Comparison of Silks from Pseudoips prasinana and Bombyx mori Shows Molecular Convergence in Fibroin Heavy Chains but Large Differences in Other Silk Components
Abstract
:1. Introduction
2. Results
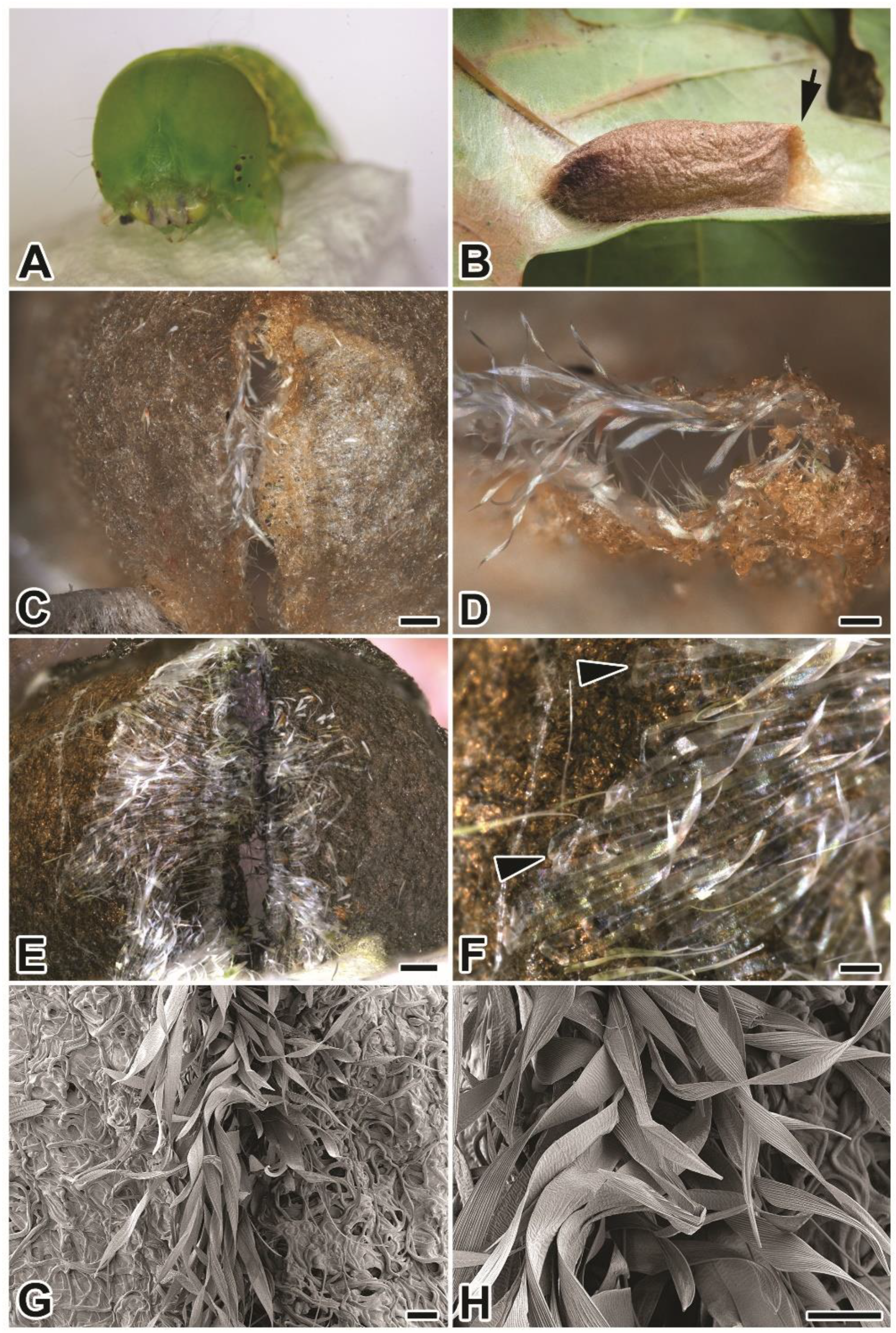
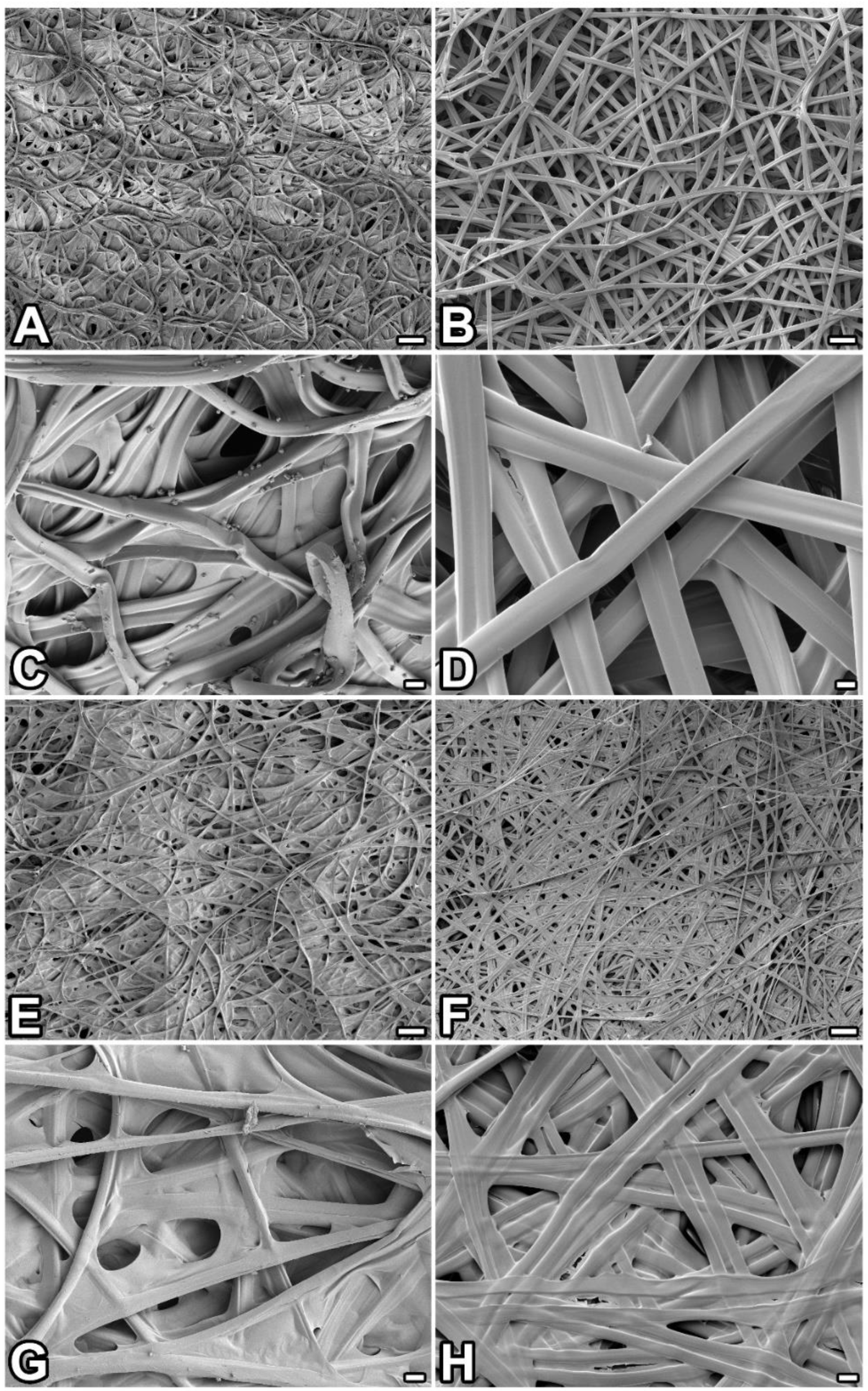
2.1. Compact Structure of P. prasinana Cocoons
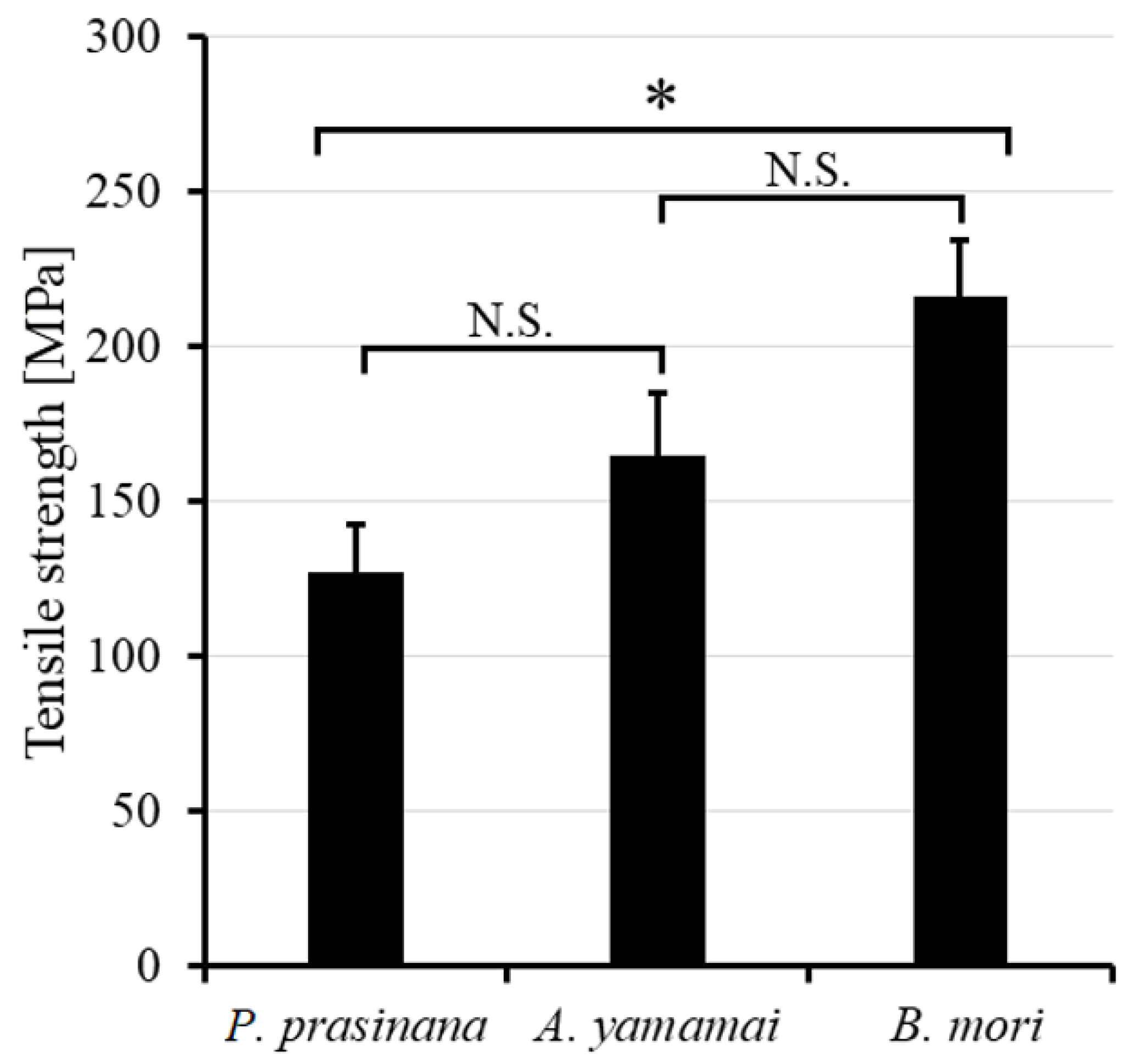
2.2. Mechanical Strength Measurement
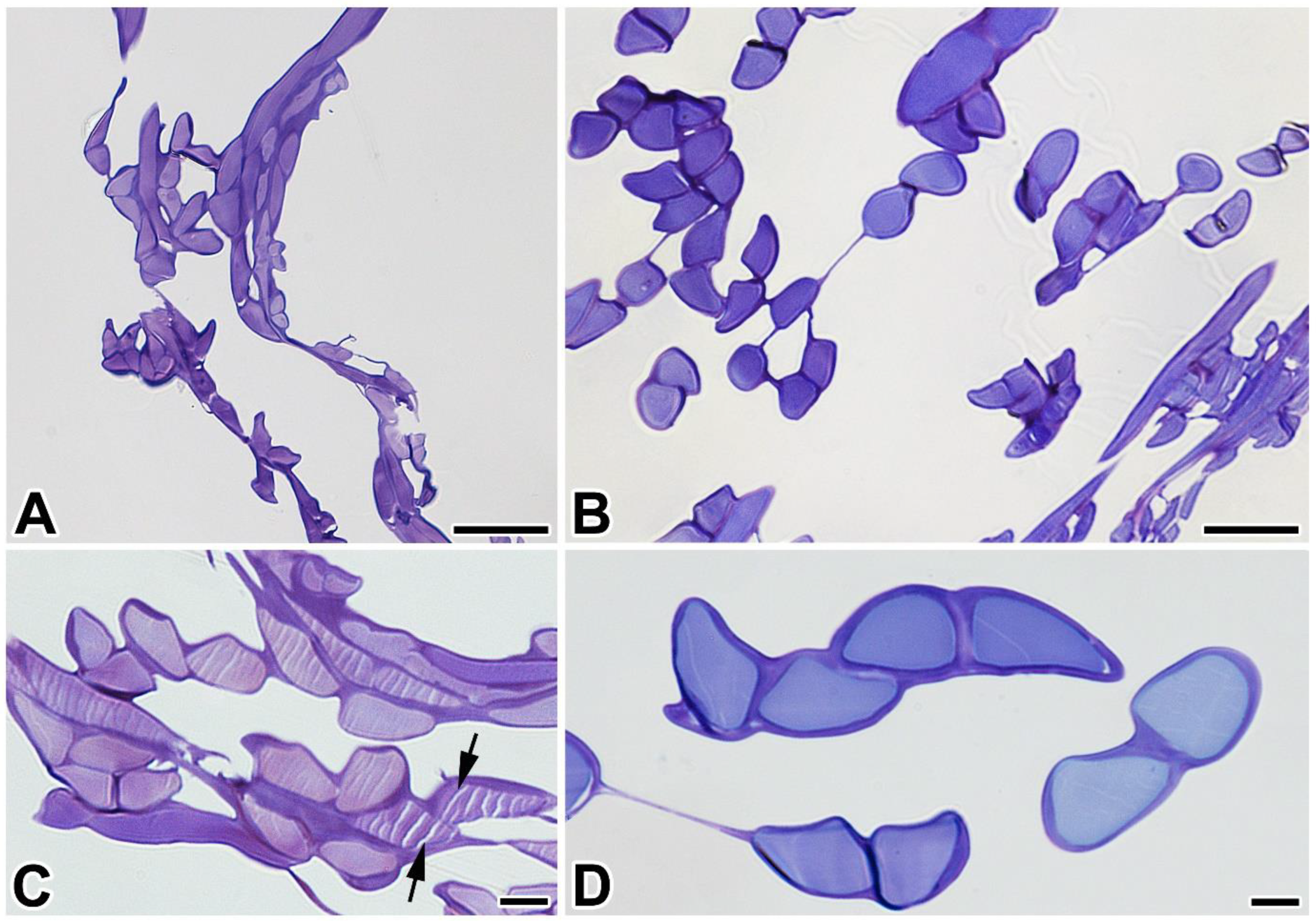
2.3. Transcriptome Construction, Proteomic Analysis
2.4. Major Silk Structural Proteins and Their Phylogenetic Relationships
3. Discussion
4. Materials and Methods
4.1. Insects
4.2. Histology and Scanning Electron Microscopy
4.3. Measurement of Silk Fiber Mechanical Strength
4.4. Transcriptome Preparation and Analysis
4.5. Protein Extraction and Identification of Protein Fragments by Mass Spectrometry
4.6. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sehnal, F.; Zurovec, M. Construction of silk fiber core in Lepidoptera. Biomacromolecules 2004, 5, 666–674. [Google Scholar] [CrossRef]
- Takei, F.; Kikuchi, Y.; Kikuchi, A.; Mizuno, S.; Shimura, K. Further evidence for importance of the subunit combination of silk fibroin in its efficient secretion from the posterior silk gland cells. J. Cell. Biol. 1987, 105, 175–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, K.; Inoue, S.; Mizuno, S. Hydrophobic interaction of P25, containing Asn-linked oligosaccharide chains, with the H-L complex of silk fibroin produced by Bombyx mori. Insect. Biochem. Mol. Biol. 1999, 29, 269–276. [Google Scholar] [CrossRef]
