Immunonutritional Protease Inhibitors from T. durum and A. sativa Display Metabolic Similarities When Assayed on Human Macrophage-like Cells
Abstract
:1. Introduction
2. Results
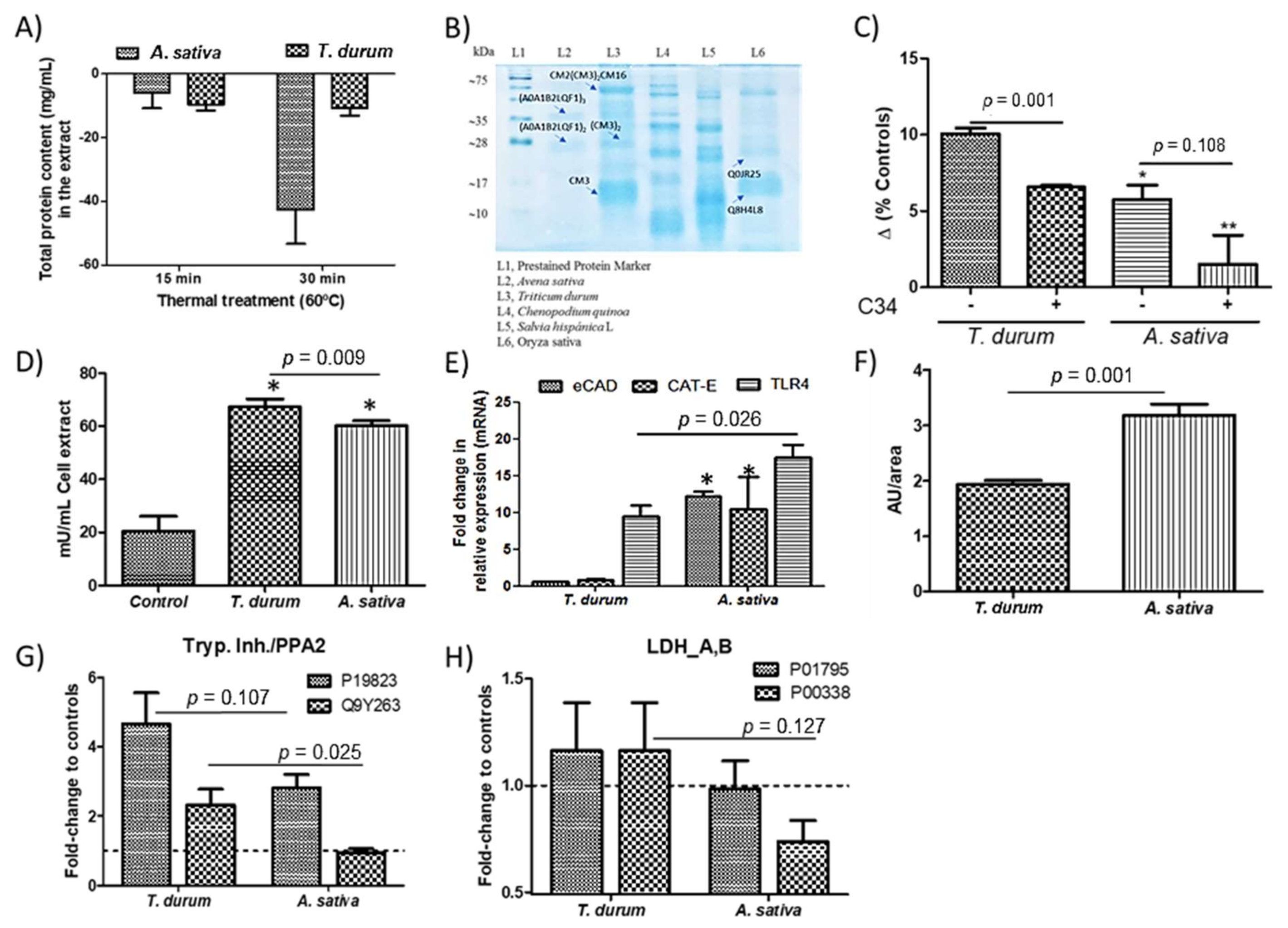
2.1. Obtention of PIs-Enriched Extracts
2.2. Metabolic Changes in Macrophages
2.3. Elicit of Inflammatory Response(s)
3. Discussion
4. Materials and Methods
4.1. Isolation of Protease Inhibitors
4.2. In Vitro Digestion
4.3. Electrophoresis
4.4. HPLC-Reverse Phase-Diode Array/Electrospray Ionization (HPLC-RP-DAD/ESI) Analyses of the Bioactive Fraction
4.5. Cell Cultures
4.6. Macrophage Cell Enzyme Activities
4.7. Reverse Transcriptase-qPCR Analyses
4.8. Protein Expression of the Aryl Hydrocarbon Receptor (AhR)
4.9. Cell Mitochondrial Stress Test Assay
4.10. Analysis of the Proteome Changes
4.11. Cytokine Production Analysis
4.12. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Junker, Y.; Zeissig, S.; Kim, S.-J.; Barisani, D.; Wieser, H.; Leffler, D.A.; Zevallos, V.; Libermann, T.A.; Dillon, S.; Freitag, T.L.; et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J. Exp. Med. 2012, 209, 2395–2408. [Google Scholar] [CrossRef]
- Zevallos, V.F.; Raker, V.; Tenzer, S.; Jimenez-Calvente, C.; Ashfaq-Khan, M.; Rüssel, N.; Pickert, G.; Schild, H.; Steinbrink, K.; Schuppan, D. Nutritional Wheat Amylase-Trypsin Inhibitors Promote Intestinal Inflammation via Activation of Myeloid Cells. Gastroenterology 2017, 152, 1100–1113.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaliszewska, A.; Martinez, V.; Laparra, J.M. Proinflammatory responses driven by non-gluten factors are masked when they appear associated to gliadins. Food Chem. Toxicol. 2016, 95, 89–95. [Google Scholar] [CrossRef]
- Laparra, J.M.; Haros, C.M. Plant seed protease inhibitors differentially affect innate immunity in a tumor microenvironment to control hepatocarcinoma. Food Funct. 2019, 10, 4210–4219. [Google Scholar] [CrossRef]
- Srdić, M.; Ovčina, I.; Fotschki, B.; Haros, C.M.; Laparra, J.M. C. quinoa and S. hispanica L. Seeds Provide Immunonutritional Agonists to Selectively Polarize Macrophages. Cells 2020, 9, 593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savelkoul, F.H.; van der Poel, A.F.; Tamminga, S. The presence and inactivation of trypsin inhibitors, tannins, lectins and amylase inhibitors in legume seeds during germination. A review. Plant Foods Hum. Nutr. 1992, 42, 71–85. [Google Scholar] [CrossRef]
- Liener, I.E. Implications of antinutritional components in soybean foods. Crit. Rev. Food Sci. Nutr. 1994, 34, 31–67. [Google Scholar] [CrossRef] [PubMed]
- Ramanarayan, K.; Swamy, G.S. Triacontanol negatively modulates the jasmonic acid-stimulated proteinase inhibitors in tomato (Lycopersicon esculentum). J. Plant Physiol. 2004, 161, 489–492. [Google Scholar] [CrossRef]
