A Proteomic Study Suggests Stress Granules as New Potential Actors in Radiation-Induced Bystander Effects
Abstract
:1. Introduction
2. Results
2.1. Secretome Analysis of the Conditioned Medium of Low Doses Irradiated Chondrosarcoma Cells
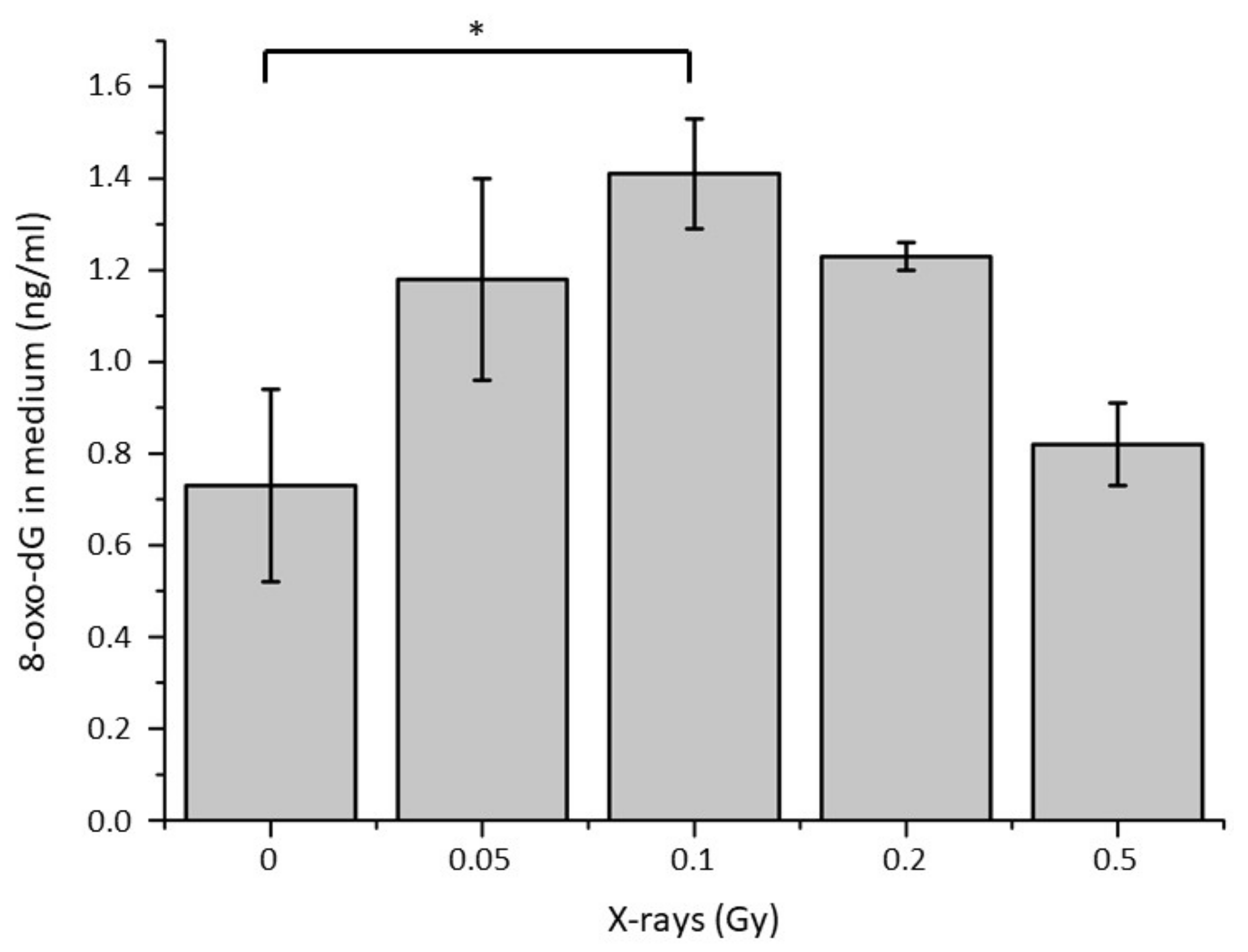
2.2. Quantification of 8-oxoG in the Conditioned Medium of Low Doses Irradiated Chondrosarcoma Cells
2.3. Whole-Cell Proteome Variations of Chondrocytes in Responses to the Conditioned Medium of Low Doses Irradiated Chondrosarcoma Cells
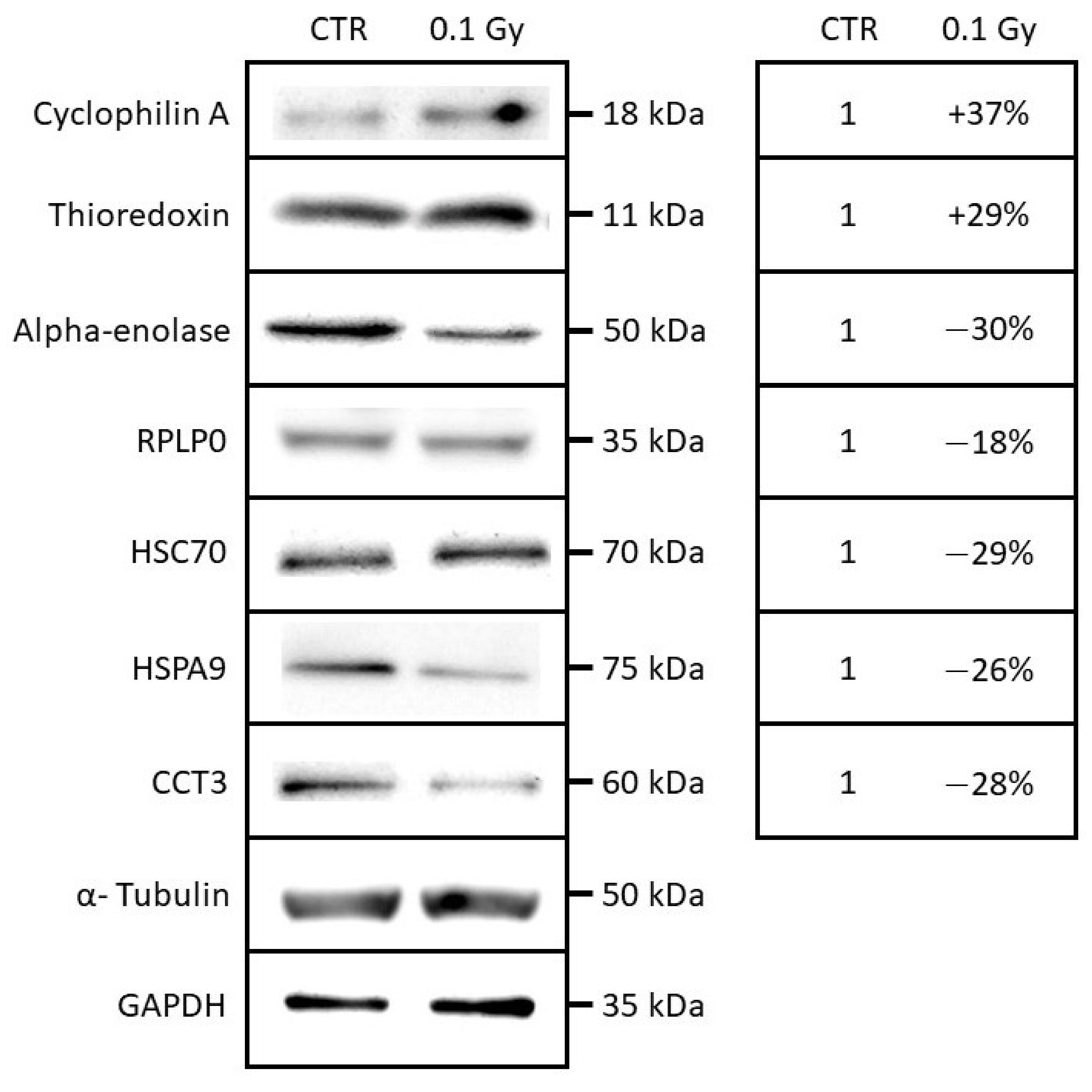
2.4. Quantification and Validation of Proteomic Biomarkers
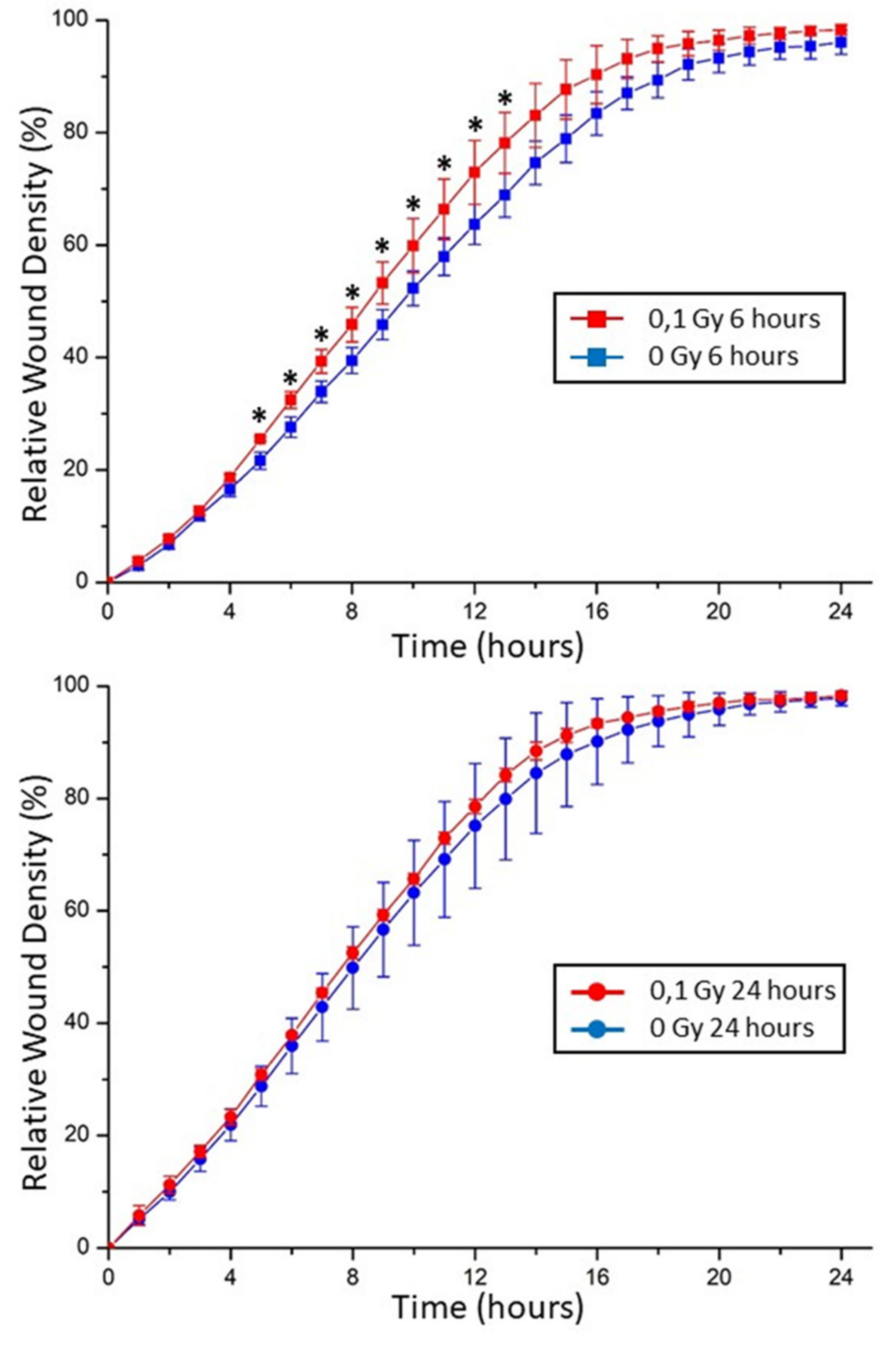
2.5. Chondrocyte Motility in Extracellular Matrix Affected by Exogenous Stresses
3. Discussion
4. Conclusions
5. Methods
5.1. Cell Culture
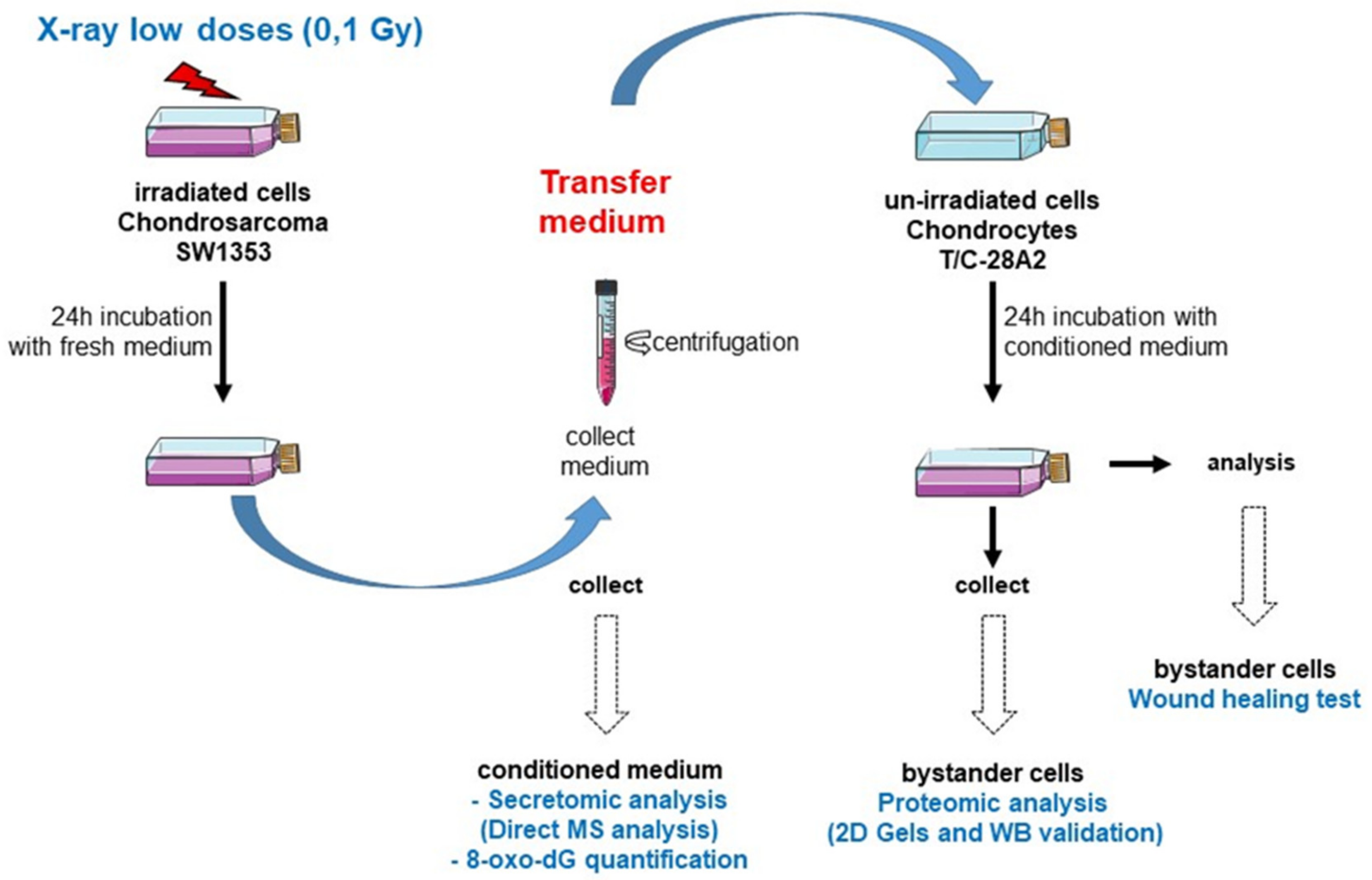
5.2. Experimental Strategy to Characterize the Bystander Effectinduced by Low Doses Irradiated Chondrosarcoma Cells
5.3. Preparation of Conditioned Medium and Shotgun Proteomics Analysis
5.4. Determination of Extracellular 8-oxo-dG in the Conditioned Media
5.5. Medium-Transfer Protocol from Irradiated Cells to Non-Irradiated Cells
5.6. Gel-Based Proteomic Study of Bystander Chondrocytes
5.6.1. Chemicals
5.6.2. Protein Extraction and Solubilisation
5.6.3. Strip Rehydration with Protein Samples: “Sample In-Gel Rehydration”
5.6.4. IPG Strips Equilibration and Second Dimension
5.6.5. Gel Staining and Picture Acquisition
5.6.6. Image Analysis
5.6.7. Mass Spectrometry Analysis of 2D-Spots
5.7. Western Blotting Analysis
5.8. Wound-Healing Assay (IncuCyte® Live-Cell Analysis Systems)
5.9. Statistical Analyses
5.10. Mass Spectrometry Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durante, M. New Challenges in High-Energy Particle Radiobiology. Br. J. Radiol. 2014, 87, 20130626. [Google Scholar] [CrossRef] [Green Version]
- Noël, G.; Feuvret, L.; Ferrand, R.; Mazeron, J.-J. Treatment with charged particles beams: Hadrontherapy part I: Physical basis and clinical experience of treatment with protons. Cancer Radiother. 2003, 7, 321–339. [Google Scholar] [CrossRef]
- Thariat, J.; Valable, S.; Laurent, C.; Haghdoost, S.; Pérès, E.A.; Bernaudin, M.; Sichel, F.; Lesueur, P.; Césaire, M.; Petit, E.; et al. Hadrontherapy Interactions in Molecular and Cellular Biology. Int. J. Mol. Sci. 2020, 21, 133. [Google Scholar] [CrossRef] [Green Version]