- Lucas, F.; Rudall, K.M. Extracellular Fibrous Proteins: The silks. In Comprehensive Biochemistry; Florkin, M., Stotz, E.H., Eds.; Elsevier: Amsterdam, The Netherlands, 1968; Volume 26, pp. 475–558. [Google Scholar]
- Craig, C.L. Evolution of arthropod silks. Annu. Rev. Entomol. 1997, 42, 231–267. [Google Scholar] [CrossRef] [PubMed]
- Warwicker, J.O. Comparative Studies of Fibroins. II. The Crystal Structures of Various Fibroins. J. Mol. Biol. 1960, 2, 350–362. [Google Scholar] [CrossRef]
- Hwang, J.S.; Lee, J.S.; Goo, T.W.; Yun, E.Y.; Lee, K.S.; Kim, Y.S.; Jin, B.R.; Lee, S.M.; Kim, K.Y.; Kang, S.W.; et al. Cloning of the fibroin gene from the oak silkworm, Antheraea yamamai and its complete sequence. Biotechnol. Lett. 2001, 23, 1321–1326. [Google Scholar] [CrossRef]
- Sezutsu, H.; Yukuhiro, K. Dynamic rearrangement within the Antheraea pernyi silk fibroin gene is associated with four types of repetitive units. J. Mol. Evol. 2000, 51, 329–338. [Google Scholar] [CrossRef]
- Zurovec, M.; Sehnal, F. Unique molecular architecture of silk fibroin in the waxmoth, Galleria mellonella. J. Biol. Chem. 2002, 277, 22639–22647. [Google Scholar] [CrossRef] [Green Version]
- Tsubota, T.; Yoshioka, T.; Jouraku, A.; Suzuki, T.K.; Yonemura, N.; Yukuhiro, K.; Kameda, T.; Sezutsu, H. Transcriptomic analysis of the bagworm moth silk gland reveals a number of silk genes conserved within Lepidoptera. Insect Sci. 2020, 28, 885–900. [Google Scholar] [CrossRef]
