The Active Mechanism of Nucleosome Depletion by Poly(dA:dT) Tracts In Vivo
Abstract
:1. Nucleosomes Are Depleted In Vivo over Poly(dA:dT) Tracts in Many Species
2. Nucleosome Depletion Is Functionally Important and Caused by Poly(dA:dT) Tracts
3. Special Intrinsic Properties of Poly(dA:dT) Tracts Were Suggested to Cause Nucleosome Depletion In Vivo by a Nucleosome-Intrinsic Mechanism
4. In Vivo Nucleosome Depletion over Poly(dA:dT) Tracts Is Not Universal
5. The Energetic Penalty for Incorporation of Poly(dA:dT) into Nucleosomes Is Not Very High
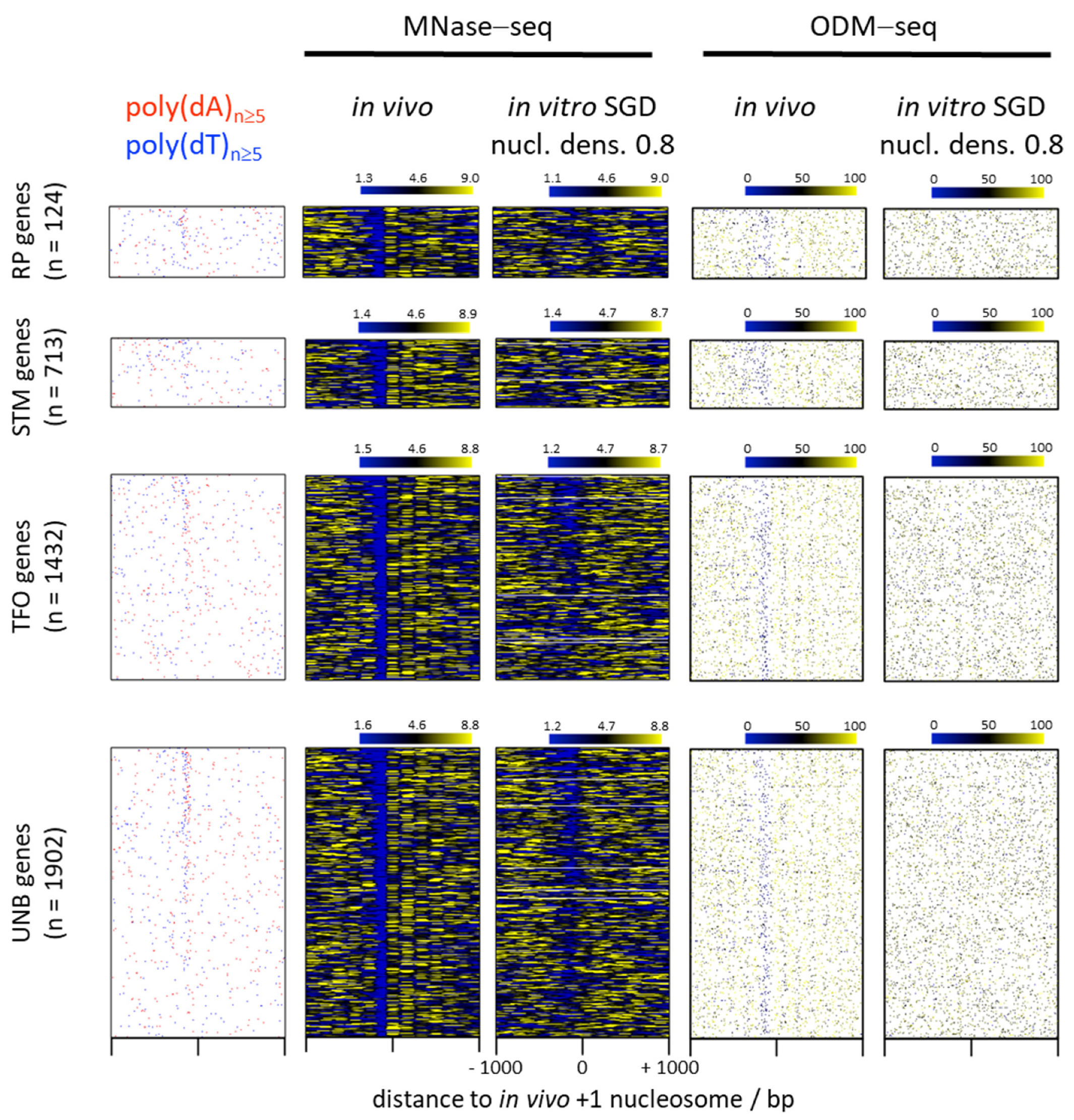
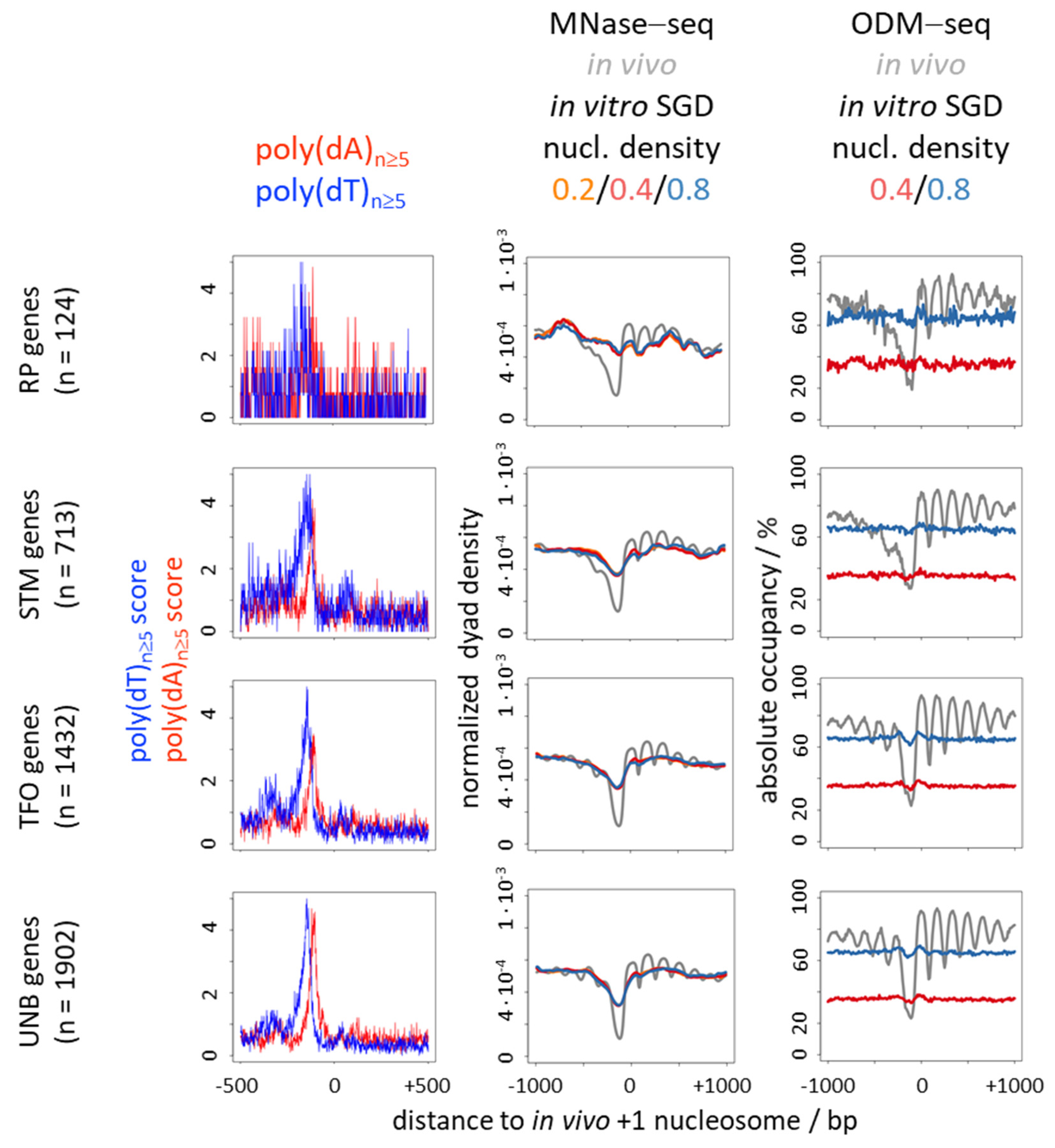
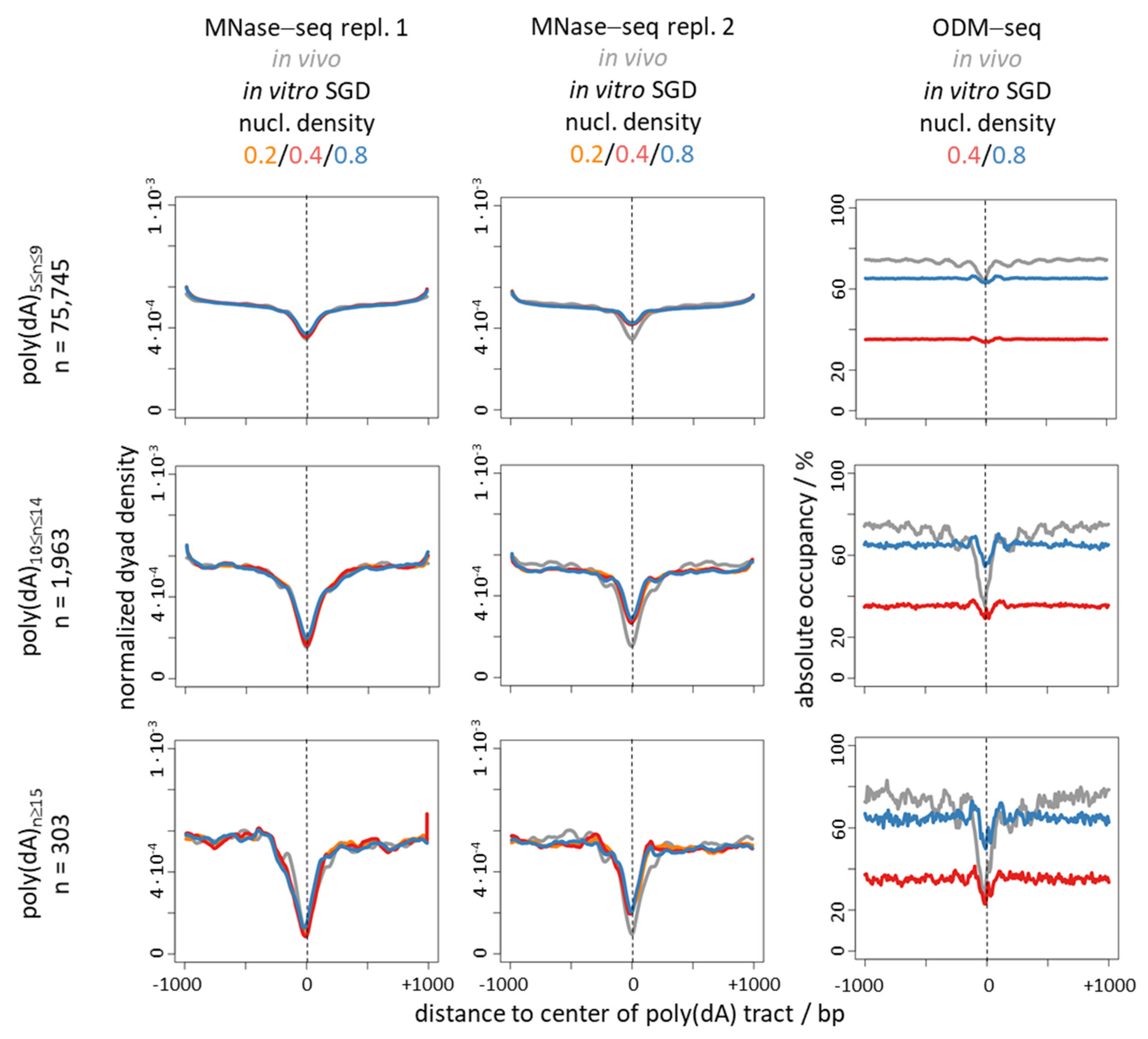
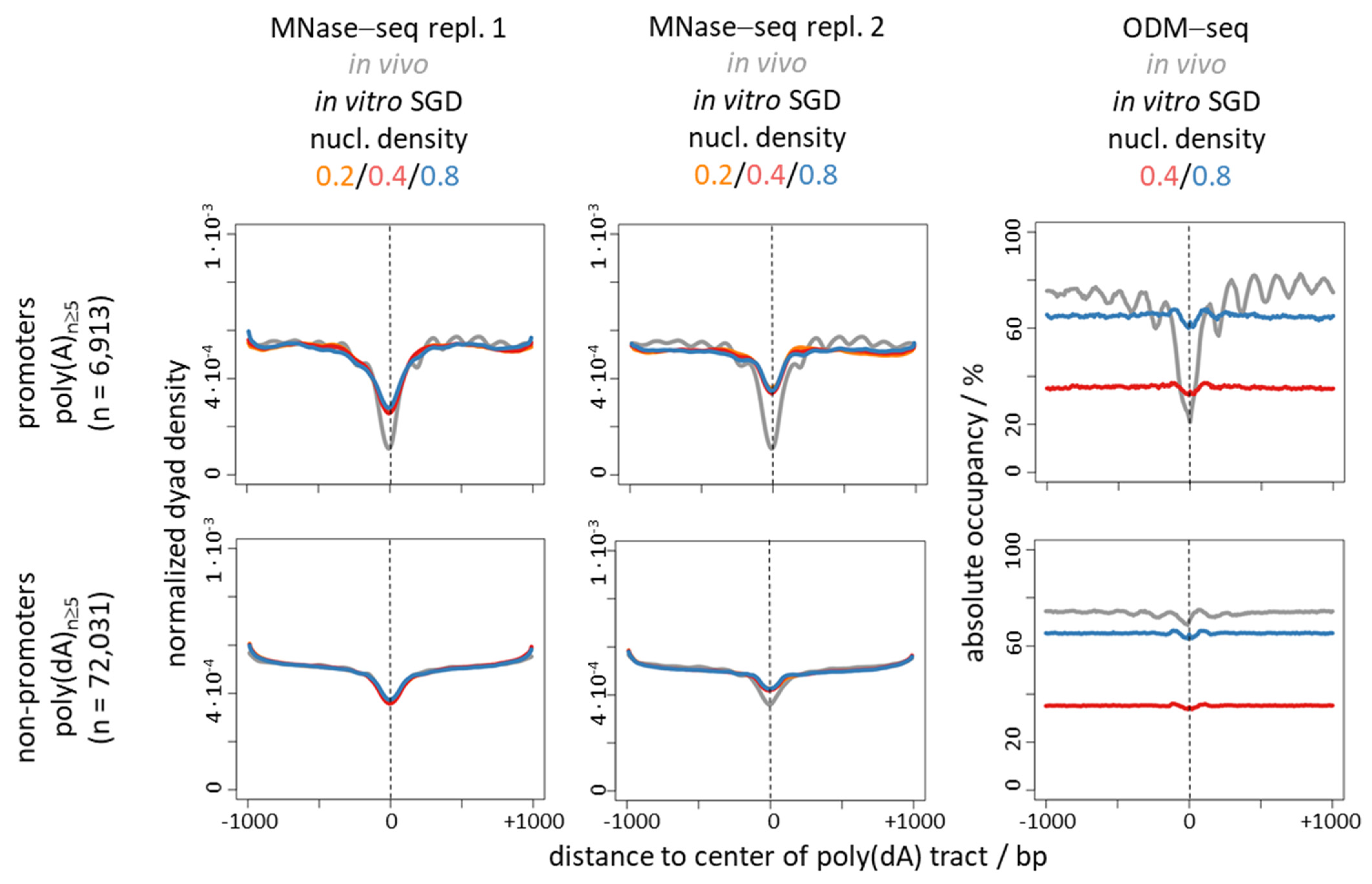
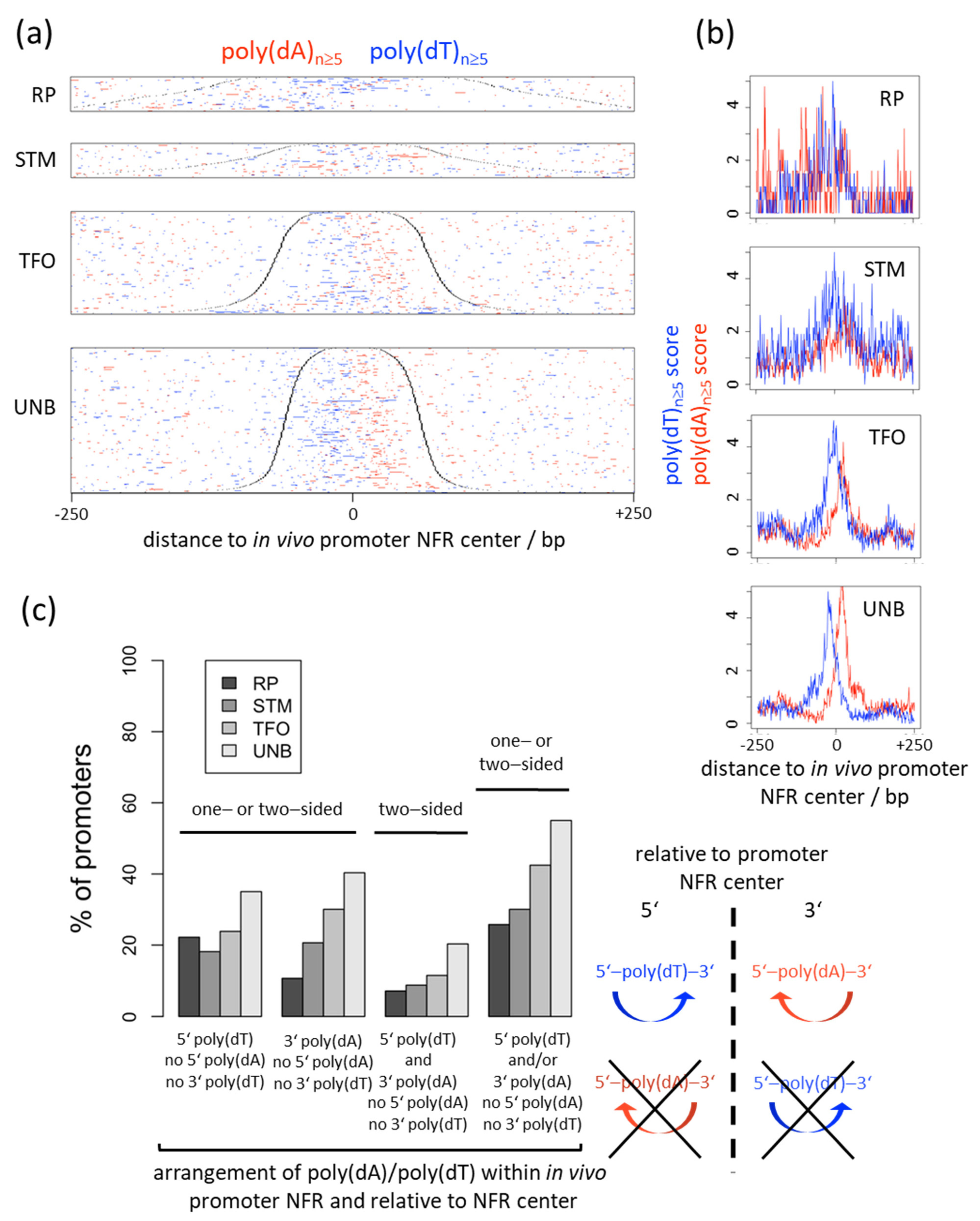
6. Nucleosome Depletion over Poly(dA) Tracts by the Intrinsic Mechanism In Vitro in SGD Is Much Weaker Than by the In Vivo Mechanism
7. Nucleosome Depletion over poly(dA:dT) Tracts Is Not an Intrinsic Default State but Actively Maintained
8. The S. cerevisiae RSC Remodeling Complex Preferentially Evicts Nucleosomes from Poly(dA:dT) Tracts
9. Genomic Strand Bias of Poly(dA) Tracts and Directional Nucleosome Displacement from Poly(dA) Tracts Argues against the Intrinsic but for an Active Nucleosome Depletion Mechanism
10. Remodelers Are Information Processing Hubs That Turn DNA Sequence Information into Nucleosome Organization
11. Species-Specific Strategies for Nucleosome Depletion
12. Afterthought on the Terminology of Nucleosome Depletion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Field, Y.; Kaplan, N.; Fondufe-Mittendorf, Y.; Moore, I.K.; Sharon, E.; Lubling, Y.; Widom, J.; Segal, E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008, 4, e1000216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsankov, A.M.; Thompson, D.A.; Socha, A.; Regev, A.; Rando, O.J. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010, 8, e1000414. [Google Scholar] [CrossRef] [Green Version]
- Segal, E.; Widom, J. Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 2009, 19, 65–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieleg, C.; Krietenstein, N.; Walker, M.; Korber, P. Nucleosome positioning in yeasts: Methods, maps, and mechanisms. Chromosoma 2015, 124, 131–151. [Google Scholar] [CrossRef]
- Jiang, C.; Pugh, B.F. Nucleosome positioning and gene regulation: Advances through genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef] [Green Version]
- Horz, W.; Altenburger, W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981, 9, 2643–2658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dingwall, C.; Lomonossoff, G.P.; Laskey, R.A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981, 9, 2659–2673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caserta, M.; Agricola, E.; Churcher, M.; Hiriart, E.; Verdone, L.; Di Mauro, E.; Travers, A. A translational signature for nucleosome positioning in vivo. Nucleic Acids Res. 2009, 37, 5309–5321. [Google Scholar] [CrossRef] [Green Version]
- Cockell, M.; Rhodes, D.; Klug, A. Location of the primary sites of micrococcal nuclease cleavage on the nucleosome core. J. Mol. Biol. 1983, 170, 423–446. [Google Scholar] [CrossRef]
- Weiner, A.; Hughes, A.; Yassour, M.; Rando, O.J.; Friedman, N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010, 20, 90–100. [Google Scholar] [CrossRef] [Green Version]
