ZmFAR1 and ZmABCG26 Regulated by microRNA Are Essential for Lipid Metabolism in Maize Anther
Abstract
:1. Introduction
2. Results
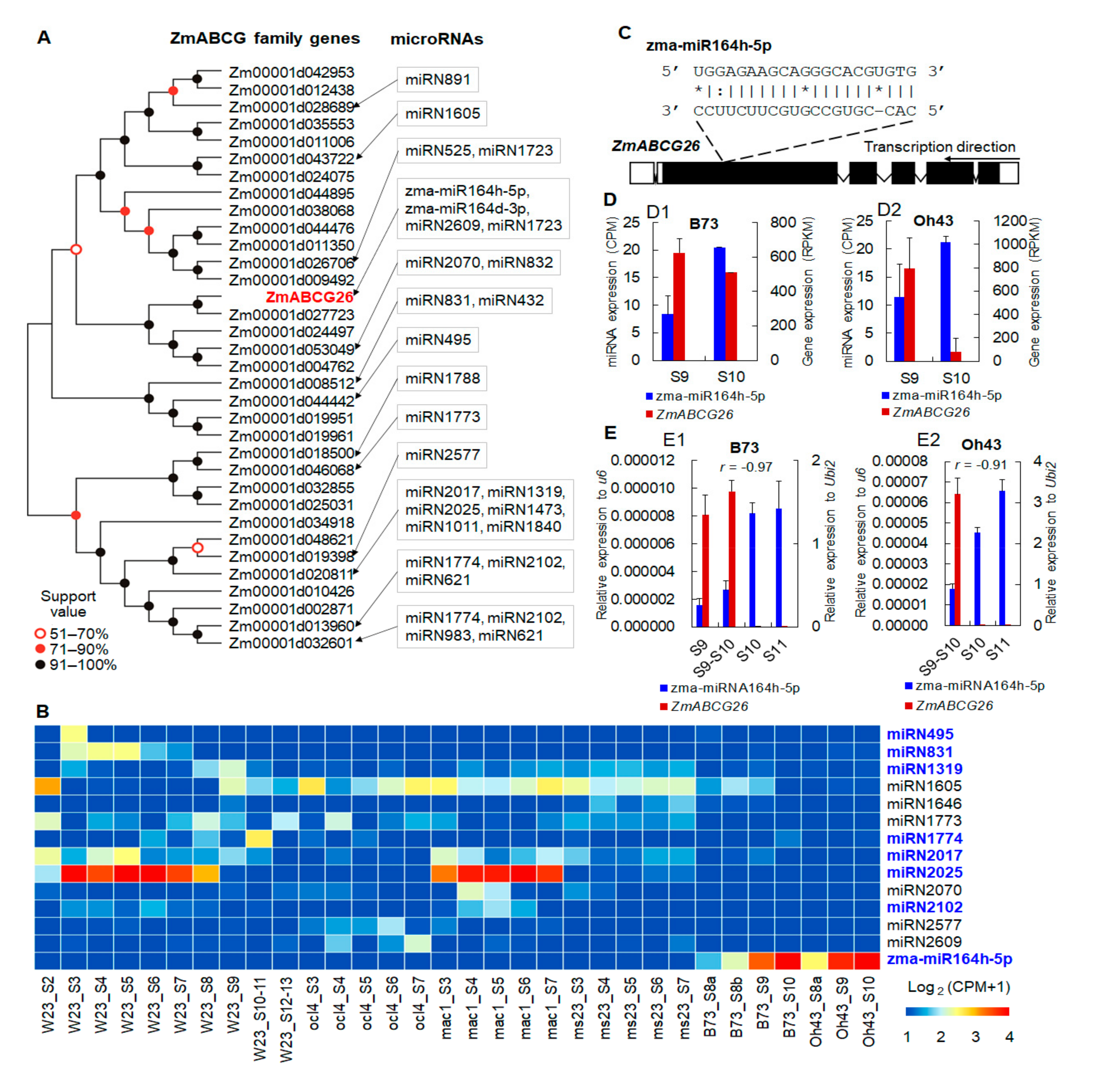
2.1. The Potential Posttranscriptional Regulation of ZmABCG26 by zma-miR164h-5p during Maize Anther Development
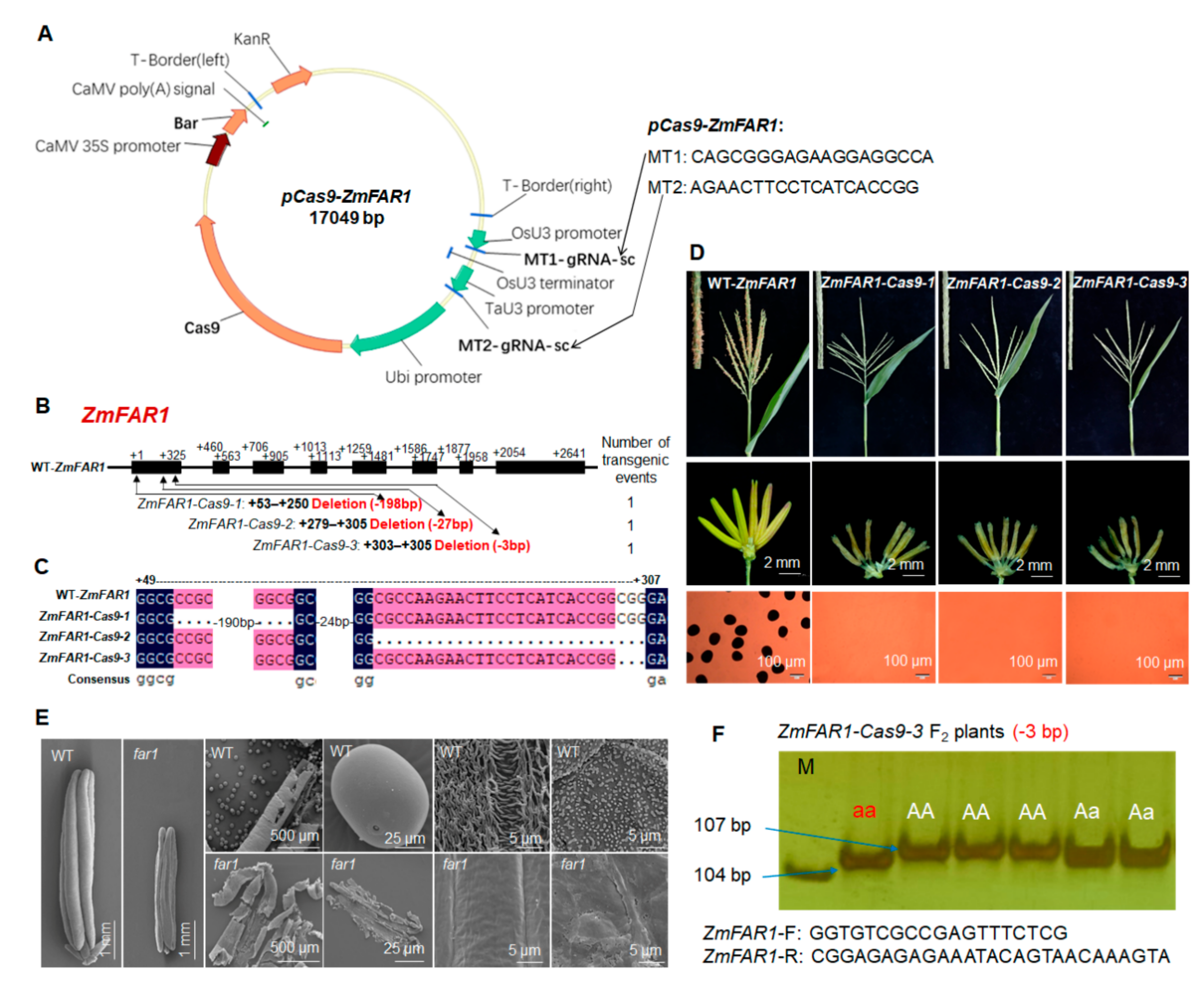
2.2. CRISPR/Cas9-Induced Loss Function of ZmABCG26 Leads to Complete Male Sterility in Maize
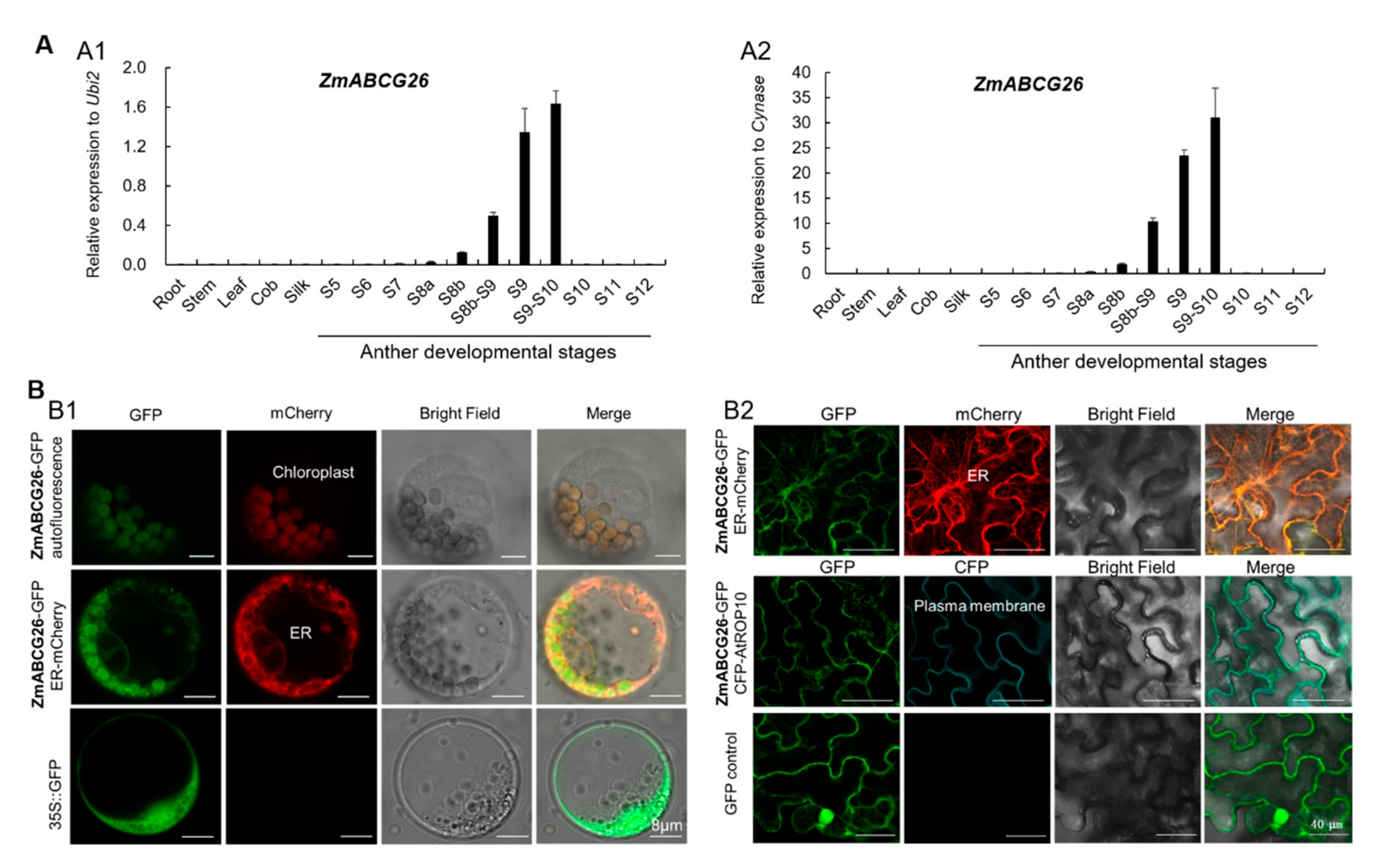
2.3. Spatiotemporal Expression Analysis of ZmABCG26 Gene and Subcellular Localization of ZmABCG26 Protein
2.4. CRISPR/Cas9-Based Confirmation of ZmFAR1 Function in Controlling Maize Male Sterility
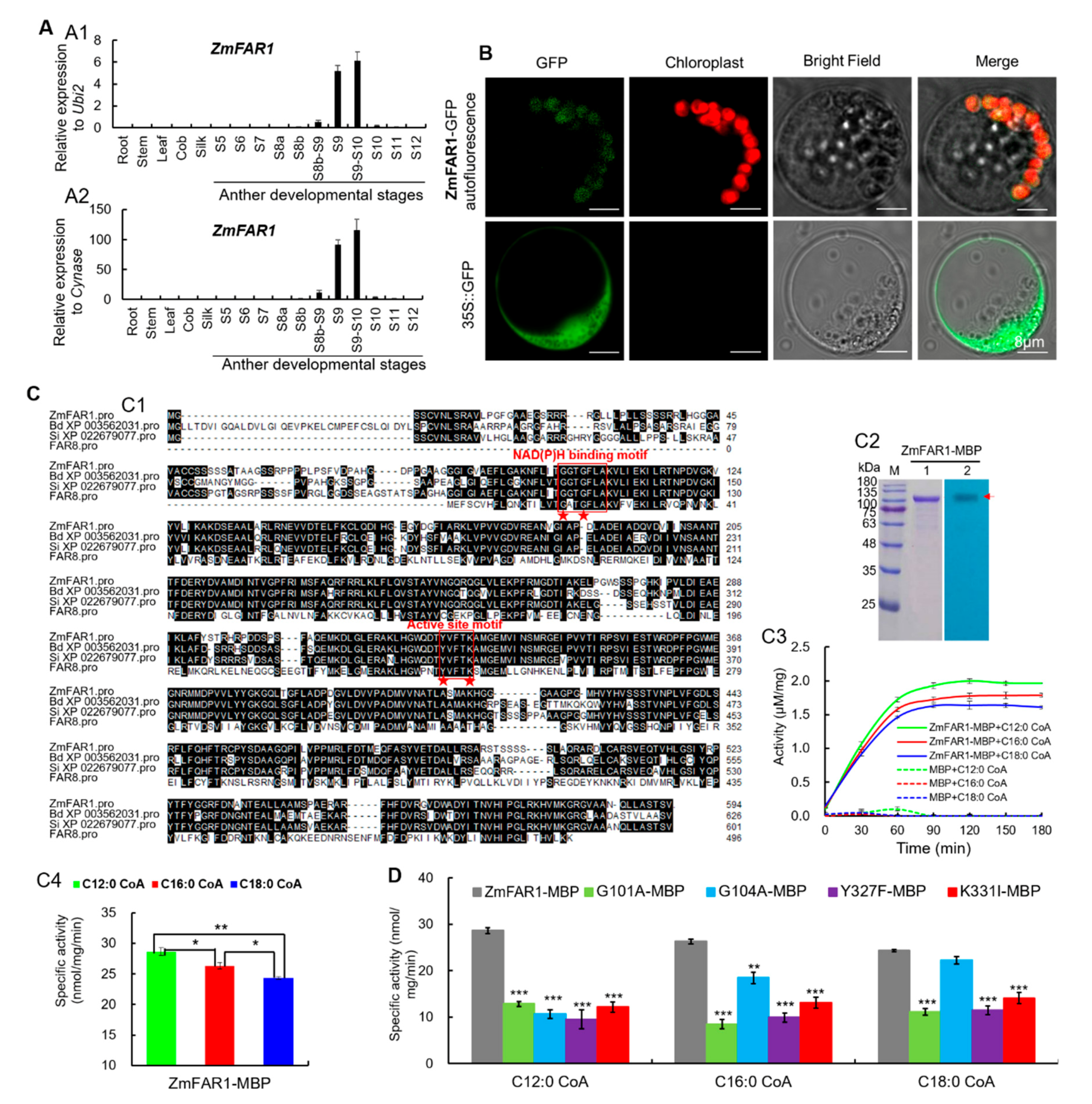
2.5. Spatiotemporal Expression Analysis of ZmFAR1 Gene and Subcellular Localization and Enzymatic Activity Analysis of ZmFAR1 Protein
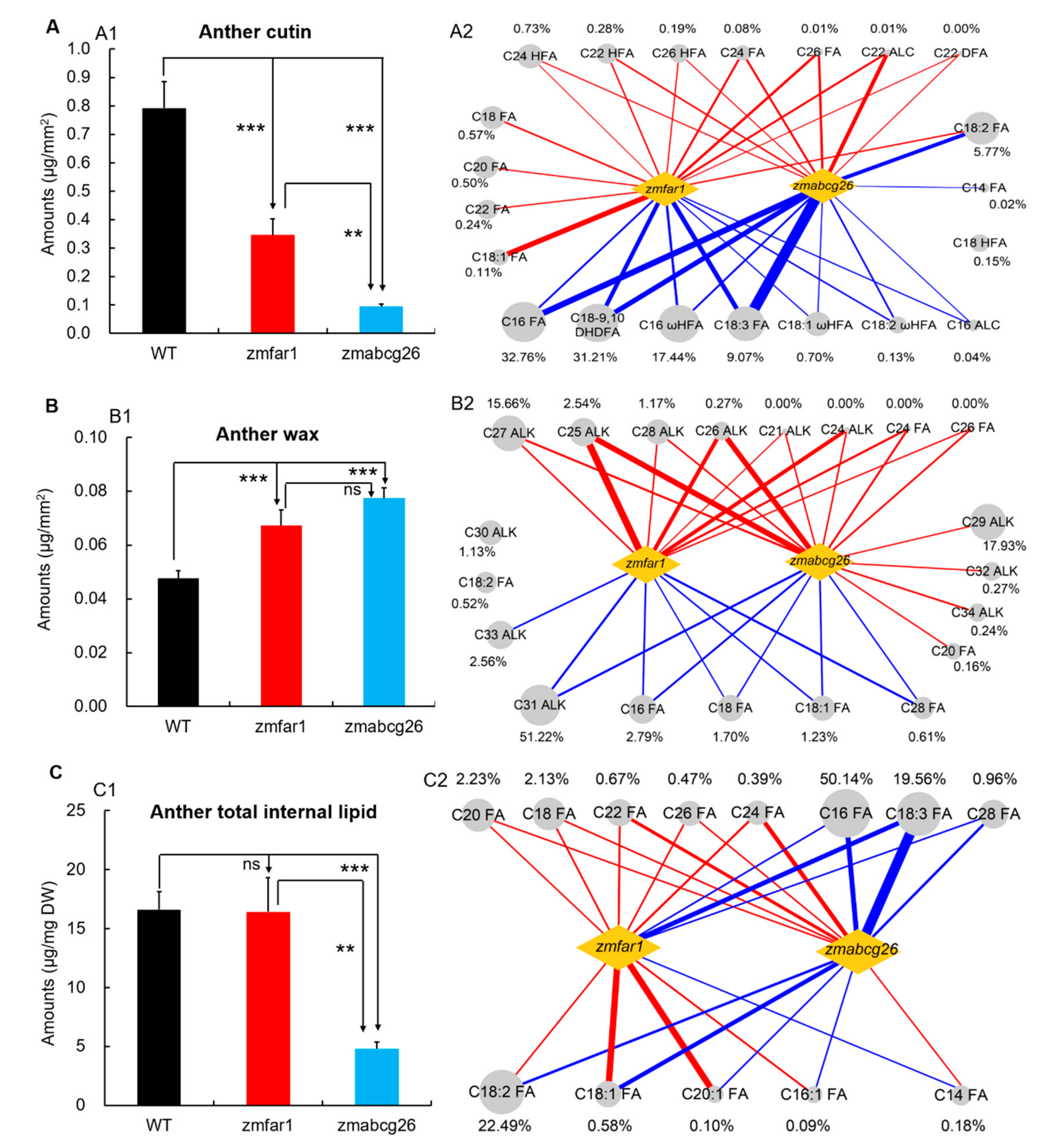
2.6. Functional Comparison of ZmABCG26 and ZmFAR1 Contributing to Lipid Metabolism in Maize Anthers
2.7. ZmFAR1 and ZmABCG26 May Be Regulated by a Complex TF Regulatory Network during Anther Development
3. Discussion
3.1. miRNA-Mediated Lipid Metabolism Is Most Probably Critical for Maize Anther Development
3.2. ZmFAR1 and ZmABCG26 Display the Functional Differences in Anther Lipid Metabolism
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Characterization of Mutant Phenotypes
4.3. RNA-Seq and Small RNA-Seq Analyses
4.4. Plasmid Construction and Maize Transformation
4.5. Genotyping Maize Plants
4.6. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.7. Phylogenetic Analysis
4.8. Subcellular Localization of ZmABCG26 and ZmFAR1
4.9. Enzyme Activity Assay of ZmFAR1 and Its Mutants
4.10. Analysis of Anther Wax, Cutin, and Internal Fatty Acids
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABCG | ATP-Binding Cassette Transporter G |
| ALC | Alcohol |
| ALK | Alkane |
| DHDFA | Docosahexaenoic Acid Fatty Acid |
| FA | Fatty Acid |
| FAR | Fatty Acyl Reductase |
| GC–MS | Gas Chromatography–Mass Spectrometry |
| GMS | Genic Male Sterility |
| HFA | Hydroxy Fatty Acid |
| miRNA | microRNA |
| miRNA-Seq | microRNA Sequencing |
| PAGE | Polyacrylamide Gel Electrophoresis |
| qRT-PCR | quantitative Reverse Transcription PCR |
| RNA-Seq | RNA Sequencing |
| SEM | Scanning Electron Microscopy |
| WBC | White Brown Complex |
| WT | Wild Type |
References
