SLIDE—Novel Approach to Apocrine Sweat Sampling for Lipid Profiling in Healthy Individuals
Abstract
:1. Introduction
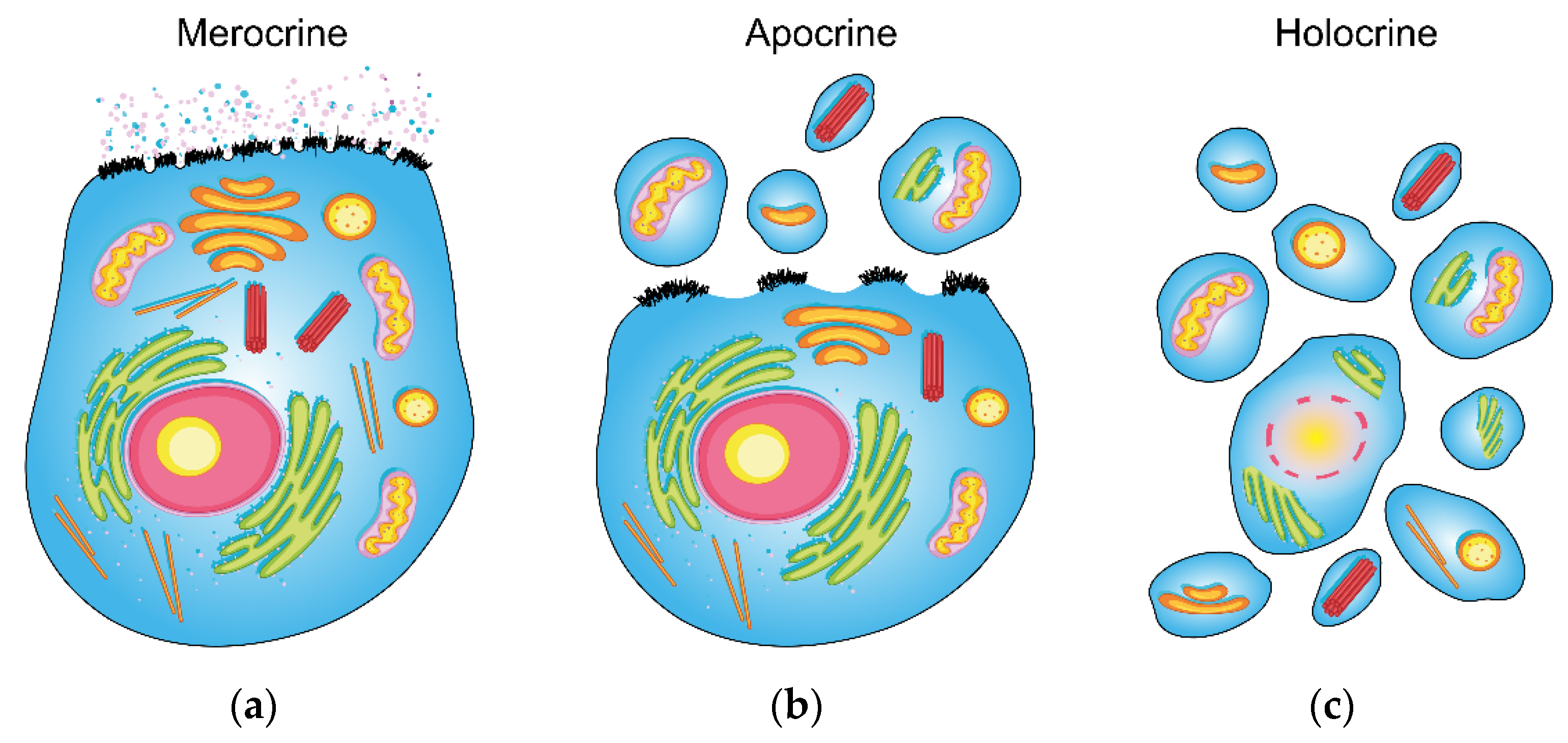
- It is a secretion that reflects the contents of the cellular cytoplasm (including cellular fragments).
- It reflects the components contained in the cell membrane of apocrine gland secretory cells.
- The components correspond to the cytoplasm composition and the cell membrane of a living cell (as opposed to a holocrine secretion, which is composed of the remnants of dead cells).