- Chereji, R.V.; Ocampo, J.; Clark, D.J. MNase-Sensitive Complexes in Yeast: Nucleosomes and Non-histone Barriers. Mol. Cell 2017, 65, 565–577.e563. [Google Scholar] [CrossRef] [Green Version]
- Chereji, R.V.; Bryson, T.D.; Henikoff, S. Quantitative MNase-seq accurately maps nucleosome occupancy levels. Genome Biol. 2019, 20, 198. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, J.; Tsukiyama, T. ATR-like kinase Mec1 facilitates both chromatin accessibility at DNA replication forks and replication fork progression during replication stress. Genes Dev. 2013, 27, 74–86. [Google Scholar] [CrossRef] [Green Version]
- Rhee, H.S.; Bataille, A.R.; Zhang, L.; Pugh, B.F. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell 2014, 159, 1377–1388. [Google Scholar] [CrossRef] [Green Version]
- Brogaard, K.; Xi, L.; Wang, J.P.; Widom, J. A map of nucleosome positions in yeast at base-pair resolution. Nature 2012, 486, 496–501. [Google Scholar] [CrossRef]
- Chereji, R.V.; Ramachandran, S.; Bryson, T.D.; Henikoff, S. Precise genome-wide mapping of single nucleosomes and linkers in vivo. Genome Biol. 2018, 19, 19. [Google Scholar] [CrossRef]
- Oberbeckmann, E.; Wolff, M.; Krietenstein, N.; Heron, M.; Ellins, J.L.; Schmid, A.; Krebs, S.; Blum, H.; Gerland, U.; Korber, P. Absolute nucleosome occupancy map for the Saccharomyces cerevisiae genome. Genome Res. 2019, 29, 1996–2009. [Google Scholar] [CrossRef] [PubMed]
- Stergachis, A.B.; Debo, B.M.; Haugen, E.; Churchman, L.S.; Stamatoyannopoulos, J.A. Single-molecule regulatory architectures captured by chromatin fiber sequencing. Science 2020, 368, 1449–1454. [Google Scholar] [CrossRef]
- Shipony, Z.; Marinov, G.K.; Swaffer, M.P.; Sinnott-Armstrong, N.A.; Skotheim, J.M.; Kundaje, A.; Greenleaf, W.J. Long-range single-molecule mapping of chromatin accessibility in eukaryotes. Nat. Methods 2020, 17, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.C.; Liu, Y.J.; Dion, M.F.; Slack, M.D.; Wu, L.F.; Altschuler, S.J.; Rando, O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 2005, 309, 626–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, V.; Struhl, K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995, 14, 2570–2579. [Google Scholar] [CrossRef] [PubMed]
- Struhl, K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc. Natl. Acad. Sci. USA 1985, 82, 8419–8423. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.; Tillo, D.; Bray, N.; Morse, R.H.; Davis, R.W.; Hughes, T.R.; Nislow, C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007, 39, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Mavrich, T.N.; Ioshikhes, I.P.; Venters, B.J.; Jiang, C.; Tomsho, L.P.; Qi, J.; Schuster, S.C.; Albert, I.; Pugh, B.F. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008, 18, 1073–1083. [Google Scholar] [CrossRef] [Green Version]
- Eaton, M.L.; Galani, K.; Kang, S.; Bell, S.P.; MacAlpine, D.M. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010, 24, 748–753. [Google Scholar] [CrossRef] [Green Version]
- Raveh-Sadka, T.; Levo, M.; Shabi, U.; Shany, B.; Keren, L.; Lotan-Pompan, M.; Zeevi, D.; Sharon, E.; Weinberger, A.; Segal, E. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat. Genet. 2012, 44, 743–750. [Google Scholar] [CrossRef]
