Development of a Novel Nanoarchitecture of the Robust Photosystem I from a Volcanic Microalga Cyanidioschyzon merolae on Single Layer Graphene for Improved Photocurrent Generation
Abstract
:1. Introduction
2. Results and Discussion
2.1. Genetic Engineering of the C. merolae His6-Tagged PsaD-PSI Biophotocatalyst
2.2. Biochemical Characterization of the His6-PsaD-PSI Complex
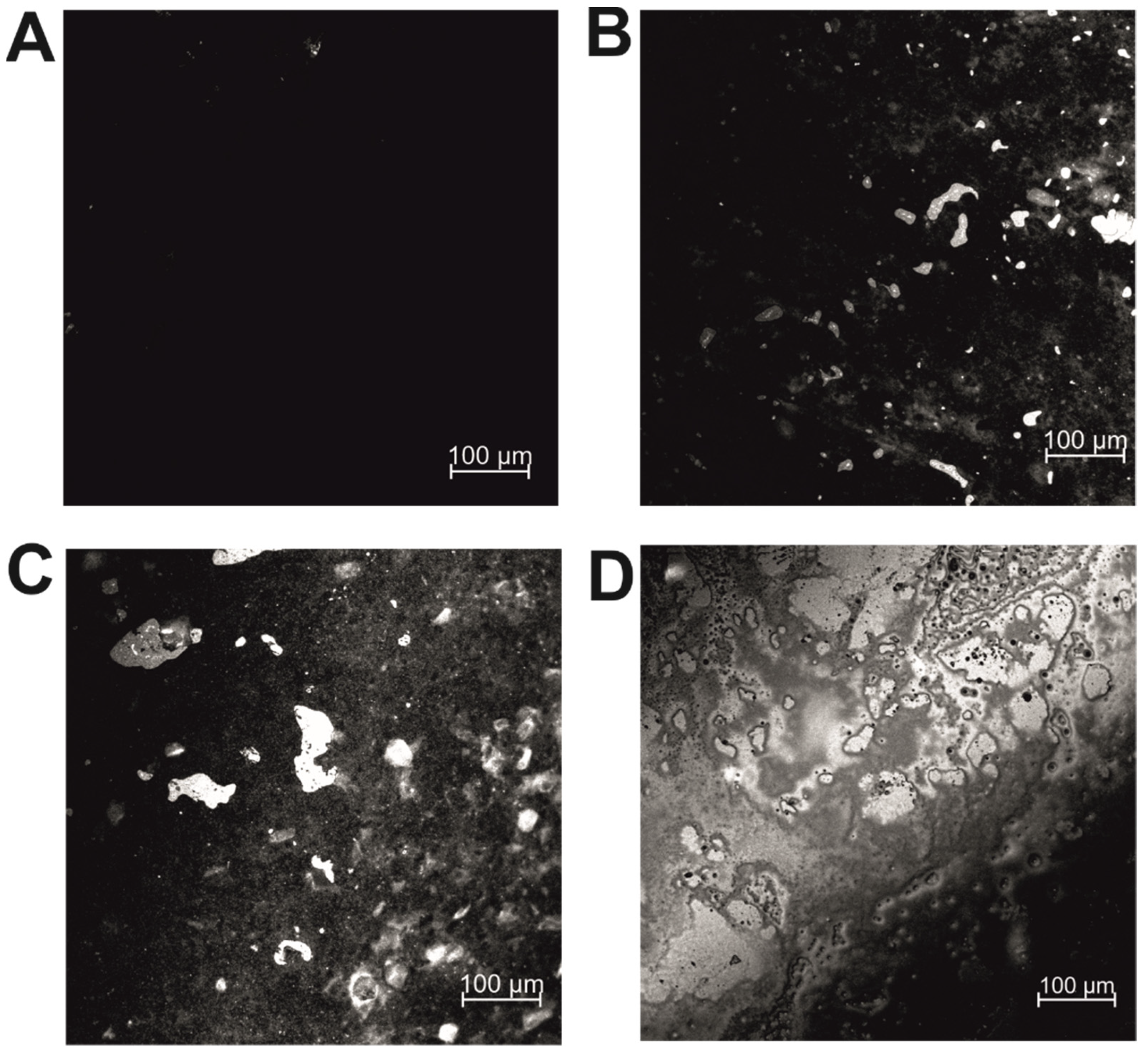
2.3. Confocal Microscopy
2.4. Analysis of the Bioelectrode Surface Morphology
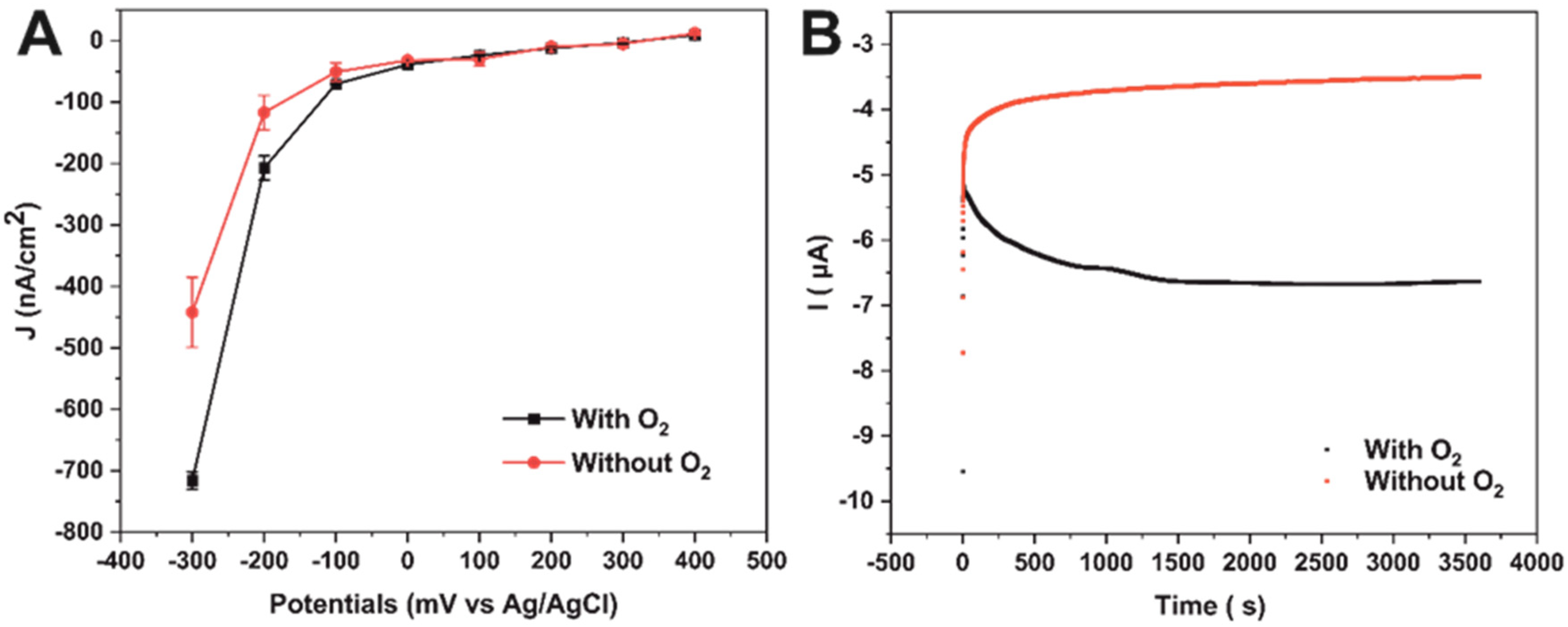
2.5. Photoelectrochemical Characterization of the FTO/SLG/pyr-NTA-M2+/His6-PsaD-PSI Biophotoelectrodes
3. Conclusions
4. Materials and Methods
4.1. Generation of the C. merolae Strain Containing His6-PsaD-PSI Biophotocatalyst
4.2. Culturing of C. merolae Cells and Isolation of Thylakoids Membranes
4.3. Purification of His6-Tagged Proteins and Their Biochemical Characterization
4.4. LC-Mass Spectrometry Analysis
4.5. Preparation of Single Layer Graphene
4.6. FTO/SLG Electrode Biofunctionalization
4.7. Confocal Microscopy Analysis
4.8. Scanning Electron Microscopy Analysis
4.9. Immunoblotting
4.10. Electrochemical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ort, D.R.; Good, N.E. Textbooks ignore photosystem II-dependent ATP formation: Is the Z scheme to blame? Trends Biochem. Sci. 1988, 13, 467–469. [Google Scholar] [CrossRef]
- Barber, J. Engine of Life and Big Bang of Evolution: A Personal Perspective. Photosynth. Res. 2004, 80, 137–155. [Google Scholar] [CrossRef]
- Olah, G.A. Beyond Oil and Gas: The Methanol Economy. Angew. Chem. Int. Ed. 2005, 44, 2636–2639. [Google Scholar] [CrossRef]
- Solar FAQs.PDF. Available online: https://old-www.sandia.gov/~jytsao/Solar%20FAQs.pdf (accessed on 9 March 2021).