- Mothersill, C.; Seymour, C. Radiation-Induced Bystander Effects: Past History and Future Directions. Radiat. Res. 2001, 155, 759–767. [Google Scholar] [CrossRef]
- Chevalier, F.; Hamdi, D.H.; Saintigny, Y.; Lefaix, J.-L. Proteomic Overview and Perspectives of the Radiation-Induced Bystander Effects. Mutat. Res./Rev. Mutat. Res. 2014, 763, 280–293. [Google Scholar] [CrossRef]
- Blyth, B.J.; Sykes, P.J. Radiation-Induced Bystander Effects: What Are They, and How Relevant Are They to Human Radiation Exposures? Radiat. Res. 2011, 176, 139–157. [Google Scholar] [CrossRef] [PubMed]
- Azzam, E.I.; de Toledo, S.M.; Little, J.B. Stress Signaling from Irradiated to Non-Irradiated Cells. Curr. Cancer Drug Targets 2004, 4, 53–64. [Google Scholar] [CrossRef]
- Prise, K.M.; O’Sullivan, J.M. Radiation-Induced Bystander Signalling in Cancer Therapy. Nat. Rev. Cancer 2009, 9, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Abramowicz, A.; Wojakowska, A.; Marczak, L.; Lysek-Gladysinska, M.; Smolarz, M.; Story, M.D.; Polanska, J.; Widlak, P.; Pietrowska, M. Ionizing Radiation Affects the Composition of the Proteome of Extracellular Vesicles Released by Head-and-Neck Cancer Cells in Vitro. J. Radiat. Res. 2019, 60, 289–297. [Google Scholar] [CrossRef]
- Desai, S.; Srambikkal, N.; Yadav, H.D.; Shetake, N.; Balla, M.M.S.; Kumar, A.; Ray, P.; Ghosh, A.; Pandey, B.N. Molecular Understanding of Growth Inhibitory Effect from Irradiated to Bystander Tumor Cells in Mouse Fibrosarcoma Tumor Model. PLoS ONE 2016, 11, e0161662. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.W.; Moccia, R.D.; Seymour, C.B.; Mothersill, C.E. Irradiation of Rainbow Trout at Early Life Stages Results in a Proteomic Legacy in Adult Gills. Part A; Proteomic Responses in the Irradiated Fish and in Non-Irradiated Bystander Fish. Environ. Res. 2018, 163, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Lepleux, C.; Marie-Brasset, A.; Temelie, M.; Boulanger, M.; Brotin, É.; Goldring, M.B.; Hirtz, C.; Varès, G.; Nakajima, T.; Saintigny, Y.; et al. Bystander Effectors of Chondrosarcoma Cells Irradiated at Different LET Impair Proliferation of Chondrocytes. J. Cell Commun. Signal. 2019, 13, 343–356. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, A.; Tudor, M.; Lepleux, C.; Rofidal, V.; Hem, S.; Almunia, C.; Armengaud, J.; Brotin, E.; Haghdoost, S.; Savu, D.; et al. Proteomic Analysis of Bystander Effects in Chondrosarcoma Cells. In Proceedings of the ERRS 45th Annual Meeting of the European Radiation Research Society, Lund, Sweden, 13–17 September 2020. p. Poster N°25. [Google Scholar]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Guzikowski, A.R.; Chen, Y.S.; Zid, B.M. Stress-Induced MRNP Granules: Form and Function of P-Bodies and Stress Granules. Wiley Interdiscip. Rev. RNA 2019, 10, e1524. [Google Scholar] [CrossRef]
- Kedersha, N.; Anderson, P. Mammalian Stress Granules and Processing Bodies. Methods Enzym. 2007, 431, 61–81. [Google Scholar] [CrossRef]
- Kedersha, N.; Ivanov, P.; Anderson, P. Stress Granules and Cell Signaling: More than Just a Passing Phase? Trends Biochem. Sci. 2013, 38, 494–506. [Google Scholar] [CrossRef] [Green Version]
- Mahboubi, H.; Stochaj, U. Cytoplasmic Stress Granules: Dynamic Modulators of Cell Signaling and Disease. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 884–895. [Google Scholar] [CrossRef]
- Moutaoufik, M.T.; Fatimy, R.E.; Nassour, H.; Gareau, C.; Lang, J.; Tanguay, R.M.; Mazroui, R.; Khandjian, E.W. UVC-Induced Stress Granules in Mammalian Cells. PLoS ONE 2014, 9, e112742. [Google Scholar] [CrossRef] [PubMed]
- Wakazono, A.; Fukao, T.; Yamaguchi, S.; Hori, T.; Orii, T.; Lambert, M.; Mitchell, G.A.; Lee, G.W.; Hashimoto, T. Molecular, Biochemical, and Clinical Characterization of Mitochondrial Acetoacetyl-Coenzyme A Thiolase Deficiency in Two Further Patients. Hum. Mutat. 1995, 5, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Haapalainen, A.M.; Meriläinen, G.; Pirilä, P.L.; Kondo, N.; Fukao, T.; Wierenga, R.K. Crystallographic and Kinetic Studies of Human Mitochondrial Acetoacetyl-CoA Thiolase: The Importance of Potassium and Chloride Ions for Its Structure and Function. Biochemistry 2007, 46, 4305–4321. [Google Scholar] [CrossRef]
- Gallego-García, A.; Monera-Girona, A.J.; Pajares-Martínez, E.; Bastida-Martínez, E.; Pérez-Castaño, R.; Iniesta, A.A.; Fontes, M.; Padmanabhan, S.; Elías-Arnanz, M. A Bacterial Light Response Reveals an Orphan Desaturase for Human Plasmalogen Synthesis. Science 2019, 366, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Mouneimne, G.; Hansen, S.D.; Selfors, L.M.; Petrak, L.; Hickey, M.M.; Gallegos, L.L.; Simpson, K.J.; Lim, J.; Gertler, F.B.; Hartwig, J.H.; et