- Hughes, A.L.; Jin, Y.; Rando, O.J.; Struhl, K. A functional evolutionary approach to identify determinants of nucleosome positioning: A unifying model for establishing the genome-wide pattern. Mol. Cell 2012, 48, 5–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Moqtaderi, Z.; Jin, Y.; Zhang, Y.; Liu, X.S.; Struhl, K. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3’-end formation. Proc. Natl. Acad. Sci. USA 2010, 107, 17945–17950. [Google Scholar] [CrossRef] [Green Version]
- Chereji, R.V.; Clark, D.J. Major Determinants of Nucleosome Positioning. Biophys. J. 2018, 114, 2279–2289. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wippo, C.J.; Wal, M.; Ward, E.; Korber, P.; Pugh, B.F. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 2011, 332, 977–980. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, N.; Moore, I.K.; Fondufe-Mittendorf, Y.; Gossett, A.J.; Tillo, D.; Field, Y.; LeProust, E.M.; Hughes, T.R.; Lieb, J.D.; Widom, J.; et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 2009, 458, 362–366. [Google Scholar] [CrossRef] [Green Version]
- Krietenstein, N.; Wal, M.; Watanabe, S.; Park, B.; Peterson, C.L.; Pugh, B.F.; Korber, P. Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell 2016, 167, 709–721.e712. [Google Scholar] [CrossRef] [Green Version]
- Oberbeckmann, E.; Niebauer, V.; Watanabe, S.; Farnung, L.; Moldt, M.; Schmid, A.; Cramer, P.; Peterson, C.L.; Eustermann, S.; Hopfner, K.P.; et al. Ruler elements in chromatin remodelers set nucleosome array spacing and phasing. Nat. Commun. 2021, 12, 3232. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.J.; Kuntala, P.K.; Lai, W.K.M.; Yamada, N.; Badjatia, N.; Mittal, C.; Kuzu, G.; Bocklund, K.; Farrell, N.P.; Blanda, T.R.; et al. A high-resolution protein architecture of the budding yeast genome. Nature 2021, 592, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Oberbeckmann, E.; Krietenstein, N.; Niebauer, V.; Wang, Y.; Schall, K.; Moldt, M.; Straub, T.; Rohs, R.; Hopfner, K.P.; Korber, P.; et al. Genome information processing by the INO80 chromatin remodeler positions nucleosomes. Nat. Commun. 2021, 12, 3231. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhao, H.; Meng, H.; Xing, Y.; Cai, L. A deformation energy model reveals sequence-dependent property of nucleosome positioning. Chromosoma 2021, 130, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Shrader, T.E.; Crothers, D.M. Effects of DNA sequence and histone-histone interactions on nucleosome placement. J. Mol. Biol. 1990, 216, 69–84. [Google Scholar] [CrossRef]
- Shrader, T.E.; Crothers, D.M. Artificial nucleosome positioning sequences. Proc. Natl. Acad. Sci. USA 1989, 86, 7418–7422. [Google Scholar] [CrossRef] [Green Version]
- Thåström, A.; Lowary, P.T.; Widom, J. Measurement of histone-DNA interaction free energy in nucleosomes. Methods 2004, 33, 33–44. [Google Scholar] [CrossRef]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Korolev, N.; Vorontsova, O.V.; Nordenskiold, L. Physicochemical analysis of electrostatic foundation for DNA-protein interactions in chromatin transformations. Prog. Biophys. Mol. Biol. 2007, 95, 23–49. [Google Scholar] [CrossRef]
- Widom, J. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 2001, 34, 269–324. [Google Scholar] [CrossRef]