- Berry, E.A.; Guergova-Kuras, M.; Huang, L.-S.; Crofts, A.R. Structure and Function of Cytochrome bc Complexes. Annu. Rev. Biochem. 2000, 69, 1005–1075. [Google Scholar] [CrossRef] [Green Version]
- Hervás, M.; Navarro, J.A.; de la Rosa, M. Electron Transfer between Membrane Complexes and Soluble Proteins in Photosynthesis. Acc. Chem. Res. 2003, 36, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Kargul, J.; Barber, J. Structure and Function of Photosynthetic Reaction Centres. In Molecular Solar Fuels; Royal Society of Chemistry: London, UK, 2011; pp. 107–142. [Google Scholar] [CrossRef]
- Kargul, J.; Kiliszek, M. Artificial Photosynthesis. In Bioelectrochemical Interface Engineering; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019; pp. 271–309. [Google Scholar] [CrossRef]
- Olmos, J.D.J.; Kargul, J. A quest for the artificial leaf. Int. J. Biochem. Cell Biol. 2015, 66, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Brettel, K.; Leibl, W. Electron transfer in photosystem I. Biochim. Biophys. Acta BBA Bioenerg. 2001, 1507, 100–114. [Google Scholar] [CrossRef] [Green Version]
- Teodor, A.; Bruce, B.D. Putting Photosystem I to Work: Truly Green Energy. Trends Biotechnol. 2020, 38, 1329–1342. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.; Bruce, B.D. Growing green electricity: Progress and strategies for use of Photosystem I for sustainable photovoltaic energy conversion. Biochim. Biophys. Acta BBA Bioenerg. 2014, 1837, 1553–1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Kalathil, S.; Reisner, E. Semi-biological approaches to solar-to-chemical conversion. Chem. Soc. Rev. 2020, 49, 4926–4952. [Google Scholar] [CrossRef]
- Witt, H.T. Primary reactions of oxygenic photosynthesis. Ber. Bunsenges. Phys. Chem. 1996, 100, 1923–1942. [Google Scholar] [CrossRef]
- Nelson, N.; Yocum, C.F. Structure and function of photosystems i and ii. Annu. Rev. Plant Biol. 2006, 57, 521–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kargul, J.; Olmos, J.D.J.; Krupnik, T. Structure and function of photosystem I and its application in biomimetic solar-to-fuel systems. J. Plant Physiol. 2012, 169, 1639–1653. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.G.; Slavov, C.; Luthra, R.; Redding, K.E.; Holzwarth, A.R. Independent initiation of primary electron transfer in the two branches of the photosystem I reaction center. Proc. Natl. Acad. Sci. USA 2010, 107, 4123–4128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisdat, F. Trends in the layer-by-layer assembly of redox proteins and enzymes in bioelectrochemistry. Curr. Opin. Electrochem. 2017, 5, 165–172. [Google Scholar] [CrossRef]
- Ocakoglu, K.; Krupnik, T.; Bosch, B.V.D.; Harputlu, E.; Gullo, M.P.; Olmos, J.J.; Yildirimcan, S.; Gupta, R.K.; Yakuphanoglu, F.; Barbieri, A.; et al. Photosystem I-based Biophotovoltaics on Nanostructured Hematite. Adv. Funct. Mater. 2014, 24, 7467–7477. [Google Scholar] [CrossRef]
- Ocampo, O.E.C.; Gordiichuk, P.; Catarci, S.; Gautier, D.A.; Herrmann, A.; Chiechi, R.C. Mechanism of Orientation-Dependent Asymmetric Charge Transport in Tunneling Junctions Comprising Photosystem I. J. Am. Chem. Soc. 2015, 137, 8419–8427. [Google Scholar] [CrossRef]