al. Differential Remodeling of Actin Cytoskeleton Architecture by Profilin Isoforms Leads to Distinct Effects on Cell Migration and Invasion. Cancer Cell 2012, 22, 615–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Martínez, E.; Ibarrola, J.; Fernández-Celis, A.; Santamaria, E.; Fernández-Irigoyen, J.; Rossignol, P.; Jaisser, F.; López-Andrés, N. Differential Proteomics Identifies Reticulocalbin-3 as a Novel Negative Mediator of Collagen Production in Human Cardiac Fibroblasts. Sci. Rep. 2017, 7, 12192. [Google Scholar] [CrossRef]
- Miotto, B.; Chibi, M.; Xie, P.; Koundrioukoff, S.; Moolman-Smook, H.; Pugh, D.; Debatisse, M.; He, F.; Zhang, L.; Defossez, P.-A. The RBBP6/ZBTB38/MCM10 Axis Regulates DNA Replication and Common Fragile Site Stability. Cell Rep. 2014, 7, 575–587. [Google Scholar] [CrossRef]
- Chibi, M.; Meyer, M.; Skepu, A.; G Rees, D.J.; Moolman-Smook, J.C.; Pugh, D.J.R. RBBP6 Interacts with Multifunctional Protein YB-1 through Its RING Finger Domain, Leading to Ubiquitination and Proteosomal Degradation of YB-1. J. Mol. Biol. 2008, 384, 908–916. [Google Scholar] [CrossRef]
- Chevalier, F.; Depagne, J.; Hem, S.; Chevillard, S.; Bensimon, J.; Bertrand, P.; Lebeau, J. Accumulation of Cyclophilin A Isoforms in Conditioned Medium of Irradiated Breast Cancer Cells. Proteomics 2012, 12, 1756–1766. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.J.; Botero, A.; Hirota, K.; Bradbury, C.M.; Markovina, S.; Laszlo, A.; Spitz, D.R.; Goswami, P.C.; Yodoi, J.; Gius, D. Thioredoxin Nuclear Translocation and Interaction with Redox Factor-1 Activates the Activator Protein-1 Transcription Factor in Response to Ionizing Radiation. Cancer Res. 2000, 60, 6688–6695. [Google Scholar]
- Ghosh, A.K.; Steele, R.; Ray, R.B. Functional Domains of C-Myc Promoter Binding Protein 1 Involved in Transcriptional Repression and Cell Growth Regulation. Mol. Cell. Biol. 1999, 19, 2880–2886. [Google Scholar] [CrossRef] [Green Version]
- Chevalier, F.; Hamdi, D.H.; Lepleux, C.; Temelie, M.; Nicol, A.; Austry, J.B.; Lesueur, P.; Vares, G.; Savu, D.; Nakajima, T.; et al. High LET Radiation Overcomes In Vitro Resistance to X-Rays of Chondrosarcoma Cell Lines. Technol. Cancer Res. Treat. 2019, 18, 1533033819871309. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, E.M.; Allain, F.; Gaillard, J.-C.; Pible, O.; Armengaud, J. Taking the Shortcut for High-Throughput Shotgun Proteomic Analysis of Bacteria. Methods Mol. Biol. 2014, 1197, 275–285. [Google Scholar] [CrossRef]