- Lussi, H.G. Chloephorinae, Noctuidae. In Die Schmetterlinge Baden-Wurttembergs; Bartsch, D., Ed.; Eugen Ulmer Verlag: Stuttgart, Germany, 1997; Volume 5, pp. 523–527. [Google Scholar]
- Fibiger, M.; Ronkay, L.; Steiner, A.; Zilli, A. Pantheinae—Bryophilinae. In Noctuidae Europaeae; Apollo Books; Entomological Press: Soro, Denmark, 2009; Volume 11, pp. 123–124. [Google Scholar]
- Davey, P.A.; Power, A.M.; Santos, R.; Bertemes, P.; Ladurner, P.; Palmowski, P.; Clarke, J.; Flammang, P.; Lengerer, B.; Hennebert, E.; et al. Omics-based molecular analyses of adhesion by aquatic invertebrates. Biol. Rev. 2021, 96, 1051–1075. [Google Scholar] [CrossRef]
- Rouhova, L.; Kludkiewicz, B.; Sehadova, H.; Sery, M.; Kucerova, L.; Konik, P.; Zurovec, M. Silk of the common clothes moth, Tineola bisselliella, a cosmopolitan pest belonging to the basal ditrysian moth line. Insect Biochem. Mol. Biol. 2021, 130, 103527. [Google Scholar] [CrossRef]
- Sehnal, F.; Craig, C. Silk Production. In Encyclopedia of Insects, 2nd ed.; Resh, V.H., Cardé, R.T., Eds.; Academic Press: Burlington, MA, USA; London, UK, 2009; p. 924. [Google Scholar]
- Peng, Z.; Yang, X.; Liu, C.; Dong, Z.; Wang, F.; Wang, X.; Hu, W.; Zhang, X.; Zhao, P.; Xia, Q. Structural and mechanical properties of silk from different instars of Bombyx mori. Biomacromolecules 2019, 20, 1203–1216. [Google Scholar] [CrossRef]