- Drew, H.R. Can one measure the free energy of binding of the histone octamer to different DNA sequences by salt-dependent reconstitution? J. Mol. Biol. 1991, 219, 391–392. [Google Scholar] [CrossRef]
- Thåström, A.; Bingham, L.M.; Widom, J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J. Mol. Biol. 2004, 338, 695–709. [Google Scholar] [CrossRef]
- Lowary, P.T.; Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998, 276, 19–42. [Google Scholar] [CrossRef]
- Zhang, Y.; Moqtaderi, Z.; Rattner, B.P.; Euskirchen, G.; Snyder, M.; Kadonaga, J.T.; Liu, X.S.; Struhl, K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 2009, 16, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Flaus, A.; Owen-Hughes, T. Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol. Cell. Biol. 2003, 23, 7767–7779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyer, P.N.; Edayathumangalam, R.S.; White, C.L.; Bao, Y.; Chakravarthy, S.; Muthurajan, U.M.; Luger, K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004, 375, 23–44. [Google Scholar]
- Andrews, A.J.; Luger, K. A coupled equilibrium approach to study nucleosome thermodynamics. Methods Enzymol. 2011, 488, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Andrews, A.J.; Luger, K. Nucleosome structure(s) and stability: Variations on a theme. Annu. Rev. Biophys. 2011, 40, 99–117. [Google Scholar] [CrossRef]
- Struhl, K.; Segal, E. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013, 20, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Segal, E.; Widom, J. What controls nucleosome positions? Trends Genet. TIG 2009, 25, 335–343. [Google Scholar] [CrossRef] [Green Version]
- Lantermann, A.B.; Straub, T.; Stralfors, A.; Yuan, G.C.; Ekwall, K.; Korber, P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2010, 17, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Moyle-Heyrman, G.; Zaichuk, T.; Xi, L.; Zhang, Q.; Uhlenbeck, O.C.; Holmgren, R.; Widom, J.; Wang, J.P. Chemical map of Schizosaccharomyces pombe reveals species-specific features in nucleosome positioning. Proc. Natl. Acad. Sci. USA 2013, 110, 20158–20163. [Google Scholar] [CrossRef] [Green Version]
- Segal, E.; Fondufe-Mittendorf, Y.; Chen, L.; Thastrom, A.; Field, Y.; Moore, I.K.; Wang, J.P.; Widom, J. A genomic code for nucleosome positioning. Nature 2006, 442, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Wippo, C.J.; Krstulovic, B.S.; Ertel, F.; Musladin, S.; Blaschke, D.; Sturzl, S.; Yuan, G.C.; Horz, W.; Korber, P.; Barbaric, S. Differential cofactor requirements for histone eviction from two nucleosomes at the yeast PHO84 promoter are determined by intrinsic nucleosome stability. Mol. Cell. Biol. 2009, 29, 2960–2981. [Google Scholar] [CrossRef] [Green Version]
- Whitehouse, I.; Rando, O.J.; Delrow, J.; Tsukiyama, T. Chromatin remodelling at promoters suppresses antisense transcription. Nature 2007, 450, 1031–1035. [Google Scholar] [CrossRef]
- Losa, R.; Omari, S.; Thoma, F. Poly(dA).poly(dT) rich sequences are not sufficient to exclude nucleosome formation in a constitutive yeast promoter. Nucleic Acids Res. 1990, 18, 3495–3502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorch, Y.; Maier-Davis, B.; Kornberg, R.D. Role of DNA sequence in chromatin remodeling and the formation of nucleosome-free regions. Genes Dev. 2014, 28, 2492–2497. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; White, C.L.; Luger, K. Nucleosome core particles containing a poly(dA.dT) sequence element exhibit a locally distorted DNA structure. J. Mol. Biol. 2006, 361, 617–624. [Google Scholar] [CrossRef]