- Zhang, D.; Yang, L. Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 2014, 17, 49–55. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, Z.; Cao, J. Pollen wall development: The associated enzymes and metabolic pathways. Plant Biol. 2013, 15, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Cui, M.; Yang, L.; Kim, Y.J.; Zhang, D. Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 2015, 20, 741–753. [Google Scholar] [CrossRef]
- Wan, X.; Wu, S.; Li, Z.; An, X.; Tian, Y. Lipid metabolism: Critical roles in male fertility and other aspects of reproductive development in plants. Mol. Plant 2020, 13, 955–983. [Google Scholar] [CrossRef]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Chen, Y.; Charnikhova, T.; Mulder, P.P.; Heijmans, J.; Hoogenboom, A.; Agalou, A.; Michel, C.; Morel, J.B.; Dreni, L.; et al. OsJAR1 is required for JA-regulated floret opening and anther dehiscence in rice. Plant Mol. Biol. 2014, 86, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Li, Z.; An, X.; Long, Y.; Xue, X.; Xie, K.; Ma, B.; Zhang, D.; Guan, Y.; Niu, C.; et al. Normal structure and function of endothecium chloroplasts maintained by ZmMs33-mediated lipid biosynthesis in tapetal cells are critical for anther development in maize. Mol. Plant 2020, 13, 1624–1643. [Google Scholar] [CrossRef]
- Wang, J.; Bao, J.L.; Zhou, B.B.; Li, M.; Li, X.Z.; Jin, J. The osa-miR164 target OsCUC1 functions redundantly with OsCUC3 in controlling rice meristem/organ boundary specification. New Phytol. 2021, 229, 1566–1581. [Google Scholar] [CrossRef]
- Millar, A.A.; Lohe, A.; Wong, G. Biology and function of miR159 in plants. Plants 2019, 8, 255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Mao, Y.; Yang, J.; He, Y. TCP24 modulates secondary cell wall thickening and anther endothecium development. Front. Plant Sci. 2015, 6, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Nagpal, P.; Villarino, G.; Trinidad, B.; Bird, L.; Huang, Y.; Reed, J.W. miR167 limits anther growth to potentiate anther dehiscence. Development 2019, 146, dev174375. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhu, T.; Liu, S.; Jiang, Y.; Liu, H.; Zhang, Y.; Xie, K.; Li, J.; An, X.; Wan, X. Genome-wide analyses on transcription factors and their potential microRNA regulators involved in maize male fertility. Crop J. 2021. [Google Scholar] [CrossRef]