- Klein, G.; Mathé, C.; Biola-Clier, M.; Devineau, S.; Drouineau, E.; Hatem, E.; Marichal, L.; Alonso, B.; Gaillard, J.-C.; Lagniel, G.; et al. RNA-Binding Proteins Are a Major Target of Silica Nanoparticles in Cell Extracts. Nanotoxicology 2016, 10, 1555–1564. [Google Scholar] [CrossRef] [Green Version]
- Dupierris, V.; Masselon, C.; Court, M.; Kieffer-Jaquinod, S.; Bruley, C. A Toolbox for Validation of Mass Spectrometry Peptides Identification and Generation of Database: IRMa. Bioinformatics 2009, 25, 1980–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakeri Manesh, S.; Sangsuwan, T.; Pour Khavari, A.; Fotouhi, A.; Emami, S.N.; Haghdoost, S. MTH1, an 8-Oxo-2′-Deoxyguanosine Triphosphatase, and MYH, a DNA Glycosylase, Cooperate to Inhibit Mutations Induced by Chronic Exposure to Oxidative Stress of Ionising Radiation. Mutagenesis 2017, 32, 389–396. [Google Scholar] [CrossRef] [Green Version]
- Dépagne, J.; Chevalier, F. Technical Updates to Basic Proteins Focalization Using IPG Strips. Proteome Sci 2012, 10, 54. [Google Scholar] [CrossRef] [Green Version]
- Chevalier, F.; Hirtz, C.; Sommerer, N.; Kelly, A.L. Use of Reducing/Nonreducing Two-Dimensional Electrophoresis for the Study of Disulfide-Mediated Interactions between Proteins in Raw and Heated Bovine Milk. J. Agric. Food Chem. 2009, 57, 5948–5955. [Google Scholar] [CrossRef]
- Chevalier, F.; Centeno, D.; Rofidal, V.; Tauzin, M.; Martin, O.; Sommerer, N.; Rossignol, M. Different Impact of Staining Procedures Using Visible Stains and Fluorescent Dyes for Large-Scale Investigation of Proteomes by MALDI-TOF Mass Spectrometry. J. Proteome Res. 2006, 5, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Saintigny, Y.; Chevalier, F.; Bravard, A.; Dardillac, E.; Laurent, D.; Hem, S.; Dépagne, J.; Radicella, J.P.; Lopez, B.S. A Threshold of Endogenous Stress Is Required to Engage Cellular Response to Protect against Mutagenesis. Sci. Rep. 2016, 6, 29412. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Sadygov, R.G.; Yates, J.R. A Model for Random Sampling and Estimation of Relative Protein Abundance in Shotgun Proteomics. Anal. Chem. 2004, 76, 4193–4201. [Google Scholar] [CrossRef]