- Mikayama, T.; Iida, K.; Suemori, Y.; Dewa, T.; Miyashita, T.; Nango, M.; Gardiner, A.T.; Cogdell, R.J. The electronic behavior of a photosynthetic reaction center monitored by conductive atomic force microscopy. J. Nanosci. Nanotechnol. 2009, 9, 97–107. [Google Scholar] [CrossRef]
- Shen, Y.-X.; Saboe, P.O.; Sines, I.T.; Erbakan, M.; Kumar, M. Biomimetic membranes: A review. J. Membr. Sci. 2014, 454, 359–381. [Google Scholar] [CrossRef]
- Badura, A.; Guschin, D.; Kothe, T.; Kopczak, M.J.; Schuhmann, W.; Rögner, M. Photocurrent generation by photosystem 1 integrated in crosslinked redox hydrogels. Energy Environ. Sci. 2011, 4, 2435–2440. [Google Scholar] [CrossRef]
- Olmos, J.D.J.; Becquet, P.; Gront, D.; Sar, J.; Dąbrowski, A.; Gawlik, G.; Teodorczyk, M.; Pawlak, D.; Kargul, J. Biofunctionalisation of p-doped silicon with cytochrome c553minimises charge recombination and enhances photovoltaic performance of the all-solid-state photosystem I-based biophotoelectrode. RSC Adv. 2017, 7, 47854–47866. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, D.; Vaughn, M.; Khomami, B.; Bruce, B.D. Modulation of cyanobacterial photosystem I deposition properties on alkanethiolate Au substrate by various experimental conditions. Colloids Surf. B Biointerfaces 2011, 88, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, S.; Giera, W.; D’Haene, S.; van Grondelle, R.; Gibasiewicz, K. Comparison of excitation energy transfer in cyanobacterial photosystem I in solution and immobilized on conducting glass. Photosynth. Res. 2016, 132, 111–126. [Google Scholar] [CrossRef] [Green Version]
- Miyachi, M.; Yamanoi, Y.; Yonezawa, T.; Nishihara, H.; Iwai, M.; Konno, M.; Iwai, M.; Inoue, Y. Surface Immobilization of PSI Using Vitamin K1-Like Molecular Wires for Fabrication of a Bio-Photoelectrode. J. Nanosci. Nanotechnol. 2009, 9, 1722–1726. [Google Scholar] [CrossRef]
- Frolov, L.; Rosenwaks, Y.; Carmeli, C.; Carmeli, I. Fabrication of a Photoelectronic Device by Direct Chemical Binding of the Photosynthetic Reaction Center Protein to Metal Surfaces. Adv. Mater. 2005, 17, 2434–2437. [Google Scholar] [CrossRef]
- Kaniber, S.; Frolov, L.; Simmel, F.C.; Holleitner, A.W.; Carmeli, C.; Carmeli, I.; Caldas, M.; Studart, N. Covalently Binding the Photosystem I to Carbon Nanotubes; American Institute of Physics: College Park, MD, USA, 2010; pp. 133–134. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Salles, R.; Miyachi, M.; Yamanoi, Y.; Tomo, T.; Takahashi, H.; Nishihara, H.H. Photoelectric Conversion System Composed of Gene-Recombined Photosystem I and Platinum Nanoparticle Nanosheet. Langmuir 2020, 36, 6429–6435. [Google Scholar] [CrossRef]
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Zhang, Y.; Dubonos, S.V.; Grigorieva, I.V.; Firsov, A.A. Electric field effect in atomically thin carbon films. Science 2004, 306, 666–669. [Google Scholar] [CrossRef] [Green Version]
- What Is Graphene? Graphene Properties and Applications. Nanowerk. Available online: https://www.nanowerk.com/what_is_graphene.php (accessed on 9 April 2021).