- Zhang, Y.; He, R.; Lian, J.; Zhou, Y.; Zhang, F.; Li, Q.; Yu, Y.; Feng, Y.; Yang, Y.; Lei, M.; et al. OsmiR528 regulates rice-pollen intine formation by targeting an uclacyanin to influence flavonoid metabolism. Proc. Natl. Acad. Sci. USA 2020, 117, 727–732. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Fang, Y.; Wu, X.; Qing, M.; Li, C.; Xie, K.; Deng, X.; Guo, W. The miR399-CsUBC24 module regulates reproductive development and male fertility in citrus. Plant Physiol. 2020, 183, 1681–1695. [Google Scholar] [CrossRef]
- Verrier, P.J.; Bird, D.; Buria, B.; Dassa, E.; Forestier, C.; Geisler, M.; Klein, M.; Kolukisaoglu, U.; Lee, Y.; Martinoia, E.; et al. Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Pang, K.; Li, Y.; Liu, M.; Meng, Z.; Yu, Y. Inventory and general analysis of the ATP-binding cassette (ABC) gene superfamily in maize (Zea mays L.). Gene 2013, 526, 411–428. [Google Scholar] [CrossRef]
- Bessire, M.; Borel, S.; Fabre, G.; Carraca, L.; Efremova, N.; Yephremov, A.; Cao, Y.; Jetter, R.; Jacquat, A.-C.; Metraux, J.-P.; et al. A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 2011, 23, 1958–1970. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Jin, J.Y.; Choi, S.; Hwang, J.U.; Kim, Y.Y.; Suh, M.C.; Lee, Y. An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J. 2011, 65, 181–193. [Google Scholar] [CrossRef]
- Quilichini, T.D.; Friedmann, M.C.; Samuels, A.L.; Douglas, C.J. ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol. 2010, 154, 678–690. [Google Scholar] [CrossRef] [Green Version]
- Yadav, V.; Molina, I.; Ranathunge, K.; Castillo, I.Q.; Rothstein, S.J.; Reed, J.W. ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell 2014, 26, 3569–3588. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Ohyama, K.; Kim, Y.-Y.; Jin, J.-Y.; Lee, S.B.; Yamaoka, Y.; Muranaka, T.; Suh, M.C.; Fujioka, S.; Lee, Y. The role of Arabidopsis ABCG9 and ABCG31 ATP binding cassette transporters in pollen fitness and the deposition of steryl glycosides on the pollen coat. Plant Cell 2014, 26, 310–324. [Google Scholar] [CrossRef] [Green Version]
- Panikashvili, D.; Shi, J.X.; Schreiber, L.; Aharoni, A. The Arabidopsis ABCG13 transporter is required for flower cuticle secretion and patterning of the petal epidermis. New Phytol. 2011, 190, 113–124. [Google Scholar] [CrossRef]
- McFarlane, H.E.; Shin, J.J.; Bird, D.A.; Samuels, A.L. Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 2010, 22, 3066–3075. [Google Scholar] [CrossRef] [Green Version]