- Tran, Q.H.; Unden, G. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur. J. Biochem. 1998, 251, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Harada, B.T.; Hwang, W.L.; Deindl, S.; Chatterjee, N.; Bartholomew, B.; Zhuang, X. Stepwise nucleosome translocation by RSC remodeling complexes. eLife 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Deindl, S.; Hwang, W.L.; Hota, S.K.; Blosser, T.R.; Prasad, P.; Bartholomew, B.; Zhuang, X. ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 2013, 152, 442–452. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.Y.; Johnson, S.L.; Gamarra, N.I.; Narlikar, G.J. Mechanisms of ATP-Dependent Chromatin Remodeling Motors. Annu. Rev. Biophys. 2016, 45, 153–181. [Google Scholar] [CrossRef] [PubMed]
- Andrews, A.J.; Chen, X.; Zevin, A.; Stargell, L.A.; Luger, K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 2010, 37, 834–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozonov, E.A.; van Nimwegen, E. Nucleosome free regions in yeast promoters result from competitive binding of transcription factors that interact with chromatin modifiers. PLoS Comput. Biol. 2013, 9, e1003181. [Google Scholar] [CrossRef] [Green Version]
- Kharerin, H.; Bai, L. Thermodynamic modeling of genome-wide nucleosome depleted regions in yeast. PLoS Comput. Biol. 2021, 17, e1008560. [Google Scholar] [CrossRef] [PubMed]
- Tsankov, A.; Yanagisawa, Y.; Rhind, N.; Regev, A.; Rando, O.J. Evolutionary divergence of intrinsic and trans-regulated nucleosome positioning sequences reveals plastic rules for chromatin organization. Genome Res. 2011, 21, 1851–1862. [Google Scholar] [CrossRef] [Green Version]
- Reja, R.; Vinayachandran, V.; Ghosh, S.; Pugh, B.F. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev. 2015, 29, 1942–1954. [Google Scholar] [CrossRef] [Green Version]
- Knight, B.; Kubik, S.; Ghosh, B.; Bruzzone, M.J.; Geertz, M.; Martin, V.; Dénervaud, N.; Jacquet, P.; Ozkan, B.; Rougemont, J.; et al. Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription. Genes Dev. 2014, 28, 1695–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Bakel, H.; Tsui, K.; Gebbia, M.; Mnaimneh, S.; Hughes, T.R.; Nislow, C. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet. 2013, 9, e1003479. [Google Scholar] [CrossRef] [Green Version]
- Parnell, T.J.; Huff, J.T.; Cairns, B.R. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2008, 27, 100–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badis, G.; Chan, E.T.; van Bakel, H.; Pena-Castillo, L.; Tillo, D.; Tsui, K.; Carlson, C.D.; Gossett, A.J.; Hasinoff, M.J.; Warren, C.L.; et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 2008, 32, 878–887. [Google Scholar] [CrossRef] [Green Version]
- Hartley, P.D.; Madhani, H.D. Mechanisms that specify promoter nucleosome location and identity. Cell 2009, 137, 445–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubik, S.; Bruzzone, M.J.; Challal, D.; Dreos, R.; Mattarocci, S.; Bucher, P.; Libri, D.; Shore, D. Opposing chromatin remodelers control transcription initiation frequency and start site selection. Nat. Struct. Mol. Biol. 2019, 26, 744–754. [Google Scholar] [CrossRef]
