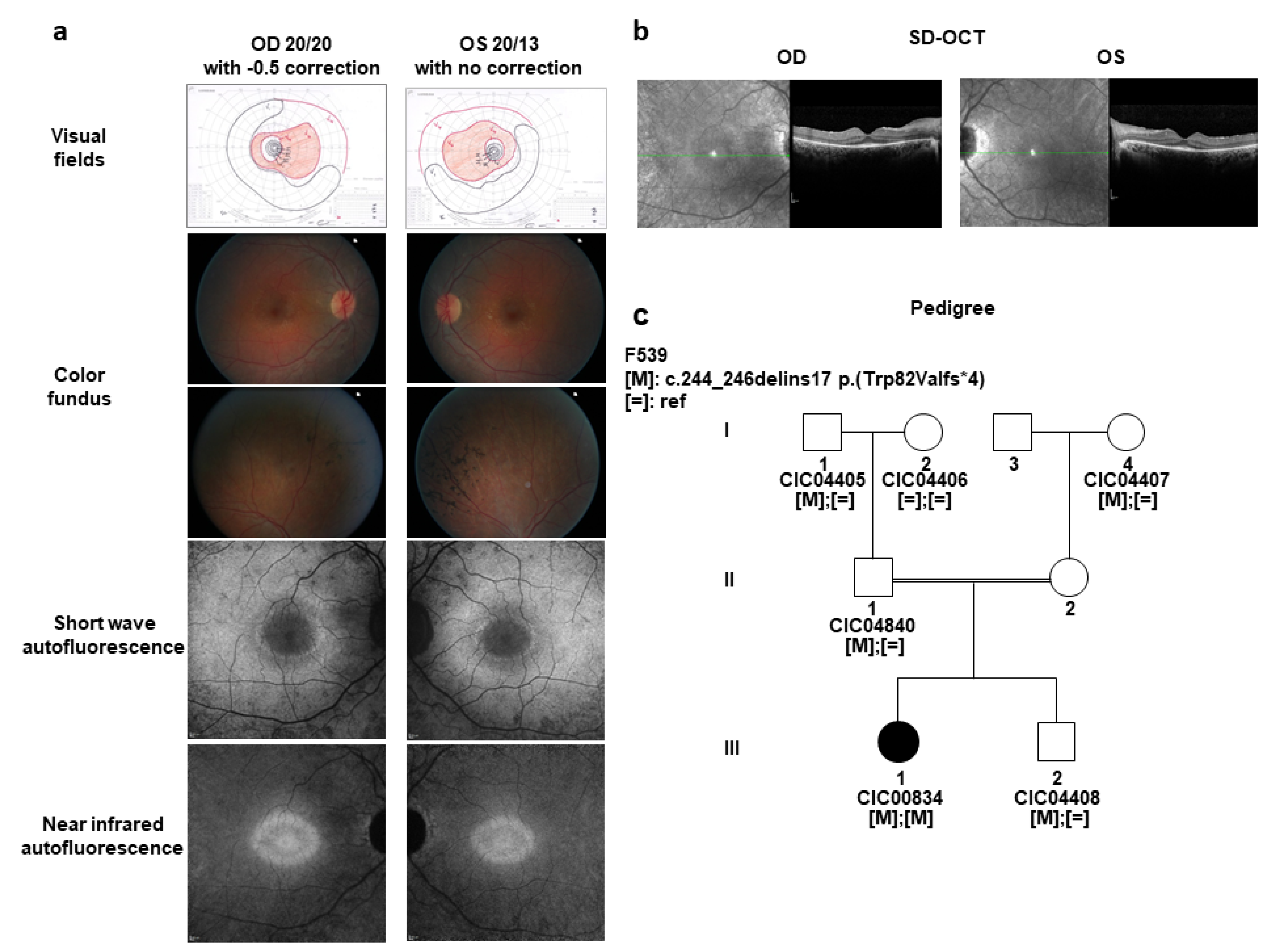





| Chromosome | Gene Name | Refseq and MIM# | Variant | Minor Allele Frequency | Retinal Expression [20] |
|---|---|---|---|---|---|
| 3 | CCDC51 | NM_001256964.1 No MIM# | c.244_246delinsGTGGAGATATGAAGATA p.(Trp82Valfs*4) | none reported | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeitz, C.; Méjécase, C.; Michiels, C.; Condroyer, C.; Wohlschlegel, J.; Foussard, M.; Antonio, A.; Démontant, V.; Emmenegger, L.; Schalk, A.; et al. Mutated CCDC51 Coding for a Mitochondrial Protein, MITOK Is a Candidate Gene Defect for Autosomal Recessive Rod-Cone Dystrophy. Int. J. Mol. Sci. 2021, 22, 7875. https://doi.org/10.3390/ijms22157875
Zeitz C, Méjécase C, Michiels C, Condroyer C, Wohlschlegel J, Foussard M, Antonio A, Démontant V, Emmenegger L, Schalk A, et al. Mutated CCDC51 Coding for a Mitochondrial Protein, MITOK Is a Candidate Gene Defect for Autosomal Recessive Rod-Cone Dystrophy. International Journal of Molecular Sciences. 2021; 22(15):7875. https://doi.org/10.3390/ijms22157875
Chicago/Turabian StyleZeitz, Christina, Cécile Méjécase, Christelle Michiels, Christel Condroyer, Juliette Wohlschlegel, Marine Foussard, Aline Antonio, Vanessa Démontant, Lisa Emmenegger, Audrey Schalk, and et al. 2021. "Mutated CCDC51 Coding for a Mitochondrial Protein, MITOK Is a Candidate Gene Defect for Autosomal Recessive Rod-Cone Dystrophy" International Journal of Molecular Sciences 22, no. 15: 7875. https://doi.org/10.3390/ijms22157875