- The lipid nature of apocrine secretion may be used in the future alongside diagnostic targets to identify/quantify lipid xenobiotics, lipophilic pharmaceuticals, and lipophilic narcotic drugs.
2. Results
2.1. Apocrine Sweat Microsampling Technique
2.2. Pseudotargeted Lipidomic Analysis
2.3. Importance of Sweat Lipidome Data Transformation
2.4. Intraindividual and Group Variability of Lipids
2.5. Quantitation of Lipid Classes
2.6. Correlation of Lipids in Apocrine Sweat
2.7. Armpit Side-Specific Differences
3. Discussion
3.1. Novel Approach for Sweat Sampling
3.2. Lipidomic Methodology
3.3. Data Processing Solution to the Problem of the Variability
3.4. Description of the Sweat Lipidome
3.5. Future Development
4. Materials and Methods
4.1. Chemicals and Reagents
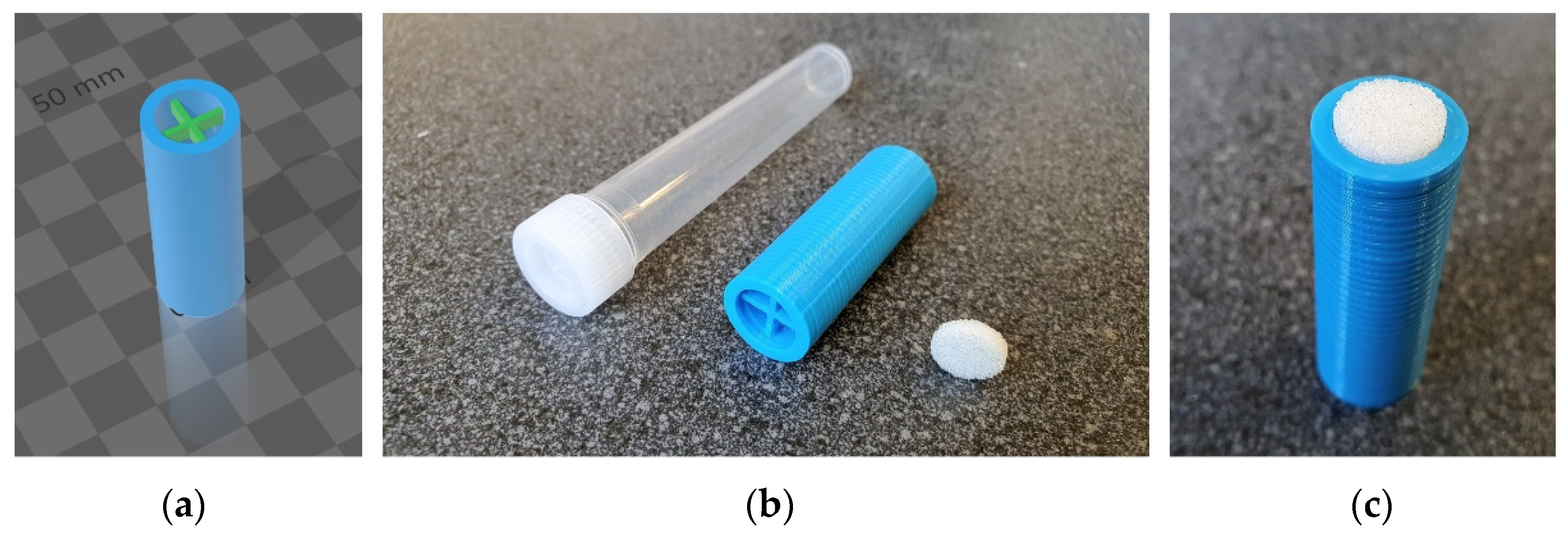
4.2. D-Printed Attachment
4.3. Sampling Technique
4.4. Sample Preparation
4.5. Pseudotargeted Lipidomic Analysis
4.6. Data Treatment and Statistical Analysis
4.7. Quantitative Evaluation of Lipid Profiles
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgment
Conflicts of Interest
References
- Wilke, K.; Martin, A.; Terstegen, L.; Biel, S.S. A short history of sweat gland biology. Int. J. Cosmet. Sci. 2007, 29, 169–179. [Google Scholar] [CrossRef]
- Montagna, W. The Structure and Function of Skin; Elsevier: Amsterdam, The Netherlands, 2012; ISBN 9780323138697. [Google Scholar]
- Sato, K. The physiology, pharmacology, and biochemistry of the eccrine sweat gland. Rev. Physiol. Biochem. Pharmacol. 1977, 79, 51–131. [Google Scholar]
- Sato, K.; Sato, F. Sweat secretion by human axillary apoeccrine sweat gland in vitro. Am. J. Physiol. 1987, 252, R181–R187. [Google Scholar] [CrossRef]
- Bovell, D.L.; Corbett, A.D.; Holmes, S.; Macdonald, A.; Harker, M. The absence of apoeccrine glands in the human axilla has disease pathogenetic implications, including axillary hyperhidrosis. Br. J. Dermatol. 2007, 156, 1278–1286. [Google Scholar] [CrossRef]
- Farkaš, R. Apocrine secretion: New insights into an old phenomenon. Biochim. Biophys. Acta 2015, 1850, 1740–1750. [Google Scholar] [CrossRef]
- Aumüller, G.; Wilhelm, B.; Seitz, J. Apocrine secretion—fact or artifact? Ann. Anat. Anat. Anz. 1999, 181, 437–446. [Google Scholar] [CrossRef]
- Stefaniak, A.B.; Harvey, C.J. Dissolution of materials in artificial skin surface film liquids. Toxicol. In Vitro 2006, 20, 1265–1283. [Google Scholar] [CrossRef]
- Baker, L.B. Physiology of sweat gland function: The roles of sweating and sweat composition in human health. Temperature (Austin) 2019, 6, 211–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Yu, W.; Wang, B.; Zhao, Y.; En, K.; Zhu, J.; Cheng, X.; Zhou, C.; Lin, H.; Wang, Z.; et al. Noninvasive wearable electroactive pharmaceutical monitoring for personalized therapeutics. Proc. Natl. Acad. Sci. USA 2020, 117, 19017–19025. [Google Scholar] [CrossRef] [PubMed]