- Kubik, S.; O’Duibhir, E.; de Jonge, W.J.; Mattarocci, S.; Albert, B.; Falcone, J.L.; Bruzzone, M.J.; Holstege, F.C.P.; Shore, D. Sequence-Directed Action of RSC Remodeler and General Regulatory Factors Modulates +1 Nucleosome Position to Facilitate Transcription. Mol. Cell 2018, 71, 89–102.e105. [Google Scholar] [CrossRef] [Green Version]
- Challal, D.; Barucco, M.; Kubik, S.; Feuerbach, F.; Candelli, T.; Geoffroy, H.; Benaksas, C.; Shore, D.; Libri, D. General Regulatory Factors Control the Fidelity of Transcription by Restricting Non-coding and Ectopic Initiation. Mol. Cell 2018, 72, 955–969.e957. [Google Scholar] [CrossRef] [Green Version]
- Ganguli, D.; Chereji, R.V.; Iben, J.R.; Cole, H.A.; Clark, D.J. RSC-dependent constructive and destructive interference between opposing arrays of phased nucleosomes in yeast. Genome Res. 2014, 24, 1637–1649. [Google Scholar] [CrossRef] [Green Version]
- Klein-Brill, A.; Joseph-Strauss, D.; Appleboim, A.; Friedman, N. Dynamics of Chromatin and Transcription during Transient Depletion of the RSC Chromatin Remodeling Complex. Cell Rep. 2019, 26, 279–292.e275. [Google Scholar] [CrossRef] [Green Version]
- Rawal, Y.; Chereji, R.V.; Qiu, H.; Ananthakrishnan, S.; Govind, C.K.; Clark, D.J.; Hinnebusch, A.G. SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast. Genes Dev. 2018, 32, 695–710. [Google Scholar] [CrossRef]
- Qiu, H.; Biernat, E.; Govind, C.K.; Rawal, Y.; Chereji, R.V.; Clark, D.J.; Hinnebusch, A.G. Chromatin remodeler Ino80C acts independently of H2A.Z to evict promoter nucleosomes and stimulate transcription of highly expressed genes in yeast. Nucleic Acids Res. 2020, 48, 8408–8430. [Google Scholar] [CrossRef]
- Yen, K.; Vinayachandran, V.; Batta, K.; Koerber, R.T.; Pugh, B.F. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 2012, 149, 1461–1473. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, S.; Henikoff, S. Transcriptional Regulators Compete with Nucleosomes Post-replication. Cell 2016, 165, 580–592. [Google Scholar] [CrossRef] [Green Version]
- Fennessy, R.T.; Owen-Hughes, T. Establishment of a promoter-based chromatin architecture on recently replicated DNA can accommodate variable inter-nucleosome spacing. Nucleic Acids Res. 2016, 44, 7189–7203. [Google Scholar] [CrossRef] [Green Version]
- Vasseur, P.; Tonazzini, S.; Ziane, R.; Camasses, A.; Rando, O.J.; Radman-Livaja, M. Dynamics of Nucleosome Positioning Maturation following Genomic Replication. Cell Rep. 2016, 16, 2651–2665. [Google Scholar] [CrossRef] [Green Version]
- Schermer, U.J.; Korber, P.; Horz, W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol. Cell 2005, 19, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Dion, M.F.; Kaplan, T.; Kim, M.; Buratowski, S.; Friedman, N.; Rando, O.J. Dynamics of replication-independent histone turnover in budding yeast. Science 2007, 315, 1405–1408. [Google Scholar] [CrossRef]
- Rufiange, A.; Jacques, P.E.; Bhat, W.; Robert, F.; Nourani, A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 2007, 27, 393–405. [Google Scholar] [CrossRef]
