Anti-Idiotype scFv Localizes an Autoepitope in the Globular Domain of C1q
Abstract
:1. Introduction
2. Results
2.1. Generation of Anti-Idyotipic scFv
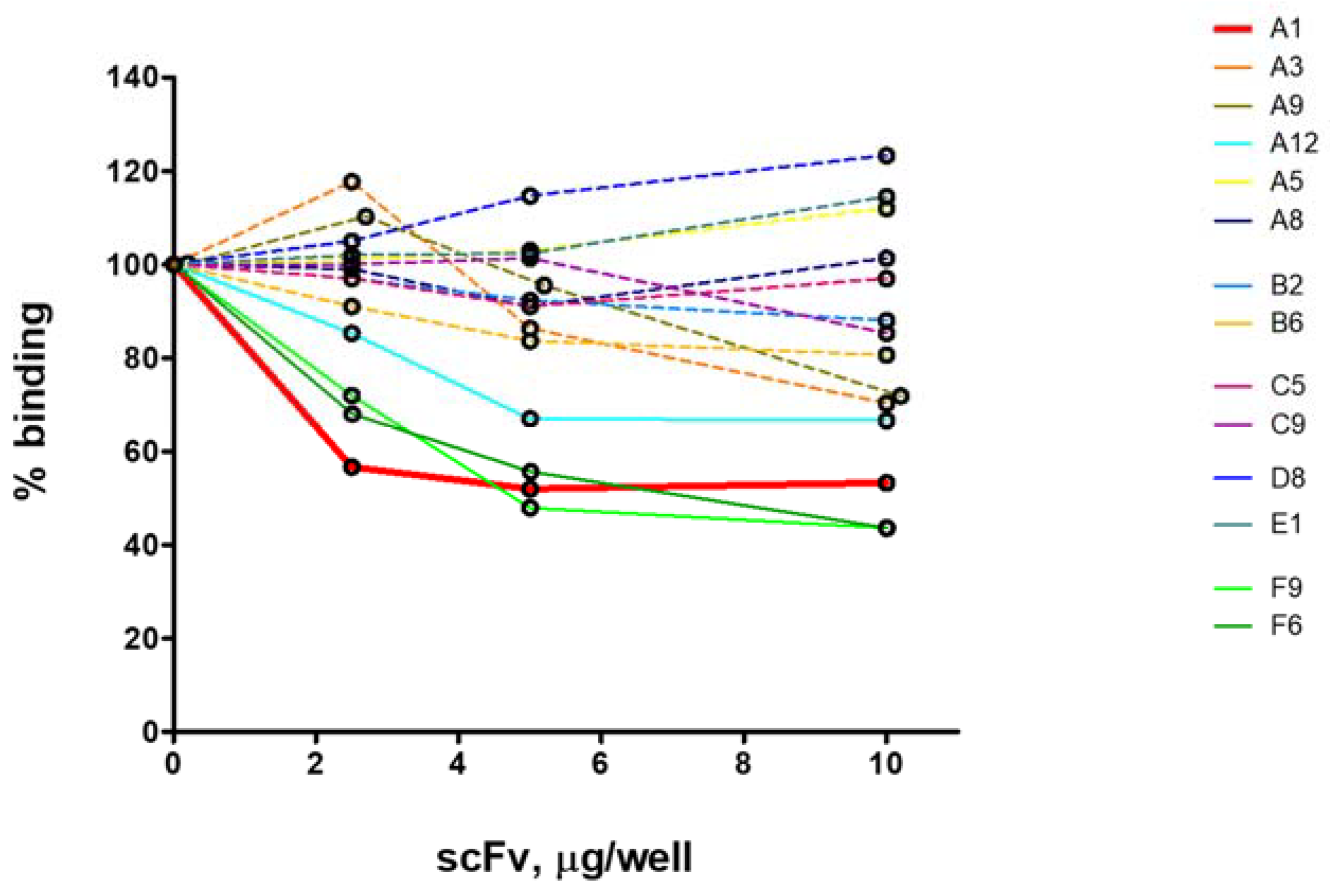
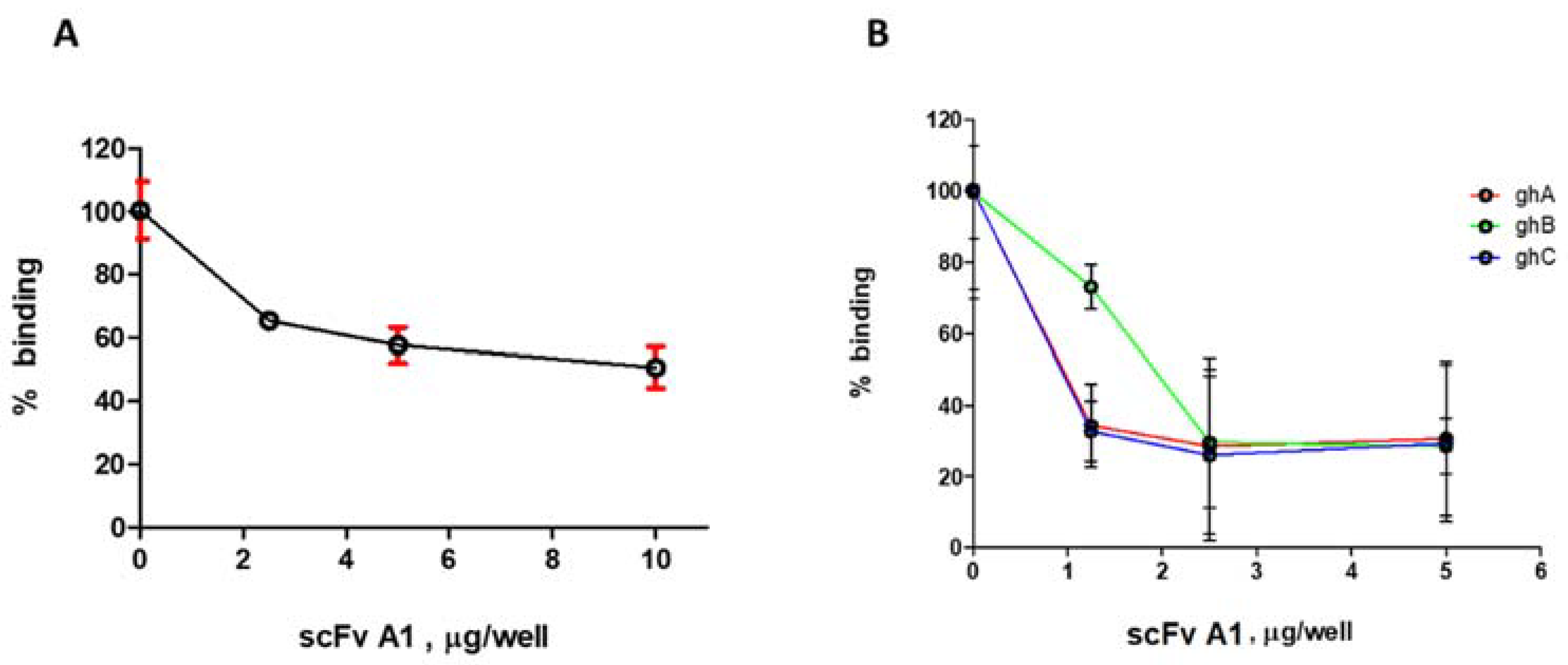
2.2. ELISA Analysis for Selection of Inhibitory scFv
2.3. Fluorescence Spectroscopy Analysis
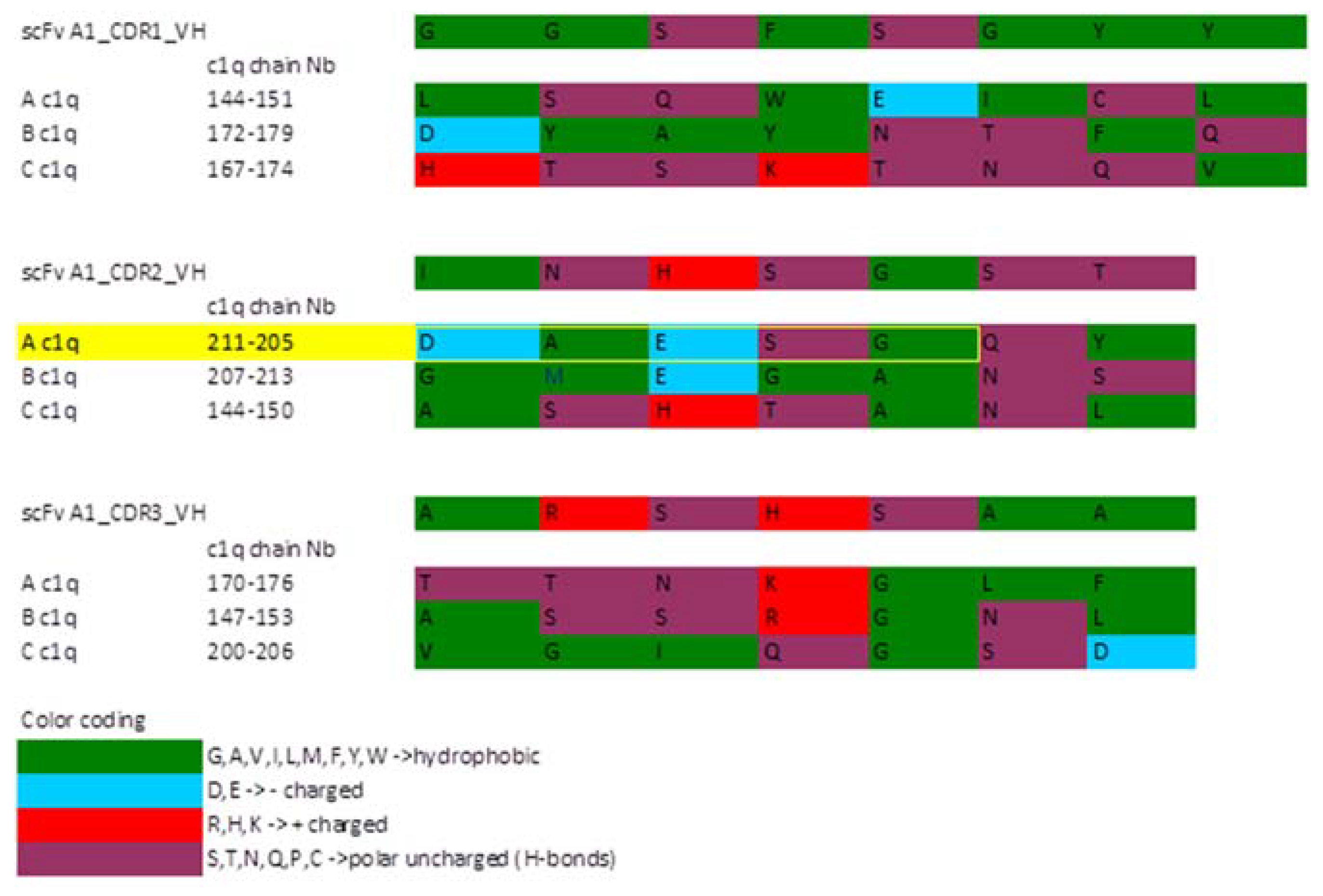
2.4. Sequence Analysis of the Selected scFv A1
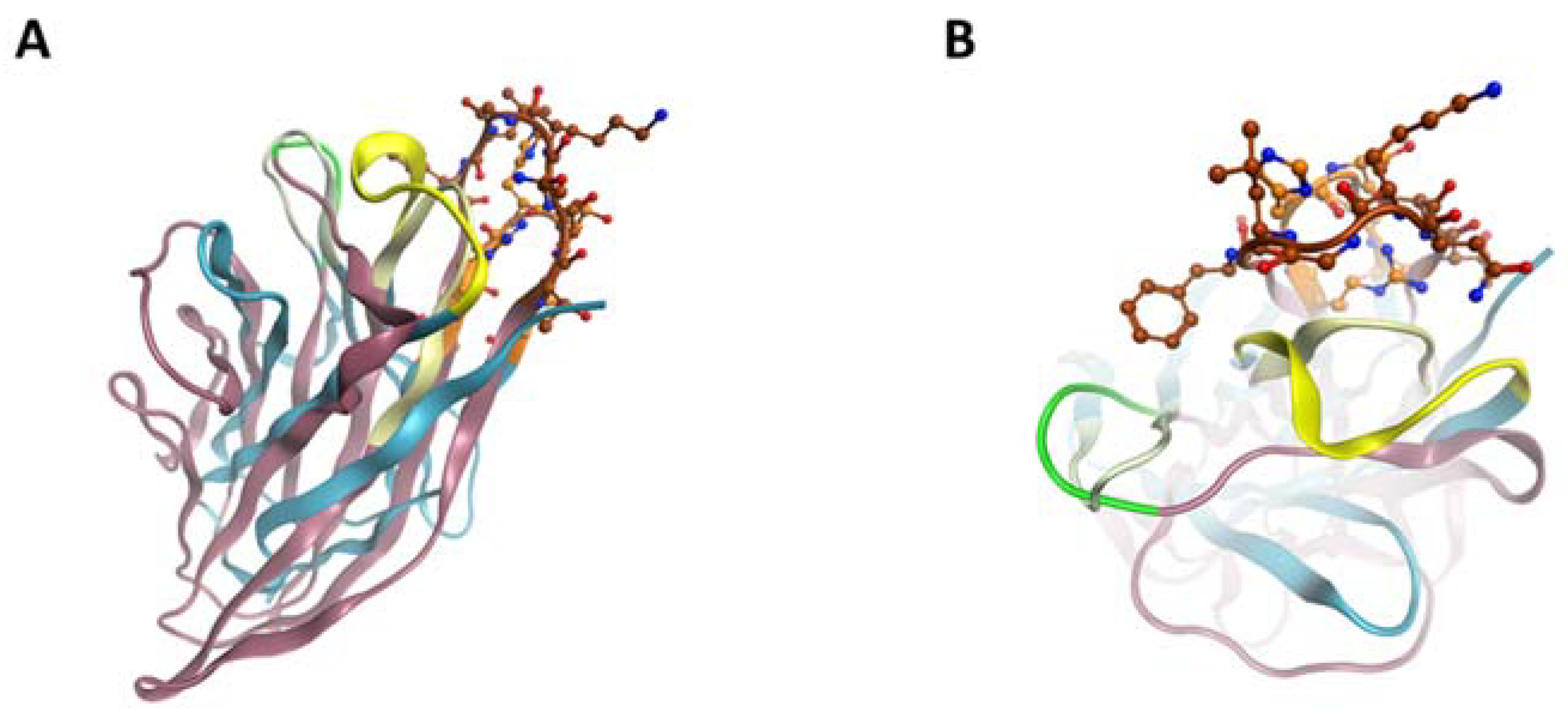
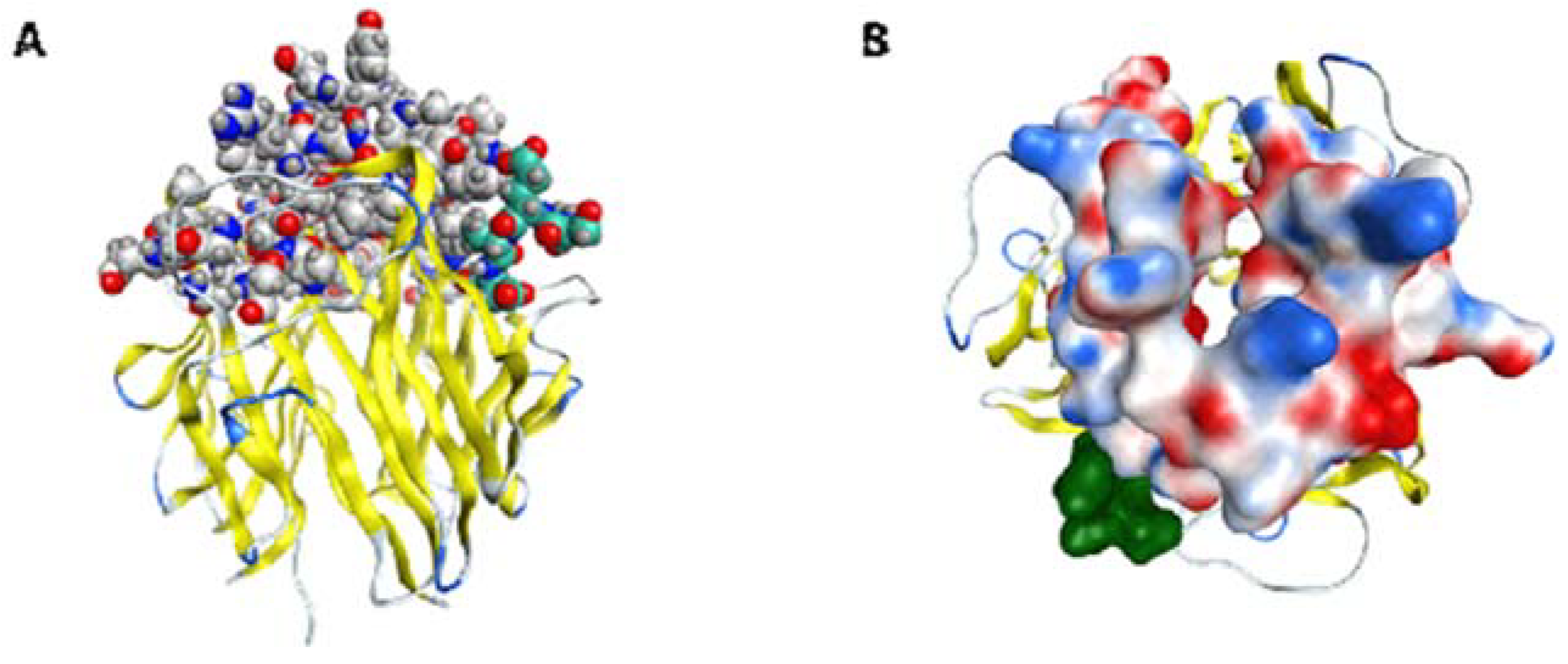
2.5. ScFv A1 Homology Model
3. Discussion
4. Materials and Methods
4.1. Buffers
4.2. Growth Media
4.3. Expression of the Globular Heads ghA, ghB and ghC Representing gC1q
4.4. Preparation of Antigens for the Selection of Anti-Idiotypic scFv
4.5. Screening of Human scFv Phagemid Library Griffin.1
4.6. Selection of Monoclonal scFv
4.7. Induction of Soluble scFv and Affinity Purification
4.8. Enzyme-Linked Immunosorbent Assays
4.9. Spectroscopy Study of the Binding of Native C1q and ghA, ghB and ghC (Recombinant Fragments)
4.10. Sequence Analysis of Selected Clones
4.11. scFv A1 Homology Model Generation
4.12. C1q Modeling
4.13. Comparative Analysis of scFv A1 Homology Model and ghC1q Chains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Agnello, V.; Koffler, D.; Eisenberg, J.W.; Winchester, R.J.; Kundel, H.G. C1q precipitins in the sera of patients with systemic lupus erythematosus and other hypocomplementemic states: Characterization of high and low molecular weight types. J. Exp. Med. 1971, 134, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Daha, M.R. Pathogenic role of auto-antibodies against complement components in systemic lupus erythematosus. Lupus 2008, 17, 385–388. [Google Scholar] [CrossRef]
- Potlukova, E.; Kralikova, P. Complement Component C1q and Anti-C1q Antibodies in Theory and in Clinical Practice. Scand. J. Immunol. 2008, 67, 423–430. [Google Scholar] [CrossRef]