- Keten, S.; Xu, Z.; Ihle, B.; Buehler, M.J. Nanoconfinement controls stiffness, strength and mechanical toughness of beta-sheet crystals in silk. Nat. Mater. 2010, 9, 359–367. [Google Scholar] [CrossRef]
- Denny, M.W. Silks-Their Properties and Functions. In The Mechanical Properties of Biological Materials; Vincent, J.F.V., Currey, J.D., Eds.; Cambridge University Press: Cambridge, UK, 1980; Volume 34, pp. 247–272. [Google Scholar]
- Numata, A.; Sato, R.; Yazawa, K.; Hikima, T.; Masunaga, H. Crystal structure and physical properties of Antheraea yamamai silk fibers: Long poly(alanine) sequences are partially in the crystalline region. Polymer 2015, 77, 87–94. [Google Scholar] [CrossRef]
- Feng, Y.; Lin, J.; Niu, L.; Wang, Y.; Cheng, Z.; Sun, X.; Li, M. High molecular weight silk fibroin prepared by papain degumming. Polymers 2020, 12, 2105. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Tanaka, K.; Arisaka, F.; Kimura, S.; Ohtomo, K.; Mizuno, S. Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain, and P25, with a 6:6:1 molar ratio. J. Biol. Chem. 2000, 275, 40517–40528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, S.; Tanaka, K.; Tanaka, H.; Ohtomo, K.; Kanda, T.; Imamura, M.; Quan, G.X.; Kojima, K.; Yamashita, T.; Nakajima, T.; et al. Assembly of the silk fibroin elementary unit in endoplasmic reticulum and a role of L-chain for protection of alpha1,2-mannose residues in N-linked oligosaccharide chains of fibrohexamerin/P25. Eur. J. Biochem. 2004, 271, 356–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, D.; Lu, W.; Zhang, Y.; Guo, Q.; Xiang, Z.; Zhao, A. New insight into the mechanism underlying fibroin secretion in silkworm, Bombyx mori. FEBS J. 2015, 282, 89–101. [Google Scholar] [CrossRef]