- Carvalho, P.C.; Fischer, J.S.G.; Chen, E.I.; Yates, J.R.; Barbosa, V.C. PatternLab for Proteomics: A Tool for Differential Shotgun Proteomics. BMC Bioinform. 2008, 9, 316. [Google Scholar] [CrossRef] [Green Version]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
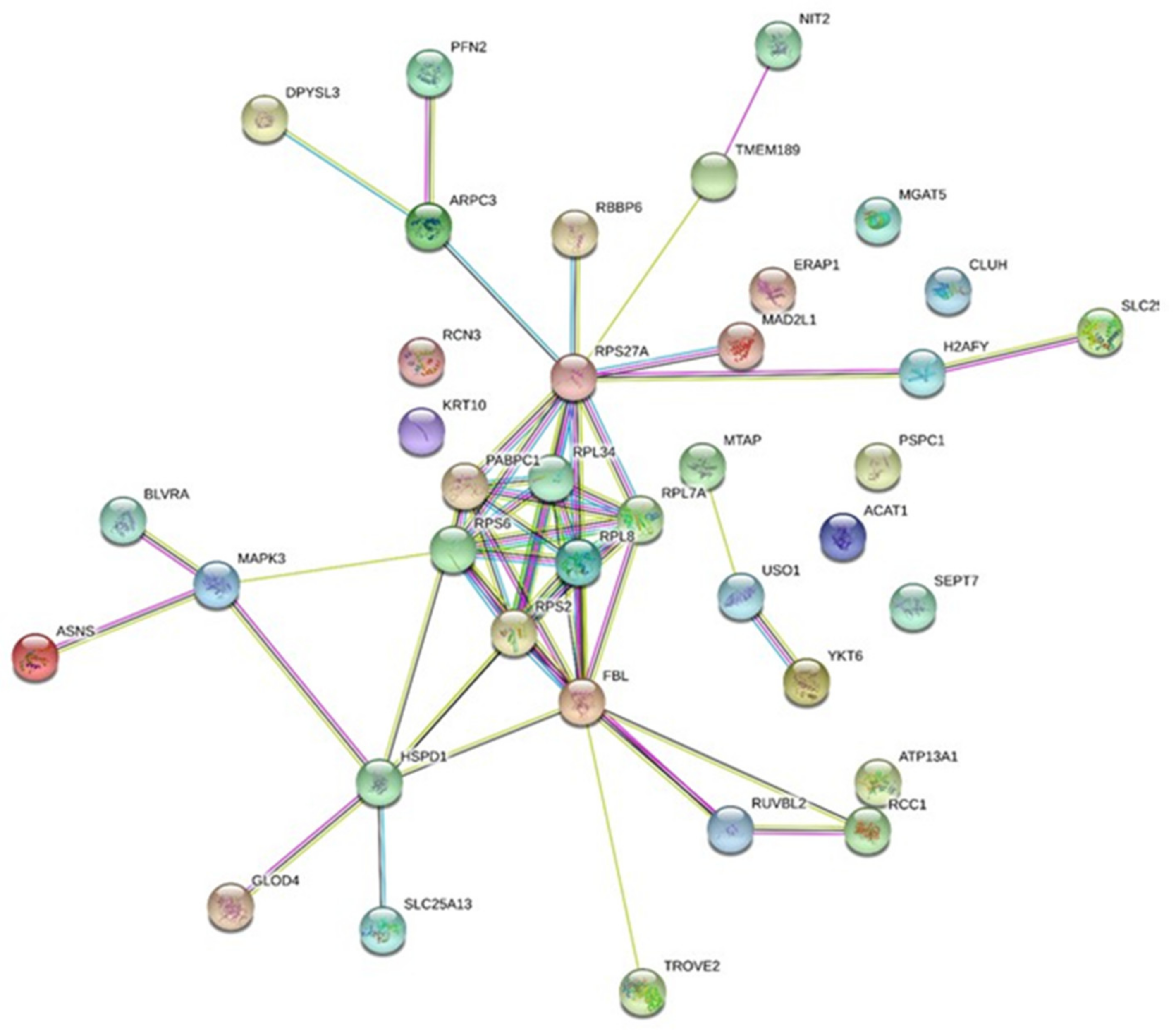

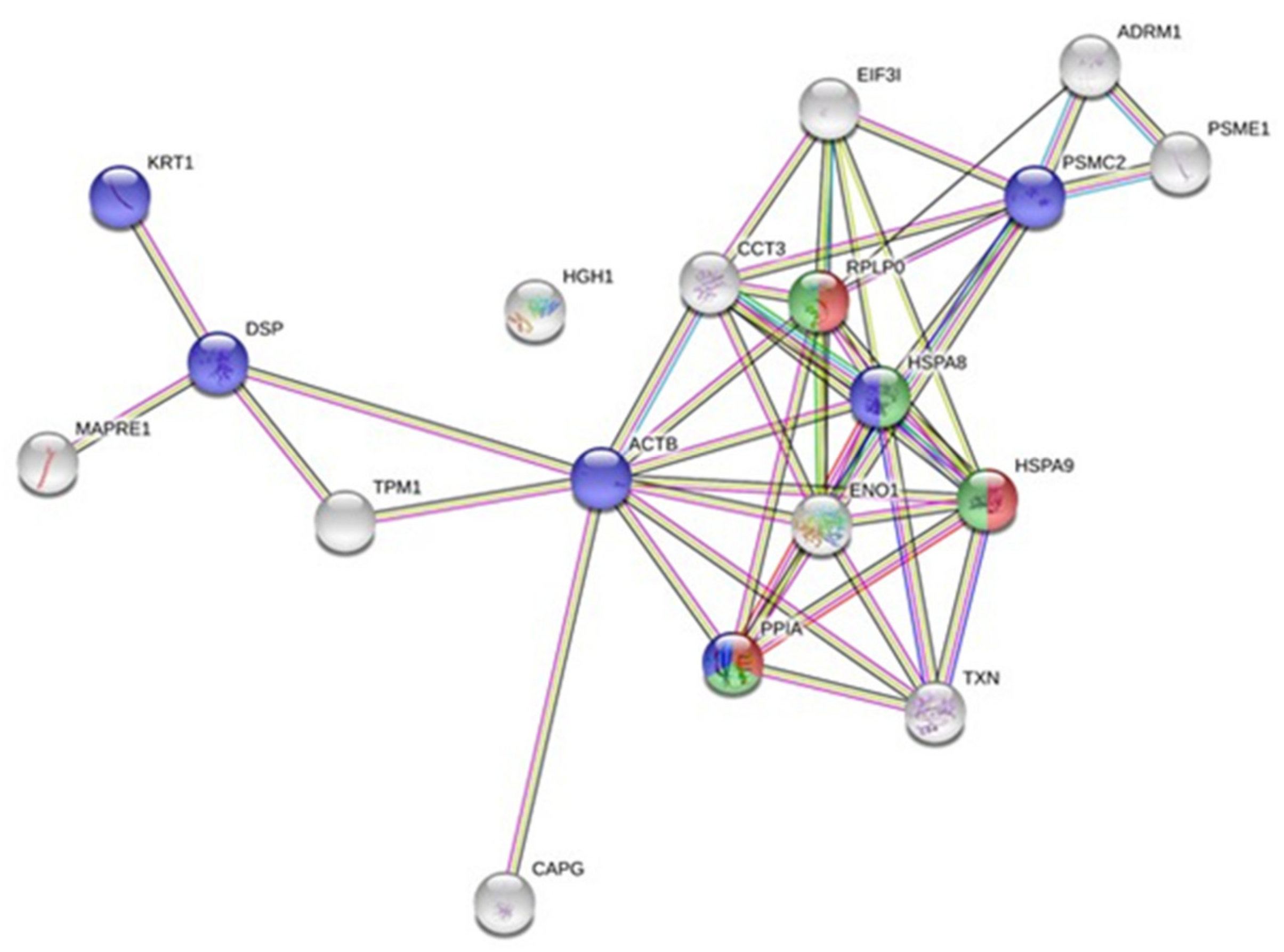
| Accession | Name | pI | Mass (Da) | Tfold * | p-Value |
|---|---|---|---|---|---|
| P11940 | Polyadenylate-binding protein 1 | 9.85 | 106.299 | 23.83 | 0.0264 |
| P24752 | Acetyl-CoAacetyltransferase, mitochondrial | 8.39 | 58.871 | 4.00 | 0.0051 |
| A5PLL7 | Transmembraneprotein 189 | 9.07 | 107.703 | 3.33 | 0.0278 |
| P08243 | Asparagine synthetase [glutamine-hydrolyzing] | 6.86 | 67.256 | 3.17 | 0.0155 |
| P35080 | Profilin-2 | 8.87 | 77.777 | 2.86 | 0.0423 |
| P49207 | 60S ribosomal protein L34 | 10.64 | 21.824 | 2.50 | 0.0011 |
| Q9HC38 | Glyoxalase domain-containing protein 4 | 7.94 | 68.961 | 2.11 | 0.0191 |
| P62424 | 60S ribosomal protein L7a | 10.54 | 33.547 | 2.00 | 0.0150 |
| P62753 | 40S ribosomal protein S6 | 10.74 | 31.799 | 2.00 | 0.0064 |
| Q16181 | Septin-7 | 8.97 | 244.012 | 2.00 | 0.0076 |
| P27361 | Mitogen-activated protein kinase 3 | 9.14 | 69.163 | 2.00 | 0.0250 |
| O60763 | General vesicular transport factor p115 | 5.88 | 159.184 | 1.91 | 0.0191 |
| P62979 | Ubiquitin-40S ribosomal protein S27a | 9.64 | 19.523 | 1.84 | 0.0356 |
| Q53SE2 | Uncharacterized protein HSPD1 | 8.32 | 70.924 | 1.83 | 0.0186 |
| P62917 | 60S ribosomal protein L8 | 11.15 | 32.789 | 1.79 | 0.0017 |
| Q9NQR4 | Omega-amidase NIT2 | 6.73 | 47.093 | 1.75 | 0.0335 |
| P15880 | 40S ribosomal protein S2 | 10.37 | 34.399 | 1.67 | 0.0156 |
| O75367 | Core histone macro-H2A.1 | 9.75 | 68.531 | 1.67 | 0.0379 |
| Q13126 | S-methyl-5’-thioadenosine phosphorylase | 9.17 | 186.699 | 1.64 | 0.0031 |
| Q96D15 | Reticulocalbin-3 | 5.05 | 46.220 | 1.64 | 0.0047 |
| Q7Z6E9 | E3 ubiquitin-protein ligase RBBP6 | 9.65 | 98.855 | 1.58 | 0.0385 |
| O00370 | LINE-1 retrotransposable element ORF2 protein | 9.51 | 5.633.488 | 1.57 | 0.0179 |
| Q9NZ08 | Endoplasmic reticulum aminopeptidase 1 | 9.00 | 202.092 | 1.50 | 0.0219 |
| P22087 | rRNA 2’-O-methyltransferase fibrillarin | 10.19 | 41.124 | 1.50 | 0.0219 |
| O15145 | Actin-related protein 2/3 complex subunit 3 | 9.60 | 28.754 | −1.67 | 0.0379 |