- Rahman, A.; Isenberg, D.A. Systemic Lupus Erythematosus. N. Engl. J. Med. 2008, 358, 929–939. [Google Scholar] [CrossRef] [Green Version]
- Rahman, A.; Hiepe, F. Anti-DNA antibodies-overview of assays and clinical correlations. Lupus 2002, 11, 770–773. [Google Scholar] [CrossRef]
- Golan, M.D.; Burger, R.; Loos, M. Conformational changes in C1q after binding to immune complexes: Detection of neoantigens with monoclonal antibodies. J. Immunol. 1982, 129, 445–447. [Google Scholar]
- Trouw, L.A.; Groeneveld, T.W.; Seelen, M.A.; Duijs, J.M.; Bajema, I.M.; Prins, F.A.; Kishore, U.; Salant, D.J.; Verbeek, J.S.; van Kooten, C.; et al. Anti-C1q autoantibodies deposit in glomeruli but are only pathogenic in combination with glomerular C1q-containing immune complexes. J. Clin. Investig. 2004, 114, 679–688. [Google Scholar] [CrossRef] [Green Version]
- Bigler, C.; Schaller, M.; Perahud, I.; Trendelenburg, M. Autoantibodies against complement C1q specifically target C1q bound on early apoptotic cells. Mol. Immunol. 2009, 183, 3512–3521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoyanova, V.; Bogoeva, V.; Petrova, L.; Tchorbadjieva, M.; Petrova, S.; Georgieva, V.; Georgiev, G.; Deliyska, B.; Vasilev, V.; Tsacheva, I. Autoantigenicity of human C1q is associated with increased hydrophobicity due to conformational transitions in the globular heads. Mol. Bio. Syst. 2015, 11, 1370–1377. [Google Scholar] [CrossRef] [PubMed]
- Eggleton, P.; Ukoumunne, O.C.; Cottrell, I.; Khan, A.; Maqsood, S.; Thornes, J.; Perry, E.; Isenberg, D. Autoantibodies against C1q as a diagnostic measure of lupus nephritis: Systematic review and meta-analysis. J. Clin. Cell Immunol. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Seshan, S.V.; Jennette, J.C. Renal disease in systemic lupus erythematosus with emphasis on classification of lupus glomerulonephritis: Advances and implications. Arch. Pathol. Lab. Med. 2009, 133, 233–248. [Google Scholar] [CrossRef]