- Li, Q.; Huang, L.; Luo, Z.; Tamer, T.M. Stability of trypsin inhibitor isolated from potato fruit juice against pH and heating treatment and in vitro gastrointestinal digestion. Food Chem. 2020, 328, 127152. [Google Scholar] [CrossRef]
- Ashfaq-Khan, M.; Aslam, M.; Qureshi, M.A.; Senkowski, M.S.; Yen-Weng, S.; Strand, S.; Kim, Y.O.; Pickert, G.; Schattenberg, J.M.; Schuppan, D. Dietary wheat amylase trypsin inhibitors promote features of murine non-alcoholic fatty liver disease. Sci. Rep. 2019, 9, 17463. [Google Scholar] [CrossRef]
- Van den Bossche, J.; Baardman, J.; Otto, N.A.; van der Velden, S.; Neele, A.E.; van den Berg, S.M.; Luque-Martin, R.; Chen, H.-J.; Boshuizen, M.C.S.; Ahmed, M.; et al. Mitochondrial Dysfunction Prevents Repolarization of Inflammatory Macrophages. Cell Rep. 2016, 17, 684–696. [Google Scholar] [CrossRef] [Green Version]
- Alisi, A.; Carpino, G.; Oliveira, F.L.; Panera, N.; Nobili, V.; Gaudio, E. The Role of Tissue Macrophage-Mediated Inflammation on NAFLD Pathogenesis and Its Clinical Implications. Mediators Inflamm. 2017, 2017, 8162421. [Google Scholar] [CrossRef]
- Laparra, J.; Fotschki, B.; Haros, C. Immunonutritional consequences of different serine-type protease inhibitors in a C57BL/6 hepatocarcinoma model. Oncotarget 2019, 10, 760–772. [Google Scholar] [CrossRef] [Green Version]
- Capocchi, A.; Muccilli, V.; Cunsolo, V.; Saletti, R.; Foti, S.; Fontanini, D. A heterotetrameric alpha-amylase inhibitor from emmer (Triticum dicoccon Schrank) seeds. Phytochemistry 2013, 88, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Serena, G.; Huynh, D.; Lima, R.S.; Vise, L.M.; Freire, R.; Ingano, L.; Leonard, M.M.; Senger, S.; Fasano, A. Intestinal Epithelium Modulates Macrophage Response to Gliadin in Celiac Disease. Front. Nutr. 2019, 6, 167. [Google Scholar] [CrossRef] [Green Version]
- Salazar, F.; Awuah, D.; Negm, O.H.; Shakib, F.; Ghaemmaghami, A.M. The role of indoleamine 2,3-dioxygenase-aryl hydrocarbon receptor pathway in the TLR4-induced tolerogenic phenotype in human DCs. Sci. Rep. 2017, 7, 43337. [Google Scholar] [CrossRef]
- Van den Bossche, J.; Laoui, D.; Naessens, T.; Smits, H.H.; Hokke, C.H.; Stijlemans, B.; Grooten, J.; De Baetselier, P.; Van Ginderachter, J.A. E-cadherin expression in macrophages dampens their inflammatory responsiveness in vitro, but does not modulate M2-regulated pathologies in vivo. Sci. Rep. 2015, 5, 12599. [Google Scholar] [CrossRef] [Green Version]
- Bertsch, T.; Aufenanger, J. Phospholipase A2 in inflammatory bowel disease. Gut 1997, 41, 859–860. [Google Scholar] [CrossRef]
- Rodríguez-Prados, J.-C.; Través, P.G.; Cuenca, J.; Rico, D.; Aragonés, J.; Martín-Sanz, P.; Cascante, M.; Boscá, L. Substrate fate in activated macrophages: A comparison between innate, classic, and alternative activation. J. Immunol. 2010, 185, 605–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, S.; Usui-Kawanishi, F.; Karasawa, T.; Kimura, H.; Kamata, R.; Komada, T.; Inoue, Y.; Mise, N.; Kasahara, T.; Takahashi, M. Glucose regulates hypoxia-induced NLRP3 inflammasome activation in macrophages. J. Cell. Physiol. 2020, 235, 7554–7566. [Google Scholar] [CrossRef] [PubMed]