- Jamai, A.; Imoberdorf, R.M.; Strubin, M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 2007, 25, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Flaus, A.; Martin, D.M.; Barton, G.J.; Owen-Hughes, T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006, 34, 2887–2905. [Google Scholar] [CrossRef] [Green Version]
- Cairns, B.R.; Lorch, Y.; Li, Y.; Zhang, M.; Lacomis, L.; Erdjument-Bromage, H.; Tempst, P.; Du, J.; Laurent, B.; Kornberg, R.D. RSC, an essential, abundant chromatin-remodeling complex. Cell 1996, 87, 1249–1260. [Google Scholar] [CrossRef] [Green Version]
- Wippo, C.J.; Israel, L.; Watanabe, S.; Hochheimer, A.; Peterson, C.L.; Korber, P. The RSC chromatin remodelling enzyme has a unique role in directing the accurate positioning of nucleosomes. EMBO J. 2011, 30, 1277–1288. [Google Scholar] [CrossRef]
- Lorch, Y.; Griesenbeck, J.; Boeger, H.; Maier-Davis, B.; Kornberg, R.D. Selective removal of promoter nucleosomes by the RSC chromatin-remodeling complex. Nat. Struct. Mol. Biol. 2011, 18, 881–885. [Google Scholar] [CrossRef] [Green Version]
- de Boer, C.G.; Hughes, T.R. Poly-dA:dT tracts form an in vivo nucleosomal turnstile. PLoS ONE 2014, 9, e110479. [Google Scholar] [CrossRef]
- Wu, R.; Li, H. Positioned and G/C-capped poly(dA:dT) tracts associate with the centers of nucleosome-free regions in yeast promoters. Genome Res. 2010, 20, 473–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lusser, A.; Urwin, D.L.; Kadonaga, J.T. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2005, 12, 160–166. [Google Scholar] [CrossRef]
- Brahma, S.; Henikoff, S. RSC-Associated Subnucleosomes Define MNase-Sensitive Promoters in Yeast. Mol. Cell 2019, 73, 238–249.e233. [Google Scholar] [CrossRef] [Green Version]
- Kubik, S.; Bruzzone, M.J.; Jacquet, P.; Falcone, J.L.; Rougemont, J.; Shore, D. Nucleosome Stability Distinguishes Two Different Promoter Types at All Protein-Coding Genes in Yeast. Mol. Cell 2015, 60, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Wittmeyer, J.; Cairns, B.R. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 2005, 12, 747–755. [Google Scholar] [CrossRef]
- Winger, J.; Bowman, G.D. The Sequence of Nucleosomal DNA Modulates Sliding by the Chd1 Chromatin Remodeler. J. Mol. Biol. 2017, 429, 808–822. [Google Scholar] [CrossRef] [Green Version]
- Korber, P. Active nucleosome positioning beyond intrinsic biophysics is revealed by in vitro reconstitution. Biochem. Soc. Trans. 2012, 40, 377–382. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Frouws, T.D.; Angst, B.; Fitzgerald, D.J.; DeLuca, C.; Schimmele, K.; Sargent, D.F.; Richmond, T.J. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature 2011, 472, 448–453. [Google Scholar] [CrossRef]
- Zhou, T.; Yang, L.; Lu, Y.; Dror, I.; Dantas Machado, A.C.; Ghane, T.; Di Felice, R.; Rohs, R. DNAshape: A method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013, 41, W56–W62. [Google Scholar] [CrossRef] [Green Version]
- Basu, A.; Bobrovnikov, D.G.; Qureshi, Z.; Kayikcioglu, T.; Ngo, T.T.M.; Ranjan, A.; Eustermann, S.; Cieza, B.; Morgan, M.T.; Hejna, M.; et al. Measuring DNA mechanics on the genome scale. Nature 2021, 589, 462–467. [Google Scholar] [CrossRef]