- Panikashvili, D.; Shi, J.X.; Bocobza, S.; Franke, R.B.; Schreiber, L.; Aharoni, A. The Arabidopsis DSO/ABCG11 transporter affects cutin metabolism in reproductive organs and suberin in roots. Mol. Plant 2010, 3, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.T.; Li, H.; Xia, X.Z.; Meng, Y.Y.; Wang, N.; Li, L.L.; Shi, J.X.; Pei, Y.X.; Lin, M.; Niu, L.F.; et al. ATP-binding cassette G transporters SGE1 and MtABCG13 control stigma exsertion. Plant Physiol. 2020, 184, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.X.; He, F.R.; He, M.; Ren, D.; Chen, L.T.; Liu, Y.G. The ATP-binding cassette transporter OsABCG15 is required for anther development and pollen fertility in rice. J. Integr. Plant Biol. 2013, 55, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Shi, J.; Liang, W.; Xue, F.; Luo, Q.; Zhu, L.; Qu, G.; Chen, M.; Schreiber, L.; Zhang, D. Two ATP binding cassette G transporters, rice ATP binding cassette G26 and ATP binding cassette G15, collaboratively regulate rice male reproduction. Plant Physiol. 2015, 169, 2064–2079. [Google Scholar] [CrossRef] [Green Version]
- Chang, Z.; Jin, M.; Yan, W.; Chen, H.; Qiu, S.; Fu, S.; Xia, J.; Liu, Y.; Chen, Z.; Wu, J.; et al. The ATP-binding cassette (ABC) transporter OsABCG3 is essential for pollen development in rice. Rice 2018, 11. [Google Scholar] [CrossRef]
- Luo, T.; Zou, T.; Yuan, G.Q.; He, Z.Y.; Li, W.J.; Tao, Y.; Liu, M.M.; Zhou, D.; Zhao, H.F.; Zhu, J.; et al. Less and shrunken pollen 1 (LSP1) encodes a member of the ABC transporter family required for pollen wall development in rice (Oryza sativa L.). Crop J. 2020, 8, 492–504. [Google Scholar] [CrossRef]
- Li, L.; Li, D.L.; Liu, S.Z.; Ma, X.L.; Dietrich, C.R.; Hu, H.C.; Zhang, G.S.; Liu, Z.Y.; Zheng, J.; Wang, G.Y.; et al. The Maize glossy13 Gene, cloned via BSR-Seq and Seq-Walking encodes a putative ABC transporter required for the normal accumulation of epicuticular waxes. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Yang, L.; Kang, D.; Ren, Z.; Liu, Y. Maize MS2 encodes an ATP-binding cassette transporter that is essential for anther development. Crop J. 2021. [Google Scholar] [CrossRef]
- Kunst, L.; Samuels, A.L. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 2003, 42, 51–80. [Google Scholar] [CrossRef]