- Hudson, M.; Stuchinskaya, T.; Ramma, S.; Patel, J.; Sievers, C.; Goetz, S.; Hines, S.; Menzies, E.; Russell, D.A. Drug screening using the sweat of a fingerprint: Lateral flow detection of Δ9-tetrahydrocannabinol, cocaine, opiates and amphetamine. J. Anal. Toxicol. 2019, 43, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Sears, M.E.; Kerr, K.J.; Bray, R.I. Arsenic, cadmium, lead, and mercury in sweat: A systematic review. J. Environ. Public Health 2012, 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genuis, S.J.; Beesoon, S.; Lobo, R.A.; Birkholz, D. Human elimination of phthalate compounds: Blood, urine, and sweat (BUS) study. Sci. World J. 2012, 2012. [Google Scholar] [CrossRef] [Green Version]
- Austin, C.; Ellis, J. Microbial pathways leading to steroidal malodour in the axilla. J. Steroid Biochem. Mol. Biol. 2003, 87, 105–110. [Google Scholar] [CrossRef]
- Mebazaa, R.; Rega, B.; Camel, V. Analysis of human male armpit sweat after fenugreek ingestion: Characterisation of odour active compounds by gas chromatography coupled to mass spectrometry and olfactometry. Food Chem. 2011, 128, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Farkaš, R.; Ďatková, Z.; Mentelová, L.; Löw, P.; Beňová-Liszeková, D.; Beňo, M.; Sass, M.; Řehulka, P.; Řehulková, H.; Raška, O.; et al. Apocrine secretion in drosophila salivary glands: Subcellular origin, dynamics, and identification of secretory proteins. PLoS ONE 2014, 9, e94383. [Google Scholar] [CrossRef] [Green Version]
- Nanjappa, V.; Thomas, J.K.; Marimuthu, A.; Muthusamy, B.; Radhakrishnan, A.; Sharma, R.; Ahmad Khan, A.; Balakrishnan, L.; Sahasrabuddhe, N.A.; Kumar, S.; et al. Plasma proteome database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 2014, 42, D959–D965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelley, W.B.; Hurley, H.J., Jr. The physiology of the human axillary apocrine sweat gland. J. Investig. Dermatol. 1953, 20, 285–297. [Google Scholar] [CrossRef] [Green Version]
- McGrath, K.G. Apocrine sweat gland obstruction by antiperspirants allowing transdermal absorption of cutaneous generated hormones and pheromones as a link to the observed incidence rates of breast and prostate cancer in the 20th century. Med. Hypotheses 2009, 72, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Jadoon, S.; Karim, S.; Akram, M.R.; Kalsoom Khan, A.; Zia, M.A.; Siddiqi, A.R.; Murtaza, G. Recent developments in sweat analysis and its applications. Int. J. Anal. Chem. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Hussain, J.N.; Mantri, N.; Cohen, M.M. Working up a good sweat-the challenges of standardising sweat collection for metabolomics analysis. Clin. Biochem. Rev. 2017, 38, 13–34. [Google Scholar]
- World Health Organization. WHO Best Practices for Injections and Related Procedures Toolkit; World Health Organization: Geneva, Switzerland, 2013; ISBN 9789241599252. [Google Scholar]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- Li, B.; Tang, J.; Yang, Q.; Cui, X.; Li, S.; Chen, S.; Cao, Q.; Xue, W.; Chen, N.; Zhu, F. Performance evaluation and online realization of data-driven normalization methods used in LC/MS based untargeted metabolomics analysis. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Liebisch, G.; Vizcaíno, J.A.; Köfeler, H.; Trötzmüller, M.; Griffiths, W.J.; Schmitz, G.; Spener, F.; Wakelam, M.J.O. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avela, H.F.; Sirén, H. Advances in lipidomics. Clin. Chim. Acta 2020, 510, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, C.; Han, X. Selection of Internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-what, how and why? Mass Spectrom. Rev. 2017, 36, 693–714. [Google Scholar] [CrossRef] [Green Version]
- Lange, M.; Fedorova, M. Evaluation of Lipid quantification accuracy using HILIC and RPLC MS on the example of NIST® SRM® 1950 metabolites in human plasma. Anal. Bioanal. Chem. 2020, 412, 3573–3584. [Google Scholar] [CrossRef] [Green Version]
- Kirk, J.M.; Keston, M.; McIntosh, I.; Al Essa, S. Variation of sweat sodium and chloride with age in cystic fibrosis and normal populations: Further investigations in equivocal cases. Ann. Clin. Biochem. 1992, 29 Pt 2, 145–152. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Shen, G.; Zhang, R.; He, J.; Zhang, Y.; Xu, J.; Yang, W.; Chen, X.; Song, Y.; Abliz, Z. Combination of injection volume calibration by creatinine and ms signals’ normalization to overcome urine variability in LC-MS-based metabolomics studies. Anal. Chem. 2013, 85, 7659–7665. [Google Scholar] [CrossRef]
- Wong, M.W.K.; Braidy, N.; Pickford, R.; Vafaee, F.; Crawford, J.; Muenchhoff, J.; Schofield, P.; Attia, J.; Brodaty, H.; Sachdev, P.; et al. Plasma lipidome variation during the second half of the human lifespan is associated with age and sex but minimally with BMI. PLoS ONE 2019, 14, e0214141. [Google Scholar] [CrossRef] [Green Version]
- Takemura, T.; Wertz, P.W.; Sato, K. Free fatty acids and sterols in human eccrine sweat. Br. J. Dermatol. 1989, 120, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Peter, G.; Schröpl, F.; Feisel, H.G.; Thürauf, W. Gaschromatographische untersuchungen von freien und gebundenen fettsäuren im ekkrinen Schweiß. Arch. Dermatol. Res. 1970, 238, 154–159. [Google Scholar] [CrossRef]
- Coderch, L.; López, O.; de la Maza, A.; Parra, J.L. Ceramides and skin function. Am. J. Clin. Dermatol. 2003, 4, 107–129. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, K.; Hassoun, L.A.; Foolad, N.; Pedersen, T.L.; Sivamani, R.K.; Newman, J.W. Sweat lipid mediator profiling: A noninvasive approach for cutaneous research. J. Lipid Res. 2017, 58, 188–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovászi, M.; Szegedi, A.; Zouboulis, C.C.; Törőcsik, D. Sebaceous-immunobiology is orchestrated by sebum lipids. Dermatoendocrinology 2017, 9, e1375636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begum, H.; Torta, F.; Narayanaswamy, P.; Mundra, P.A.; Ji, S.; Bendt, A.K.; Saw, W.-Y.; Teo, Y.Y.; Soong, R.; Little, P.F.; et al. Lipidomic profiling of plasma in a healthy singaporean population to identify ethnic specific differences in lipid levels and associations with disease risk factors. Clin. Mass Spectrom. 2017, 6, 25–31. [Google Scholar] [CrossRef]