- Ortega, L.M.; Schultz, D.R.; Lenz, O.; Pardo, V.; Contreras, G.N. Review: Lupus nephritis: Pathologic features, epidemiology and a guide to therapeutic decisions. Lupus 2010, 19, 557–574. [Google Scholar] [CrossRef]
- Trendelenburg, M. Antibodies against C1q in patients with systemic lupus erythematosus. Springer Semin. Immunopathol. 2005, 27, 276–285. [Google Scholar] [CrossRef]
- Schaller, M.; Bigler, C.; Danner, D.; Ditzel, H.J.; Trendelenburg, M. Autoantibodies against C1q in systemic lupus erythematosus are antigen-driven. J. Immunol. 2009, 183, 8225–8231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoyanova, V.; Tchorbadjieva, M.; Deliyska, B.; Vasilev, V.; Tsacheva, I. Biochemical analysis of the epitope specificities of anti-C1q autoantibodies accompanying human Lupus Nephrites reveals them as a dynamic population in the course of the disease. Immunol. Lett. 2012, 148, 69–76. [Google Scholar] [CrossRef]
- Wener, M.H.; Uwatoko, S.; Mannik, M. Antibodies to the collagen-like region of C1q in sera of patients with autoimmune rheumatic diseases. Arth. Rheum. 1989, 32, 544–551. [Google Scholar] [CrossRef]
- Tsacheva, I.; Radanova, M.; Todorova, N.; Argirova, T.; Kishore, U. Detection of autoantibodies against the globular domain of human C1q in the sera of systemic lupus erythematosus patients. Mol. Immunol. 2007, 44, 2157–2161. [Google Scholar] [CrossRef]
- Tan, Y.; Zhou, W.; Yu, F.; Fang, Q.; Yang, H.-z.; Zhao, M.-h. Detection of anti-C1q antibodies and anti-C1q globular head domain antibodies in sera from Chinese patients with lupus nephritis. Mol. Immunol. 2009, 46, 2178–2182. [Google Scholar] [CrossRef]
- Vanhecke, D.; Roumenina, L.T.; Wan, H.; Osthoff, M.; Schaller, M.; Trendelenburg, M. Identification of a major linear C1q epitope allows detection of systemic lupus erythematosus anti-C1q antibodies by a specific peptide-based enzyme-linked immunosorbent assay. Arth. Rheum. 2012, 64, 3706–3714. [Google Scholar] [CrossRef] [PubMed]