- Aarts, M.; Hodge, R.; Kalantidis, K.; Florack, D.; Wilson, Z.; Mulligan, B.; Stiekema, W.; Scott, R.; Pereira, A. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 1997, 12, 615–623. [Google Scholar] [CrossRef]
- Chen, W.; Yu, X.H.; Zhang, K.; Shi, J.; De Oliveira, S.; Schreiber, L.; Shanklin, J.; Zhang, D. Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 2011, 157, 842–853. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Guo, Z.; Zhou, W.; Zhang, C.; Zhang, Z.; Lou, Y.; Xiong, S.; Yao, X.; Fan, J.; Zhu, J.; et al. The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation. Plant Physiol. 2018, 178, 283–294. [Google Scholar] [CrossRef] [Green Version]
- Rowland, O.; Zheng, H.; Hepworth, S.R.; Lam, P.; Jetter, R.; Kunst, L. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Tan, H.; Yu, X.H.; Liu, Y.; Liang, W.; Ranathunge, K.; Franke, R.B.; Schreiber, L.; Wang, Y.; Kai, G.; et al. Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 2011, 23, 2225–2246. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Yan, W.; Chang, Z.; Xu, Y.; Luo, M.; Xu, C.; Chen, Z.; Wu, J.; Tang, X. OsMYB80 regulates anther development and pollen fertility by targeting multiple biological pathways. Plant Cell Physiol. 2020, 61, 988–1004. [Google Scholar] [CrossRef]
- Wang, A.M.; Xia, Q.; Xie, W.S.; Dumonceaux, T.; Zou, J.T.; Datla, R.; Selvaraj, G. Male gametophyte development in bread wheat (Triticum aestivum L.): Molecular, cellular, and biochemical analyses of a sporophytic contribution to pollen wall ontogeny. Plant J. 2002, 30, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Xiao, S.; Liu, J.; Somaratne, Y.; Zhang, H.; Wang, M.; Zhang, H.; Zhao, L.; Chen, H. MALE STERILE6021 (MS6021) is required for the development of anther cuticle and pollen exine in maize. Sci. Rep. 2017, 7, 16736. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, S.; Niu, C.; Liu, D.; Yan, T.; Tian, Y.; Liu, S.; Xie, K.; Li, Z.; Wang, Y.; et al. ZmMs25 encoding a plastid-localized fatty acyl reductase is critical for anther and pollen development in maize. J. Exp. Bot. 2021, 72, 4298–4318. [Google Scholar] [CrossRef]