- Geim, A.K.; Novoselov, K. The rise of graphene. Nat. Mater. 2007, 6, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.R.; Blake, P.; Grigorenko, A.N.; Novoselov, K.; Booth, T.; Stauber, T.; Peres, N.M.R.; Geim, A.K. Fine Structure Constant Defines Visual Transparency of Graphene. Science 2008, 320, 1308. [Google Scholar] [CrossRef] [Green Version]
- Gunther, D.; Leblanc, G.; Prasai, D.; Zhang, J.R.; Cliffel, D.E.; Bolotin, K.I.; Jennings, G.K. Photosystem I on Graphene as a Highly Transparent, Photoactive Electrode. Langmuir 2013, 29, 4177–4180. [Google Scholar] [CrossRef]
- LeBlanc, G.; Winter, K.M.; Crosby, W.B.; Jennings, G.K.; Cliffel, D.E. Integration of Photosystem I with Graphene Oxide for Photocurrent Enhancement. Adv. Energy Mater. 2014, 4, 1301953. [Google Scholar] [CrossRef]
- Feifel, S.C.; Stieger, K.R.; Lokstein, H.; Lux, H.; Lisdat, F. High photocurrent generation by photosystem I on artificial interfaces composed of π-system-modified graphene. J. Mater. Chem. A 2015, 3, 12188–12196. [Google Scholar] [CrossRef]
- Kiliszek, M.; Harputlu, E.; Szalkowski, M.; Kowalska, D.; Ünlü, G.; Haniewicz, P.; Abram, M.; Wiwatowski, K.; Niedziółka-Jönsson, J.; Mackowski, S.; et al. Orientation of photosystem I on graphene through cytochrome c553 leads to improvement in photocurrent generation. J. Mater. Chem. A 2018, 6, 18615–18626. [Google Scholar] [CrossRef]
- Kuroiwa, T. The primitive red algae Cyanidium caldarium and Cyanidioschyzon merolae as model system for investigating the dividing apparatus of mitochondria and plastids. BioEssays 1998, 20, 344–354. [Google Scholar] [CrossRef]
- Miyagishima, S.-Y.; Nishida, K.; Mori, T.; Matsuzaki, M.; Higashiyama, T.; Kuroiwa, H.; Kuroiwa, T. A Plant-Specific Dynamin-Related Protein Forms a Ring at the Chloroplast Division Site. Plant Cell 2003, 15, 655–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haniewicz, P.; Abram, M.; Nosek, L.; Kirkpatrick, J.; El-Mohsnawy, E.; Olmos, J.D.J.; Kouřil, R.; Kargul, J.M. Molecular Mechanisms of Photoadaptation of Photosystem I Supercomplex from an Evolutionary Cyanobacterial/Algal Intermediate. Plant Physiol. 2018, 176, 1433–1451. [Google Scholar] [CrossRef] [Green Version]
- Osella, S.; Kiliszek, M.; Harputlu, E.; Ünlü, G.; Ocakoglu, K.; Kargul, J.; Trzaskowski, B. Controlling the charge transfer flow at the graphene/pyrene–nitrilotriacetic acid interface. J. Mater. Chem. C 2018, 6, 5046–5054. [Google Scholar] [CrossRef] [Green Version]
- Szalkowski, M.; Harputlu, E.; Kiliszek, M.; Ünlü, G.; Maćkowski, S.; Ocakoglu, K.; Kargul, J.; Kowalska, D. Plasmonic enhancement of photocurrent generation in a photosystem I-based hybrid electrode. J. Mater. Chem. C 2020, 8, 5807–5814. [Google Scholar] [CrossRef]
- Jacquet, M.; Izzo, M.; Osella, S.; Kozdra, S.; Michałowski, P.P.; Gołowicz, D.; Kazimierczuk, K.; Gorzkowski, M.T.; Lewera, A.; Teodorczyk, M.; et al. Development of a universal conductive platform for anchoring photo- and electroactive proteins using organometallic terpyridine molecular wires. Nanoscale 2021, 13, 9773–9787. [Google Scholar] [CrossRef] [PubMed]
- Izzo, M.; Osella, S.; Jacquet, M.; Kiliszek, M.; Harputlu, E.; Starkowska, A.; Łasica, A.; Unlu, C.G.; Uśpieński, T.; Niewiadomski, P.; et al. Enhancement of direct electron transfer in graphene bioelectrodes containing novel cytochrome c variants with optimized heme orientation. Bioelectrochemistry 2021, 140, 107818. [Google Scholar] [CrossRef]