| Q13257 | Mitotic spindle assembly checkpoint protein MAD2A | 6.30 | 55.269 | −1.67 | 0.0250 |
| O15498 | Synaptobrevin homolog YKT6 | 8.79 | 96.462 | −1.90 | 0.0171 |
| Q9UJS0 | Calcium-binding mitochondrial carrier protein Aralar2 | 9.33 | 119.145 | −2.00 | 0.0011 |
| Q00325 | Phosphate carrier protein, mitochondrial | 9.34 | 63.151 | −2.00 | 0.0409 |
| Q8WXF1 | Paraspeckle component 1 | 8.96 | 74.454 | −2.08 | 0.0003 |
| P10155 | 60 kDa SS-A/Ro ribonucleoprotein | 9.63 | 350.653 | −2.10 | 0.0160 |
| Q5JXB2 | Putative ubiquitin-conjugating enzyme E2 N-like | 9.15 | 95.474 | −2.11 | 0.0030 |
| P53004 | Biliverdin reductase A | 6.47 | 41.158 | −2.18 | 0.0405 |
| Q9Y230 | RuvB-like 2 | 5.40 | 54.097 | −2.24 | 0.0169 |
| O75153 | Clustered mitochondria protein homolog | 6.34 | 190.902 | −2.29 | 0.0427 |
| Q9HD20 | Manganese-transporting ATPase 13A1 | 9.48 | 70.012 | −2.50 | 0.0029 |
| P18754 | Regulator of chromosome condensation | 5.79 | 42.947 | −2.67 | 0.0108 |
| Q09328 | Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyl transferase A | 9.09 | 255.616 | −2.75 | 0.0003 |
| Q14195 | Dihydropyrimidinase-related protein 3 | 8.69 | 171.819 | −2.85 | 0.0245 |
| P13645 | Keratin, type I cytoskeletal 10 | 6.00 | 75.115 | −2.87 | 0.0138 |
| Spot Number | Fold | pI * | MW * | Accession | Names | HighestMean | GO—Biological Process |
|---|---|---|---|---|---|---|---|
| 16 | 1.31 | 4.79 | 20 | P60709 | Actin | Bystander 0.1 Gy | Cell junction assembly |
| 39 | 1.62 | 6.69 | 26 | P62937 | Cyclophilin A | Bystander 0.1 Gy | positive regulation of protein secretion; interleukin-12-mediated signaling pathway |
| 85 | 1.38 | 5.11 | 63 | Q15691 | Microtubule-associated protein RP | Bystander 0.1 Gy | cell migration |
| 93 | 1.31 | 5.99 | 57 | Q06323 | PSME1 | Bystander 0.1 Gy | interleukin-1-mediated signaling pathway; tumor necrosis factor-mediated signaling pathway |
| 116 | 1.27 | 4.87 | 23 | P15924 | Desmoplakin | Bystander 0.1 Gy | adherent junction organization; cell-cell adhesion |
| 116 | P10599 | Thioredoxin | cell redox homeostasis; | ||||
| 126 | 1.63 | 4.67 | 56 | P09493 | Tropomyosin alpha-1 chain | Bystander 0.1 Gy | negative regulation of cell migration |
| 138 | 1.98 | 4.18 | 145 | P04264 | Keratin, type II | Bystander 0.1 Gy | Extracellular exosome |