- Li, Z.; An, X.; Zhu, T.; Yan, T.; Wu, S.; Tian, Y.; Li, J.; Wan, X. Discovering and constructing ceRNA-miRNA-target gene regulatory networks during anther development in maize. Int. J. Mol. Sci. 2019, 20, 3480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, J.; Zhang, H.; Arikit, S.; Huang, K.; Nan, G.-L.; Walbot, V.; Meyers, B.C. Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. USA 2015, 112, 3146–3151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, X.; Ma, B.; Duan, M.; Dong, Z.; Liu, R.; Yuan, D.; Hou, Q.; Wu, S.; Zhang, D.; Liu, D.; et al. Molecular regulation of ZmMs7 required for maize male fertility and development of a dominant male-sterility system in multiple species. Proc. Natl. Acad. Sci. USA 2020, 117, 23499–23509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wu, S.; An, X.; Xie, K.; Dong, Z.; Zhou, Y.; Xu, L.; Fang, W.; Liu, S.; Liu, S.; et al. Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 2018, 16, 459–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, X.; Dong, Z.; Tian, Y.; Xie, K.; Wu, S.; Zhu, T.; Zhang, D.; Zhou, Y.; Niu, C.; Ma, B.; et al. ZmMs30 encoding a novel GDSL lipase is essential for male fertility and valuable for hybrid breeding in maize. Mol. Plant 2019, 12, 343–359. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; Wu, S.; Li, Z.; Zhou, Y.; Zhang, D.; Dong, Z.; An, X.; Zhu, T.; Zhang, S.; Liu, S.; et al. Map-based cloning and characterization of Zea mays male sterility33 (ZmMs33) gene, encoding a glycerol-3-phosphate acyltransferase. Theor. Appl. Genet. 2018, 131, 1363–1378. [Google Scholar] [CrossRef] [Green Version]
- Zhu, T.; Wu, S.; Zhang, D.; Li, Z.; Xie, K.; An, X.; Ma, B.; Hou, Q.; Dong, Z.; Tian, Y.; et al. Genome-wide analysis of maize GPAT gene family and cytological characterization and breeding application of ZmMs33/ZmGPAT6 gene. Theor. Appl. Genet. 2019, 132, 2137–2154. [Google Scholar] [CrossRef]
- Somaratne, Y.; Tian, Y.; Zhang, H.; Wang, M.; Huo, Y.; Cao, F.; Zhao, L.; Chen, H. ABNORMAL POLLEN VACUOLATION1 (APV1) is required for male fertility by contributing to anther cuticle and pollen exine formation in maize. Plant J. 2017, 90, 96–110. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, H.; Sun, H.; Luo, H.; Zhao, L.; Dong, Z.; Yan, S.; Zhao, C.; Liu, R.; Xu, C.; et al. IRREGULAR POLLEN EXINE1 is a novel factor in anther cuticle and pollen exine formation. Plant Physiol. 2017, 173, 307–325. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, D.; Tian, Y.; Wu, S.; An, X.; Dong, Z.; Zhang, S.; Bao, J.; Li, Z.; Li, J.; et al. Map-based cloning, phylogenetic, and microsynteny analyses of ZmMs20 gene regulating male fertility in maize. Int. J. Mol. Sci. 2019, 20, 1411. [Google Scholar] [CrossRef] [Green Version]