- Jacquet, M.; Kiliszek, M.; Osella, S.; Izzo, M.; Sar, J.; Harputlu, E.; Unlu, C.G.; Trzaskowski, B.; Ocakoglu, K.; Kargul, J. Molecular mechanism of direct electron transfer in the robust cytochrome-functionalised graphene nanosystem. RSC Adv. 2021, 11, 18860–18869. [Google Scholar] [CrossRef]
- Abram, M.; Białek, R.; Szewczyk, S.; Karolczak, J.; Gibasiewicz, K.; Kargul, J. Remodeling of excitation energy transfer in extremophilic red algal PSI-LHCI complex during light adaptation. Biochim. Biophys. Acta BBA Bioenerg. 2020, 1861, 148093. [Google Scholar] [CrossRef]
- Kubota, H.; Sakurai, I.; Katayama, K.; Mizusawa, N.; Ohashi, S.; Kobayashi, M.; Zhang, P.; Aro, E.-M.; Wada, H. Purification and characterization of photosystem I complex from Synechocystis sp. PCC 6803 by expressing histidine-tagged subunits. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1797, 98–105. [Google Scholar] [CrossRef] [Green Version]
- Gulis, G.; Narasimhulu, K.V.; Fox, L.N.; Redding, K.E. Purification of His6-tagged Photosystem I from Chlamydomonas reinhardtii. Photosynth. Res. 2008, 96, 51–60. [Google Scholar] [CrossRef]
- Jordan, P.; Fromme, P.; Witt, H.T.; Klukas, O.; Saenger, W.; Krauß, N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Natture 2001, 411, 909–917. [Google Scholar] [CrossRef]
- Ciesielski, P.N.; Scott, A.M.; Faulkner, C.J.; Berron, B.J.; Cliffel, D.; Jennings, G.K. Functionalized Nanoporous Gold Leaf Electrode Films for the Immobilization of Photosystem I. ACS Nano 2008, 2, 2465–2472. [Google Scholar] [CrossRef]
- Morlock, S.; Subramanian, S.K.; Zouni, A.; Lisdat, F. Scalable Three-Dimensional Photobioelectrodes Made of Reduced Graphene Oxide Combined with Photosystem I. ACS Appl. Mater. Interfaces 2021, 13, 11237–11246. [Google Scholar] [CrossRef]
- Kim, I.; Jo, N.; Yang, M.Y.; Kim, J.; Jun, H.; Lee, G.Y.; Shin, T.; Kim, S.O.; Nam, Y.S. Directed Nanoscale Self-Assembly of Natural Photosystems on Nitrogen-Doped Carbon Nanotubes for Solar-Energy Harvesting. ACS Appl. Bio Mater. 2019, 2, 2109–2115. [Google Scholar] [CrossRef]
- Singh, A.; Mandal, S.; Chen, S.; Liu, M.; Gisriel, C.J.; Carey, A.-M.; Yan, H.; Seo, D.-K.; Lin, S.; Woodbury, N.W. Interfacing Photosystem I Reaction Centers with a Porous Antimony-Doped Tin Oxide Electrode to Perform Light-Driven Redox Chemistry. ACS Appl. Electron. Mater. 2021, 3, 2087–2096. [Google Scholar] [CrossRef]
- Fujiwara, T.; Ohnuma, M.; Yoshida, M.; Kuroiwa, T.; Hirano, T. Gene Targeting in the Red Alga Cyanidioschyzon merolae: Single- and Multi-Copy Insertion Using Authentic and Chimeric Selection Markers. PLoS ONE 2013, 8, e73608. [Google Scholar] [CrossRef] [PubMed]
- Minoda, A.; Sakagami, R.; Yagisawa, F.; Kuroiwa, T.; Tanaka, K. Improvement of Culture Conditions and Evidence for Nuclear Transformation by Homologous Recombination in a Red Alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2004, 45, 667–671. [Google Scholar] [CrossRef] [Green Version]