| 145 | 1.47 | 4.67 | 98 | Q9BTY7 | Protein HGH1 homolog | Bystander 0.1 Gy | unknown (interact with Peptidyl-prolyl cis-trans isomerase and Heat shock protein 90) |
| 65 | 1.81 | 5.86 | 72 | P06733 | Alpha-enolase | Bystander 0 Gy | negative regulation of cell growth |
| 65 | P05388 | 60S acidic ribosomal protein P0 | interleukin-12-mediated signaling pathway | ||||
| 77 | 1.56 | 5.79 | 130 | P01876 | IGHA1 protein | Bystander 0 Gy | Extracellular exosome |
| 91 | 1.29 | 5.63 | 140 | P11142 | HSC 70 protein | Bystander 0 Gy | cytokine-mediated signaling pathway |
| 97 | 1.32 | 5.64 | 141 | P38646 | Hspa9 | Bystander 0 Gy | interleukin-12-mediated signaling pathway |
| 113 | 1.27 | 6.25 | 79 | P40121 | CAP-G protein | Bystander 0 Gy | Protein motility |
| 121 | 1.47 | 6.15 | 94 | P35998 | 26S proteasome regulatory subunit 7 | Bystander 0 Gy | interleukin-1-mediated signaling pathway |
| 123 | 1.31 | 6.56 | 136 | P49368 | T-complex protein 1 subunit gamma | Bystander 0 Gy | Extracellular exosome |
| 127 | 1.31 | 4.91 | 87 | P60709 | Actin | Bystander 0 Gy | Cell junction assembly |
| 128 | 1.36 | 5.03 | 87 | Q16186 | Proteasomal ubiquitin receptor ADRM1 | Bystander 0 Gy | Proteasome complex |
| 134 | 1.26 | 5.68 | 73 | Q13347 | Eukaryotic translation initiation factor 3 subunit I | Bystander 0 Gy | Extracellular exosome |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tudor, M.; Gilbert, A.; Lepleux, C.; Temelie, M.; Hem, S.; Armengaud, J.; Brotin, E.; Haghdoost, S.; Savu, D.; Chevalier, F. A Proteomic Study Suggests Stress Granules as New Potential Actors in Radiation-Induced Bystander Effects. Int. J. Mol. Sci. 2021, 22, 7957. https://doi.org/10.3390/ijms22157957
Tudor M, Gilbert A, Lepleux C, Temelie M, Hem S, Armengaud J, Brotin E, Haghdoost S, Savu D, Chevalier F. A Proteomic Study Suggests Stress Granules as New Potential Actors in Radiation-Induced Bystander Effects. International Journal of Molecular Sciences. 2021; 22(15):7957. https://doi.org/10.3390/ijms22157957
Chicago/Turabian StyleTudor, Mihaela, Antoine Gilbert, Charlotte Lepleux, Mihaela Temelie, Sonia Hem, Jean Armengaud, Emilie Brotin, Siamak Haghdoost, Diana Savu, and François Chevalier. 2021. "A Proteomic Study Suggests Stress Granules as New Potential Actors in Radiation-Induced Bystander Effects" International Journal of Molecular Sciences 22, no. 15: 7957. https://doi.org/10.3390/ijms22157957