- Weir, J.M.; Wong, G.; Barlow, C.K.; Greeve, M.A.; Kowalczyk, A.; Almasy, L.; Comuzzie, A.G.; Mahaney, M.C.; Jowett, J.B.M.; Shaw, J.; et al. Plasma lipid profiling in a large population-based cohort. J. Lipid Res. 2013, 54, 2898–2908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skandalakis, J.E.; Skandalakis, L.J.; Skandalakis, P.N. Anatomy of the lymphatics. surg. Oncol. Clin. N. Am. 2007, 16, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zane, P.; Emmons, G.T. (Eds.) Microsampling in Pharmaceutical Bioanalysis; Future Science: London, UK, 2013; ISBN 9781909453586. [Google Scholar]
- Moyer, J.; Wilson, D.; Finkelshtein, I.; Wong, B.; Potts, R. Correlation between sweat glucose and blood glucose in subjects with diabetes. Diabetes Technol. Ther. 2012, 14, 398–402. [Google Scholar] [CrossRef]
- Xuan, Q.; Hu, C.; Yu, D.; Wang, L.; Zhou, Y.; Zhao, X.; Li, Q.; Hou, X.; Xu, G. Development of a high coverage pseudotargeted lipidomics method based on ultra-high performance liquid chromatography-mass spectrometry. Anal. Chem. 2018, 90, 7608–7616. [Google Scholar] [CrossRef]
- Drotleff, B.; Roth, S.R.; Henkel, K.; Calderón, C.; Schlotterbeck, J.; Neukamm, M.A.; Lämmerhofer, M. Lipidomic profiling of non-mineralized dental plaque and biofilm by untargeted UHPLC-QTOF-MS/MS and swath acquisition. Anal. Bioanal. Chem. 2020, 412, 2303–2314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AlzbetaG. AlzbetaG/Metabol: First Version (Version v1.0.0). Zenodo. 30 May 2019. Available online: https://zenodo.org/record/3235775#.YP4UukARWUk (accessed on 1 July 2021).
- Wei, R.; Wang, J.; Su, M.; Jia, E.; Chen, S.; Chen, T.; Ni, Y. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci. Rep. 2018, 8, 663. [Google Scholar] [CrossRef] [PubMed] [Green Version]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kvasnička, A.; Friedecký, D.; Tichá, A.; Hyšpler, R.; Janečková, H.; Brumarová, R.; Najdekr, L.; Zadák, Z. SLIDE—Novel Approach to Apocrine Sweat Sampling for Lipid Profiling in Healthy Individuals. Int. J. Mol. Sci. 2021, 22, 8054. https://doi.org/10.3390/ijms22158054
Kvasnička A, Friedecký D, Tichá A, Hyšpler R, Janečková H, Brumarová R, Najdekr L, Zadák Z. SLIDE—Novel Approach to Apocrine Sweat Sampling for Lipid Profiling in Healthy Individuals. International Journal of Molecular Sciences. 2021; 22(15):8054. https://doi.org/10.3390/ijms22158054
Chicago/Turabian StyleKvasnička, Aleš, David Friedecký, Alena Tichá, Radomír Hyšpler, Hana Janečková, Radana Brumarová, Lukáš Najdekr, and Zdeněk Zadák. 2021. "SLIDE—Novel Approach to Apocrine Sweat Sampling for Lipid Profiling in Healthy Individuals" International Journal of Molecular Sciences 22, no. 15: 8054. https://doi.org/10.3390/ijms22158054
APA StyleKvasnička, A., Friedecký, D., Tichá, A., Hyšpler, R., Janečková, H., Brumarová, R., Najdekr, L., & Zadák, Z. (2021). SLIDE—Novel Approach to Apocrine Sweat Sampling for Lipid Profiling in Healthy Individuals. International Journal of Molecular Sciences, 22(15), 8054. https://doi.org/10.3390/ijms22158054