- Xia, Q.; Zhou, Z.; Lu, C.; Cheng, D.; Dai, F.; Li, B.; Zhao, P.; Zha, X.; Cheng, T.; Chai, C.; et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 2004, 306, 1937–1940. [Google Scholar]
- Sutherland, T.D.; Young, J.H.; Weisman, S.; Hayashi, C.Y.; Merritt, D.J. Insect silk: One name, many materials. Annu. Rev. Entomol. 2010, 55, 171–188. [Google Scholar] [CrossRef]
- Kludkiewicz, B.; Kucerova, L.; Konikova, T.; Strnad, H.; Hradilova, M.; Zaloudikova, A.; Sehadova, H.; Konik, P.; Sehnal, F.; Zurovec, M. The expansion of genes encoding soluble silk components in the greater wax moth, Galleria mellonella. Insect Biochem. Mol. Biol. 2019, 106, 28–38. [Google Scholar] [CrossRef]
- Tanaka, K.; Mizuno, S. Homologues of fibroin L-chain and P25 of Bombyx mori are present in Dendrolimus spectabilis and Papilio xuthus but not detectable in Antheraea yamamai. Insect Biochem. Mol. Biol. 2001, 31, 665–677. [Google Scholar] [CrossRef]
- Zurovec, M.; Yonemura, N.; Kludkiewicz, B.; Sehnal, F.; Kodrik, D.; Vieira, L.C.; Kucerova, L.; Strnad, H.; Konik, P.; Sehadova, H. Sericin composition in the silk of Antheraea yamamai. Biomacromolecules 2016, 17, 1776–1787. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Hughes, C.S.; Foehr, S.; Garfield, D.A.; Furlong, E.E.; Steinmetz, L.M.; Krijgsveld, J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014, 10, 757. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Kludkiewicz, B.; Takasu, Y.; Fedic, R.; Tamura, T.; Sehnal, F.; Zurovec, M. Structure and expression of the silk adhesive protein Ser2 in Bombyx mori. Insect Biochem. Mol. Biol. 2009, 39, 938–946. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart model selection in PhyML. Mol. Biol Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rindos, M.; Kucerova, L.; Rouhova, L.; Sehadova, H.; Sery, M.; Hradilova, M.; Konik, P.; Zurovec, M. Comparison of Silks from Pseudoips prasinana and Bombyx mori Shows Molecular Convergence in Fibroin Heavy Chains but Large Differences in Other Silk Components. Int. J. Mol. Sci. 2021, 22, 8246. https://doi.org/10.3390/ijms22158246
Rindos M, Kucerova L, Rouhova L, Sehadova H, Sery M, Hradilova M, Konik P, Zurovec M. Comparison of Silks from Pseudoips prasinana and Bombyx mori Shows Molecular Convergence in Fibroin Heavy Chains but Large Differences in Other Silk Components. International Journal of Molecular Sciences. 2021; 22(15):8246. https://doi.org/10.3390/ijms22158246
Chicago/Turabian StyleRindos, Michal, Lucie Kucerova, Lenka Rouhova, Hana Sehadova, Michal Sery, Miluse Hradilova, Peter Konik, and Michal Zurovec. 2021. "Comparison of Silks from Pseudoips prasinana and Bombyx mori Shows Molecular Convergence in Fibroin Heavy Chains but Large Differences in Other Silk Components" International Journal of Molecular Sciences 22, no. 15: 8246. https://doi.org/10.3390/ijms22158246