- Djukanovic, V.; Smith, J.; Lowe, K.; Yang, M.; Gao, H.; Jones, S.; Nicholson, M.G.; West, A.; Lape, J.; Bidney, D.; et al. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J. 2013, 76, 888–899. [Google Scholar] [CrossRef]
- Yang, X.; Wu, D.; Shi, J.; He, Y.; Pinot, F.; Grausem, B.; Yin, C.; Zhu, L.; Chen, M.; Luo, Z.; et al. Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integra. Plant Biol. 2014, 56, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pinot, F.; Sauveplane, V.; Werck-Reichhart, D.; Diehl, P.; Schreiber, L.; Franke, R.; Zhang, P.; Chen, L.; Gao, Y.; et al. Cytochrome P450 family member CYP704B2 catalyzes the omega-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 2010, 22, 173–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, K.H.; Han, M.J.; Lee, D.Y.; Lee, Y.S.; Schreiber, L.; Franke, R.; Faust, A.; Yephremov, A.; Saedler, H.; Kim, Y.W.; et al. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 2006, 18, 3015–3032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurdyukov, S.; Faust, A.; Nawrath, C.; Bar, S.; Voisin, D.; Efremova, N.; Franke, R.; Schreiber, L.; Saedler, H.; Metraux, J.P.; et al. The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 2006, 18, 321–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, R.K.; Jain, M. NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [Green Version]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic. Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic. Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [Green Version]
- Friedlaender, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic. Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Bonnet, E.; He, Y.; Billiau, K.; Van de Peer, Y. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 2010, 26, 1566–1568. [Google Scholar] [CrossRef]
- Wu, H.-J.; Ma, Y.-K.; Chen, T.; Wang, M.; Wang, X.-J. PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic. Acids Res. 2012, 40, W22–W28. [Google Scholar] [CrossRef]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [Green Version]