- Lampropoulou, V.; Sergushichev, A.; Bambouskova, M.; Nair, S.; Vincent, E.E.; Loginicheva, E.; Cervantes-Barragan, L.; Ma, X.; Huang, S.C.-C.; Griss, T.; et al. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab. 2016, 24, 158–166. [Google Scholar] [CrossRef] [Green Version]
- Maglio, M.; Mazzarella, G.; Barone, M.V.; Gianfrani, C.; Pogna, N.; Gazza, L.; Stefanile, R.; Camarca, A.; Colicchio, B.; Nanayakkara, M.; et al. Immunogenicity of two oat varieties, in relation to their safety for celiac patients. Scand. J. Gastroenterol. 2011, 46, 1194–1205. [Google Scholar] [CrossRef]
- Comino, I.; Real, A.; de Lorenzo, L.; Cornell, H.; López-Casado, M.Á.; Barro, F.; Lorite, P.; Torres, M.I.; Cebolla, A.; Sousa, C. Diversity in oat potential immunogenicity: Basis for the selection of oat varieties with no toxicity in coeliac disease. Gut 2011, 60, 915–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.-T.; Gao, F.; Gu, K.; Chen, D.-K. The Role of Monocytes and Macrophages in Autoimmune Diseases: A Comprehensive Review. Front. Immunol. 2019, 10, 1140. [Google Scholar] [CrossRef] [Green Version]
- Diosdado, B.; Wijmenga, C. Molecular mechanisms of the adaptive, innate and regulatory immune responses in the intestinal mucosa of celiac disease patients. Expert Rev. Mol. Diagn. 2005, 5, 681–700. [Google Scholar] [CrossRef]
- Maiuri, L.; Ciacci, C.; Ricciardelli, I.; Vacca, L.; Raia, V.; Auricchio, S.; Picard, J.; Osman, M.; Quaratino, S.; Londei, M. Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet 2003, 362, 30–37. [Google Scholar] [CrossRef]
- Van den Bossche, J.; Malissen, B.; Mantovani, A.; De Baetselier, P.; Van Ginderachter, J.A. Regulation and function of the E-cadherin/catenin complex in cells of the monocyte-macrophage lineage and DCs. Blood 2012, 119, 1623–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Leeuwen, M.; Costes, L.; van Berkel, L.; Simons-Oosterhuis, Y.; du Pré, F.; Kozijn, A.; Raatgeep, R.; Lindenbergh-Kortleve, D.; van Rooijen, N.; Koning, F.; et al. Macrophage-mediated gliadin degradation and concomitant IL-27 production drive IL-10- and IFN-γ 3-secreting Tr1-like-cell differentiation in a murine model for gluten tolerance. Mucosal Immunol. 2017, 10, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Laparra, J.M.; Sanz, Y. Bifidobacteria inhibit the inflammatory response induced by gliadins in intestinal epithelial cells via modifications of toxic peptide generation during digestion. J. Cell. Biochem. 2010, 109, 801–807. [Google Scholar] [CrossRef] [Green Version]