- Kishore, U.; Ghai, R.; Greenhough, T.J.; Shrive, A.K.; Bonifati, D.M.; Gadjeva, M.G.; Waters, P.; Kojouharova, M.S.; Chakraboty, T.; Agrawal, A. Structural and functional anatomy of the globular domain of complement protein C1q. Immunol. Lett. 2004, 95, 113–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marks, J.D.; Hoogenboom, H.R.; Bonnert, T.P.; McCafferty, J.; Griffiths, A.D.; Winter, G. By-passing immunization: Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991, 222, 581–597. [Google Scholar] [CrossRef]
- Retter, I.; Althaus, H.H.; Münch, R.; Müller, W. VBASE2, an integrative V gene database. Nucleic Acids Res. 2005, 33 (Suppl. S1), D671–D674. [Google Scholar] [CrossRef] [Green Version]
- Gaboriaud, C.; Juanhuix, J.; Gruez, A.; Lacroix, M.; Darnault, C.; Pignol, D.; Verger, D.; Fontecilla-Camps, J.C.; Arlaud, G.J. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J. Biol. Chem. 2003, 278, 46974–46982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franchin, G.; Son, M.; Kim, S.J.; Ben-Zvi, I.; Zhang, J.; Diamond, B. Anti-DNA antibodies cross-react with C1q. J. Autoimmun. 2013, 44, 34–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahler, M.; van Schaarenburg, R.A.; Trouw, L.A. Anti-C1q autoantibodies, novel tests and clinical consequences. Front. Immunol. 2013, 4, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moroni, G.; Trendelenburg, M.; Del Papa, N.; Quaglini, S.; Raschi, E.; Panzeri, P.; Testoni, C.; Tincani, A.; Banfi, G.; Balestrieri, G.; et al. Anti-C1q antibodies may help in diagnosing a renal flare in lupus nephritis. Am. J. Kidney Dis. 2001, 37, 490–498. [Google Scholar] [CrossRef]
- Trendelenburg, M.; Lopez-Trascasa, M.; Potlukova, E.; Moll, S.; Regenass, S.; Fremeaux-Bacchi, V.; Martinez-Ara, J.; Jancova, E.; Picazo, M.L.; Honsova, E.; et al. High prevalence of anti-C1q antibodies in biopsy-proven active lupus nephritis. Nephrol. Dial. Transpl. 