- Krupnik, T.; Kotabová, E.; van Bezouwen, L.S.; Mazur, R.; Garstka, M.; Nixon, P.; Barber, J.; Kana, R.; Boekema, E.J.; Kargul, J. A Reaction Center-dependent Photoprotection Mechanism in a Highly Robust Photosystem II from an Extremophilic Red Alga, Cyanidioschyzon merolae. J. Biol. Chem. 2013, 288, 23529–23542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumak, I.; Mazur, R.; Gieczewska, K.; Kozioł-Lipińska, J.; Kierdaszuk, B.; Michalski, W.P.; Shiell, B.J.; Venema, J.H.; Vredenberg, W.J.; Mostowska, A.; et al. Correlation between spatial (3D) structure of pea and bean thylakoid membranes and arrangement of chlorophyll-protein complexes. BMC Plant Biol. 2012, 12, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Kanesaki, Y.; Ehirooka, S.; Eera, A.; Sumiya, N.; Eyoshikawa, H.; Tanaka, K.; Miyagishima, S.-Y. A nitrogen source-dependent inducible and repressible gene expression system in the red alga Cyanidioschyzon merolae. Front. Plant Sci. 2015, 6, 657. [Google Scholar] [CrossRef]






| Domain | Amino Acid Sequence | Molecular Weight (kDa) |
|---|---|---|
| Chloroplast-targeting peptide (Chl-TP) | MFVQTSFFGTGVKASAKSAESQRCLAHSSWSVRMTGYDMNGSSAGNLGPRRIKSSGVAND | 6.33 |
| His6 epitope | MRGSHHHHHH | 1.27 |
| CMV144CT (psaD gene) | MLNLKMPSPSFLGSTGGWLRCAETEEKYAMTWSSDQQHIFEMPTGGAAVMNSGDNLLYLARKEQALALATQLRTQFKIQDYKIYRIFPSGEVQYLHPKDGVLPYQVNKGREQVGRVKSTIGKNVNPAQVKFTSKATYDR | 15.68 |
| Electrode Material | Photocurrent Density (nA·cm−2) | Presence of Redox Mediators | Applied Potential (V) (vs. Ag/AgCl) | Light Intensity (mW·cm−2) | Reference |
|---|---|---|---|---|---|
| Planar gold Electrode/TPDA-modified SAM/PSI | 100 | Yes | −0.1 | 80 | [51] |
| Nanoporous gold leaf electrodes/TPDA-modified SAM/PSI | 810 | Yes | −0.1 | 80 | [51] |
| FTO/SLG/pyr-NTA-Ni/His6-cyt c553/PSI | 370 | No | −0.3 | 100 | [38] |
| 3D reduced Graphene Oxide/PSI | 480 | No | −0.15 | 100 | [52] |
| PS/Nitrogen-doped Carbon Nanotubes | 625 | Yes | +0.3 | 78 | [53] |
| FTO/SLG/pyr-NTA-Ni/His6-PsaD-PSI | 774 | No | −0.3 | 100 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Izzo, M.; Jacquet, M.; Fujiwara, T.; Harputlu, E.; Mazur, R.; Wróbel, P.; Góral, T.; Unlu, C.G.; Ocakoglu, K.; Miyagishima, S.; et al. Development of a Novel Nanoarchitecture of the Robust Photosystem I from a Volcanic Microalga Cyanidioschyzon merolae on Single Layer Graphene for Improved Photocurrent Generation. Int. J. Mol. Sci. 2021, 22, 8396. https://doi.org/10.3390/ijms22168396
Izzo M, Jacquet M, Fujiwara T, Harputlu E, Mazur R, Wróbel P, Góral T, Unlu CG, Ocakoglu K, Miyagishima S, et al. Development of a Novel Nanoarchitecture of the Robust Photosystem I from a Volcanic Microalga Cyanidioschyzon merolae on Single Layer Graphene for Improved Photocurrent Generation. International Journal of Molecular Sciences. 2021; 22(16):8396. https://doi.org/10.3390/ijms22168396
Chicago/Turabian StyleIzzo, Miriam, Margot Jacquet, Takayuki Fujiwara, Ersan Harputlu, Radosław Mazur, Piotr Wróbel, Tomasz Góral, C. Gokhan Unlu, Kasim Ocakoglu, Shinya Miyagishima, and et al. 2021. "Development of a Novel Nanoarchitecture of the Robust Photosystem I from a Volcanic Microalga Cyanidioschyzon merolae on Single Layer Graphene for Improved Photocurrent Generation" International Journal of Molecular Sciences 22, no. 16: 8396. https://doi.org/10.3390/ijms22168396
APA StyleIzzo, M., Jacquet, M., Fujiwara, T., Harputlu, E., Mazur, R., Wróbel, P., Góral, T., Unlu, C. G., Ocakoglu, K., Miyagishima, S., & Kargul, J. (2021). Development of a Novel Nanoarchitecture of the Robust Photosystem I from a Volcanic Microalga Cyanidioschyzon merolae on Single Layer Graphene for Improved Photocurrent Generation. International Journal of Molecular Sciences, 22(16), 8396. https://doi.org/10.3390/ijms22168396