- Mao, R.; Wu, L.; Zhu, N.; Liu, X.; Liu, R.; Li, Y. Naked Oat (Avena nuda L.) Oligopeptides:Immunomodulatory Effects on Innate and AdaptiveImmunity in Mice via Cytokine Secretion, AntibodyProduction, and Th Cells Stimulation. Nutrients 2019, 11, 927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, L.L.Y.; Cheung, B.K.W.; Li, J.C.B.; Lau, A.S.Y. A role for STAT3 and cathepsin S in IL-10 down-regulation of IFN-gamma-induced MHC class II molecule on primary human blood macrophages. J. Leukoc. Biol. 2010, 88, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Peters, T.J.; Heath, J.R.; Wansbrough-Jones, M.H.; Foe, W.F. Enzyme activities and properties of lysosomes and brush borders in jejunal biopsies from control subjects and patients with coeliac disease. Clin. Sci. Mol. Med. 1975, 48, 259–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laparra, J.M.; Alfonso-García, A.; Alegría, A.; Barberá, R.; Cilla, A. 7keto-stigmasterol and 7keto-cholesterol induce differential proteome changes to intestinal epitelial (Caco-2) cells. Food Chem. Toxicol. 2015, 84, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Moreno, M.-L.; Escobar, J.; Izquierdo-Álvarez, A.; Gil, A.; Pérez, S.; Pereda, J.; Zapico, I.; Vento, M.; Sabater, L.; Marina, A.; et al. Disulfide stress: A novel type of oxidative stress in acute pancreatitis. Free Radic. Biol. Med. 2014, 70, 265–277. [Google Scholar] [CrossRef]
- Alonso, R.; Pisa, D.; Marina, A.I.; Morato, E.; Rábano, A.; Rodal, I.; Carrasco, L. Evidence for fungal infection in cerebrospinal fluid and brain tissue from patients with amyotrophic lateral sclerosis. Int. J. Biol. Sci. 2015, 11, 546–558. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fotschki, B.; Garcia Tejedor, A.; Nieto Fuentes, J.A.; Laparra Llopis, J.M. Immunonutritional Protease Inhibitors from T. durum and A. sativa Display Metabolic Similarities When Assayed on Human Macrophage-like Cells. Int. J. Mol. Sci. 2021, 22, 8307. https://doi.org/10.3390/ijms22158307
Fotschki B, Garcia Tejedor A, Nieto Fuentes JA, Laparra Llopis JM. Immunonutritional Protease Inhibitors from T. durum and A. sativa Display Metabolic Similarities When Assayed on Human Macrophage-like Cells. International Journal of Molecular Sciences. 2021; 22(15):8307. https://doi.org/10.3390/ijms22158307
Chicago/Turabian StyleFotschki, Bartosz, Aurora Garcia Tejedor, Juan Antonio Nieto Fuentes, and Jose Moisés Laparra Llopis. 2021. "Immunonutritional Protease Inhibitors from T. durum and A. sativa Display Metabolic Similarities When Assayed on Human Macrophage-like Cells" International Journal of Molecular Sciences 22, no. 15: 8307. https://doi.org/10.3390/ijms22158307
APA StyleFotschki, B., Garcia Tejedor, A., Nieto Fuentes, J. A., & Laparra Llopis, J. M. (2021). Immunonutritional Protease Inhibitors from T. durum and A. sativa Display Metabolic Similarities When Assayed on Human Macrophage-like Cells. International Journal of Molecular Sciences, 22(15), 8307. https://doi.org/10.3390/ijms22158307