2006, 21, 3115–3121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, C.G.; Lima, I.; Barbosa, L.; Athanazio, D.; Reis, E.; Reis, M.; Burlingame, R.W.; Santiago, M.B. Anti-C1q Antibodies: Association with Nephritis and Disease Activity in Systemic Lupus Erythematosus. J. Clin. Lab. Anal. 2009, 23, 19–23. [Google Scholar] [CrossRef]
- Akhter, E.; Burlingame, R.; Seaman, A.; Magder, L.; Petri, M. Anti-C1q antibodies have higher correlation with flares of lupus nephritis than other serum markers. Lupus 2011, 20, 1267–1274. [Google Scholar] [CrossRef]
- Yin, Y.; Wu, X.; Shan, G.; Zhang, X. Diagnostic value of serum anti-C1q antibodies in patients with lupus nephritis: A meta-analysis. Lupus 2012, 21, 1088–1097. [Google Scholar] [CrossRef] [PubMed]
- Wener, M.H. Autoantibodies to C1q. In Autoantibodies; Elsevier: Oxford, UK, 2014; pp. 707–715. [Google Scholar]
- Stoyanova, V.; Petrova, S.; Tchorbadjieva, M.; Deliyska, B.; Vasilev, V.; Tsacheva, I. New insight into the autoimmunogenicity of the complement protein C1q. Mol. Immunol. 2011, 48, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Rosen, A.; Casciola-Rosen, L. Autoantigens as partners in initiation and propagation of autoimmune rheumatic diseases. Ann. Rev. Immunol. 2016, 34, 395–420. [Google Scholar] [CrossRef]
- Arbuckle, M.R.; McClain, M.T.; Rubertone, M.V.; Scofield, R.H.; Dennis, G.J.; James, J.A.; Harley, J.B. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N. Engl. J. Med. 2003, 349, 1526–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, C.; Kokkonen, H.; Johansson, M.; Hallmans, G.; Wadell, G.; Rantapää-Dahlqvist, S. Autoantibodies predate the onset of systemic lupus erythematosus in northern Sweden. Arthritis Res. Ther. 2011, 13, R30. [Google Scholar] [CrossRef] [Green Version]