- Frame, B.R.; Shou, H.; Chikwamba, R.K.; Zhang, Z.; Xiang, C.; Fonger, T.M.; Pegg, S.E.; Li, B.; Nettleton, D.S.; Pei, D.; et al. Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 2002, 129, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic. Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic. Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gupta, B.B.; Selter, L.L.; Baranwal, V.K.; Arora, D.; Mishra, S.K.; Sirohi, P.; Poonia, A.K.; Chaudharya, R.; Kumar, R.; Krattinger, S.G.; et al. Updated inventory, evolutionary and expression analyses of G, (PDR) type ABC transporter genes of rice. Plant Physiol. Bioch. 2019, 142, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Grafe, K.; Schmitt, L. The ABC transporter G subfamily in Arabidopsis thaliana. J. Exp. Bot. 2021, 72, 92–106. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Y.; Liu, D.; Niu, X.L.; Wang, J.Z.; Qian, L.C.; Han, L.; Liu, N.; Zhao, J.P.; Hong, Y.G.; Liu, Y.L. Antiviral resistance protein Tm-2(2) functions on the plasma membrane. Plant Physiol. 2017, 173, 2399–2410. [Google Scholar] [CrossRef] [Green Version]
- Chacon, M.G.; Fournier, A.E.; Tran, F.; Dittrich-Domergue, F.; Pulsifer, I.P.; Domergue, F.; Rowland, O. Identification of amino acids conferring chain length substrate specificities on fatty alcohol-forming reductases FAR5 and FAR8 from Arabidopsis thaliana. J. Biol. Chem. 2013, 288, 30345–30355. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Li, Z.; Liu, X.; Zhu, T.; Xie, K.; Hou, Q.; Yan, T.; Niu, C.; Zhang, S.; Yang, M.; et al. ZmFAR1 and ZmABCG26 Regulated by microRNA Are Essential for Lipid Metabolism in Maize Anther. Int. J. Mol. Sci. 2021, 22, 7916. https://doi.org/10.3390/ijms22157916
Jiang Y, Li Z, Liu X, Zhu T, Xie K, Hou Q, Yan T, Niu C, Zhang S, Yang M, et al. ZmFAR1 and ZmABCG26 Regulated by microRNA Are Essential for Lipid Metabolism in Maize Anther. International Journal of Molecular Sciences. 2021; 22(15):7916. https://doi.org/10.3390/ijms22157916
Chicago/Turabian StyleJiang, Yilin, Ziwen Li, Xinze Liu, Taotao Zhu, Ke Xie, Quancan Hou, Tingwei Yan, Canfang Niu, Shaowei Zhang, Mengbing Yang, and et al. 2021. "ZmFAR1 and ZmABCG26 Regulated by microRNA Are Essential for Lipid Metabolism in Maize Anther" International Journal of Molecular Sciences 22, no. 15: 7916. https://doi.org/10.3390/ijms22157916
APA StyleJiang, Y., Li, Z., Liu, X., Zhu, T., Xie, K., Hou, Q., Yan, T., Niu, C., Zhang, S., Yang, M., Xie, R., Wang, J., Li, J., An, X., & Wan, X. (2021). ZmFAR1 and ZmABCG26 Regulated by microRNA Are Essential for Lipid Metabolism in Maize Anther. International Journal of Molecular Sciences, 22(15), 7916. https://doi.org/10.3390/ijms22157916