- Rippe, K.; Schrader, A.; Riede, P.; Strohner, R.; Lehmann, E.; Langst, G. DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc. Natl. Acad. Sci. USA 2007, 104, 15635–15640. [Google Scholar] [CrossRef] [Green Version]
- van Vugt, J.J.; de Jager, M.; Murawska, M.; Brehm, A.; van Noort, J.; Logie, C. Multiple aspects of ATP-dependent nucleosome translocation by RSC and Mi-2 are directed by the underlying DNA sequence. PLoS ONE 2009, 4, e6345. [Google Scholar] [CrossRef]
- Eustermann, S.; Schall, K.; Kostrewa, D.; Lakomek, K.; Strauss, M.; Moldt, M.; Hopfner, K.P. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 2018, 556, 386–390. [Google Scholar] [CrossRef]
- Monahan, B.J.; Villen, J.; Marguerat, S.; Bahler, J.; Gygi, S.P.; Winston, F. Fission yeast SWI/SNF and RSC complexes show compositional and functional differences from budding yeast. Nat. Struct. Mol. Biol. 2008, 15, 873–880. [Google Scholar] [CrossRef] [Green Version]
- Ozsolak, F.; Song, J.S.; Liu, X.S.; Fisher, D.E. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007, 25, 244–248. [Google Scholar] [CrossRef]
- Haberle, V.; Lenhard, B. Promoter architectures and developmental gene regulation. Semin. Cell Dev. Biol. 2016, 57, 11–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tillo, D.; Kaplan, N.; Moore, I.K.; Fondufe-Mittendorf, Y.; Gossett, A.J.; Field, Y.; Lieb, J.D.; Widom, J.; Segal, E.; Hughes, T.R. High nucleosome occupancy is encoded at human regulatory sequences. PLoS ONE 2010, 5, e9129. [Google Scholar] [CrossRef]
- Tompitak, M.; Vaillant, C.; Schiessel, H. Genomes of Multicellular Organisms Have Evolved to Attract Nucleosomes to Promoter Regions. Biophys. J. 2017, 112, 505–511. [Google Scholar] [CrossRef] [Green Version]
- Morse, R.H. Transcription factor access to promoter elements. J. Cell. Biochem. 2007, 102, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Oruba, A.; Saccani, S.; van Essen, D. Role of cell-type specific nucleosome positioning in inducible activation of mammalian promoters. Nat. Commun. 2020, 11, 1075. [Google Scholar] [CrossRef] [PubMed]
- Reinke, H.; Horz, W. Anatomy of a hypersensitive site. Biochimica Biophysica Acta 2004, 1677, 24–29. [Google Scholar] [CrossRef]
- Elgin, S.C. Anatomy of hypersensitive sites. Nature 1984, 309, 213–214. [Google Scholar] [CrossRef]
- Wu, C. The 5’ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 1980, 286, 854–860. [Google Scholar] [CrossRef]
- Jin, C.; Zang, C.; Wei, G.; Cui, K.; Peng, W.; Zhao, K.; Felsenfeld, G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 2009, 41, 941–945. [Google Scholar] [CrossRef] [Green Version]
- Lai, W.K.M.; Pugh, B.F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 2017, 18, 548–562. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barnes, T.; Korber, P. The Active Mechanism of Nucleosome Depletion by Poly(dA:dT) Tracts In Vivo. Int. J. Mol. Sci. 2021, 22, 8233. https://doi.org/10.3390/ijms22158233
Barnes T, Korber P. The Active Mechanism of Nucleosome Depletion by Poly(dA:dT) Tracts In Vivo. International Journal of Molecular Sciences. 2021; 22(15):8233. https://doi.org/10.3390/ijms22158233
Chicago/Turabian StyleBarnes, Toby, and Philipp Korber. 2021. "The Active Mechanism of Nucleosome Depletion by Poly(dA:dT) Tracts In Vivo" International Journal of Molecular Sciences 22, no. 15: 8233. https://doi.org/10.3390/ijms22158233