- Csorba, K.; Schirmbeck, L.A.; Tuncer, E.; Ribi, C.; Roux-Lombard, P.; Chizzolini, C.; Huynh-Do, U.; Vanhecke, D.; Trendelenburg, M. Anti-C1q antibodies as occurring in systemic lupus erythematosus could be induced by an epstein-barr virus-derived antigenic site. Front. Immunol. 2019, 10, 2619. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.Y.; Chiang, C.Y.; Pan, Y.R.; Tse, K.P.; Chang, Y.S.; Chang, H.Y. Modulation of Epstein-Barr virus latent membrane protein 1 activity by intrabodies. Intervirology 2007, 50, 254–263. [Google Scholar] [CrossRef]
- Botto, M.; Walport, M.J. C1q, autoimmunity and apoptosis. Immunobiology 2002, 205, 395–406. [Google Scholar] [CrossRef]
- Navratil, J.S.; Watkins, S.C.; Wisnieski, J.J.; Ahearn, J.M. The globular heads of C1q specifically recognize surface blebs of apoptotic vascular endothelial cells. J. Immunol. 2001, 166, 3231–3239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nauta, A.J.; Trouw, L.A.; Daha, M.R.; Tijsma, O.; Nieuwland, R.; Schwaeble, W.J.; Gingras, A.R.; Mantovani, A.; Hack, E.C.; Roos, A. Direct binding of C1q to apoptotic cells and cell blebs induces complement activation. Eur. J. Immunol. 2002, 32, 1726–1736. [Google Scholar] [CrossRef]
- Zwart, B.; Ciurana, C.; Rensink IManoe, R.; Hack, C.E.; Aarden, L.A. Complement activation by apoptotic cells occurs predominantly via IgM and is limited to late apoptotic (secondary necrotic) cells. Autoimmunity 2004, 37, 95–102. [Google Scholar] [CrossRef]
- Quartier, P.; Potte, P.K.; Ehrenstein, M.R.; Walport, M.J.; Botto, M. Predominant role of IgM-dependent activation of the classical pathway in the clearance of dying cells by murine bone marrow-derived macrophages in vitro. Eur. J. Immunol. 2005, 35, 252–260. [Google Scholar] [CrossRef]
- Ogden, C.A.; Kowalewski, R.; Peng, Y.; Montenegro, V.; Elkon, K.B. IgM is required for efficient complement mediated phagocytosis of apoptotic cells in vivo. Autoimmunity 2005, 38, 259–264. [Google Scholar] [CrossRef]
- Chen, Y.; Park, Y.B.; Patel, E.; Silverman, G.J. IgM antibodies to apoptosis-associated determinants recruit C1q and enhance dendritic cell phagocytosis of apoptotic cells. J. Immunol. 2009, 182, 6031–6043. [Google Scholar] [CrossRef] [Green Version]
- Kojouharova, M.S.; Tsacheva, I.G.; Tchorbadjieva, M.I.; Reid, K.B.; Kishore, U. Localization of ligand-binding sites on human C1q globular head region using recombinant globular head fragments and single-chain antibodies. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2003, 1652, 64–74. [Google Scholar] [CrossRef]
- Botto, M.; Dell’Agnola, C.; Bygrave, A.E.; Thompson, E.M.; Cook, H.T.; Petry, F.; Loos, M.; Pandolfi, P.P.; Walport, M.J. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet. 1998, 19, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Fossati-Jimack, L.; Cortes-Hernandez, J.; Norsworthy, P.J.; Walport, M.J.; Cook, H.T.; Botto, M. C1q deficiency promotes the production of transgenic-derived IgM and IgG3 autoantibodies in anti-DNA knock-in transgenic mice. Mol. Immunol. 2008, 45, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Yang, X.W.; Song, Y.; Yu, F.; Zhao, M.H. Anti-C1q autoantibodies from active lupus nephritis patients could inhibit the clearance of apoptotic cells and complement classical pathway activation mediated by C1q in vitro. Immunobiology 2014, 219, 980–989. [Google Scholar] [CrossRef]
- Nikolova, G.; Georgieva, Y.; Atanasova, A.; Radulova, G.; Kapogianni, A.; Tsacheva, I. Autoinduction as Means forOptimization of the Heterologous Expression of Recombinant Single-Chain Fv (scFv) Antibodies. Mol. Biotechnol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Liners, F.; Helbert, W.; Van Cutsem, P. Production and characterization of a phage-display recombinant antibody against carrageenans: Evidence for the recognition of a secondary structure of carrageenan chains present in red algae tissues. Glycobiology 2005, 15, 849–860. [Google Scholar] [CrossRef] [Green Version]
- MOE (Molecular Operating Environment) Version 20170530; Chemical Computing Group ULC: Montreal, QC, Canada, 2018.
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- GROMACS version 2016.5. In Software Versatile Package to Perform Molecular Dynamics; free software; University of Groningen: Groningen, Netherlands; Royal Institute of Technology: Stockholm, Sweden; Uppsala University: Netherlands, Sweden, 1991.
- Emsley, P. COOT Version 0.8.8 Software, Crystallographic Object-Oriented Toolkit; Free Software; MRC Laboratory of Molecular Biology: Cambridge, UK, 2004. [Google Scholar]


| Sera Group/ Anti-C1q Fraction | Sera, Positive for Autoantibodies against: |
|---|---|
| A | intact C1q |
| B | intact C1q and CLR |
| C | intact C1q and CLR and ghC |
| D | ghC |
| E | CLR and/or ghA and/or ghB and ghC |
| F | ghA and/or ghB and ghC |
| Anti-C1q Fraction | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | + | − | + | − | + | − | − | + | + | − | + | |
| B | − | + | − | − | − | + | − | − | + | − | − | − |
| C | + | − | − | − | + | − | − | − | + | − | − | − |
| D | − | + | − | − | − | − | + | + | − | − | − | |
| E | + | − | − | − | − | +/− | − | − | +/− | − | − | |
| F | − | − | +/− | − | − | + | − | − | + | − | − |
| Antigen for Screening | C myc | His-Tag | Human IgGfrom Healthy Donors | Potentially Anti-Idiotypic scFv | |
|---|---|---|---|---|---|
| Anti-C1q | A1 | +/− | + | − | * |
| A3 | +/− | + | − | * | |
| A5 | +/− | + | − | * | |
| A8 | +/− | + | − | * | |
| A9 | +/− | + | − | * | |
| Anti-C1q and Anti-CLR | B2 | +/− | + | − | * |
| B6 | +/− | + | − | * | |
| B9 | +/− | + | +/− | ||
| A12 | +/− | + | − | * | |
| Anti-C1q and Anti-CLR and Anti-ghC | C1 | +/− | + | +/− | |
| C5 | +/− | + | − | * | |
| C9 | +/− | + | − | * | |
| Anti-ghC | D2 | +/− | + | +/− | |
| D7 | +/− | + | +/− | ||
| D8 | +/− | + | − | * | |
| Anti-CLR and/or Anti-ghA and/or Anti-ghB and Anti-ghC | E1 | +/− | + | − | * |
| E6 | +/− | + | +/− | ||
| E9 | +/− | + | +/− | ||
| Anti-ghA and/or Anti-ghB and Anti-ghC | F3 | +/− | +/− | ||
| F6 | +/− | + | − | * | |
| F9 | +/− | + | − | * | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Todorova, N.; Rangelov, M.; Bogoeva, V.; Stoyanova, V.; Yordanova, A.; Nikolova, G.; Georgiev, H.; Dimitrova, D.; Mohedin, S.; Stoyanova, K.; et al. Anti-Idiotype scFv Localizes an Autoepitope in the Globular Domain of C1q. Int. J. Mol. Sci. 2021, 22, 8288. https://doi.org/10.3390/ijms22158288
Todorova N, Rangelov M, Bogoeva V, Stoyanova V, Yordanova A, Nikolova G, Georgiev H, Dimitrova D, Mohedin S, Stoyanova K, et al. Anti-Idiotype scFv Localizes an Autoepitope in the Globular Domain of C1q. International Journal of Molecular Sciences. 2021; 22(15):8288. https://doi.org/10.3390/ijms22158288
Chicago/Turabian StyleTodorova, Nadezhda, Miroslav Rangelov, Vanya Bogoeva, Vishnya Stoyanova, Anna Yordanova, Ginka Nikolova, Hristo Georgiev, Daniela Dimitrova, Safa Mohedin, Katerina Stoyanova, and et al. 2021. "Anti-Idiotype scFv Localizes an Autoepitope in the Globular Domain of C1q" International Journal of Molecular Sciences 22, no. 15: 8288. https://doi.org/10.3390/ijms22158288
APA StyleTodorova, N., Rangelov, M., Bogoeva, V., Stoyanova, V., Yordanova, A., Nikolova, G., Georgiev, H., Dimitrova, D., Mohedin, S., Stoyanova, K., & Tsacheva, I. (2021). Anti-Idiotype scFv Localizes an Autoepitope in the Globular Domain of C1q. International Journal of Molecular Sciences, 22(15), 8288. https://doi.org/10.3390/ijms22158288







