Plant DNA Repair and Agrobacterium T−DNA Integration
Abstract
:1. Introduction
2. Are Agrobacterium Proteins Involved in T-DNA Integration into the Plant Genome?
3. Where in the Plant Genome Does T-DNA Integrate?
4. Give Me a Break (?)
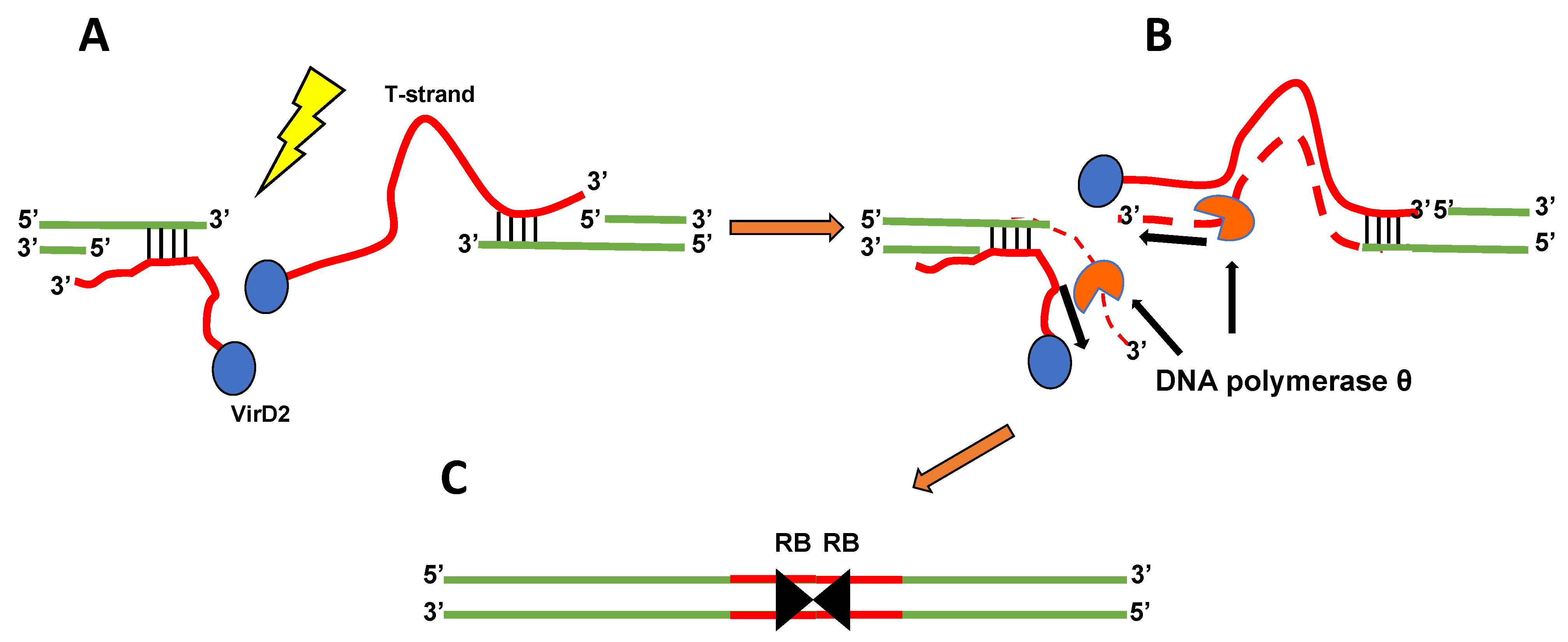
5. What Is the Mechanism of T-DNA Integration?
6. The Importance of DNA Polymerase θ for Agrobacterium-mediated Transformation and T-DNA Integration
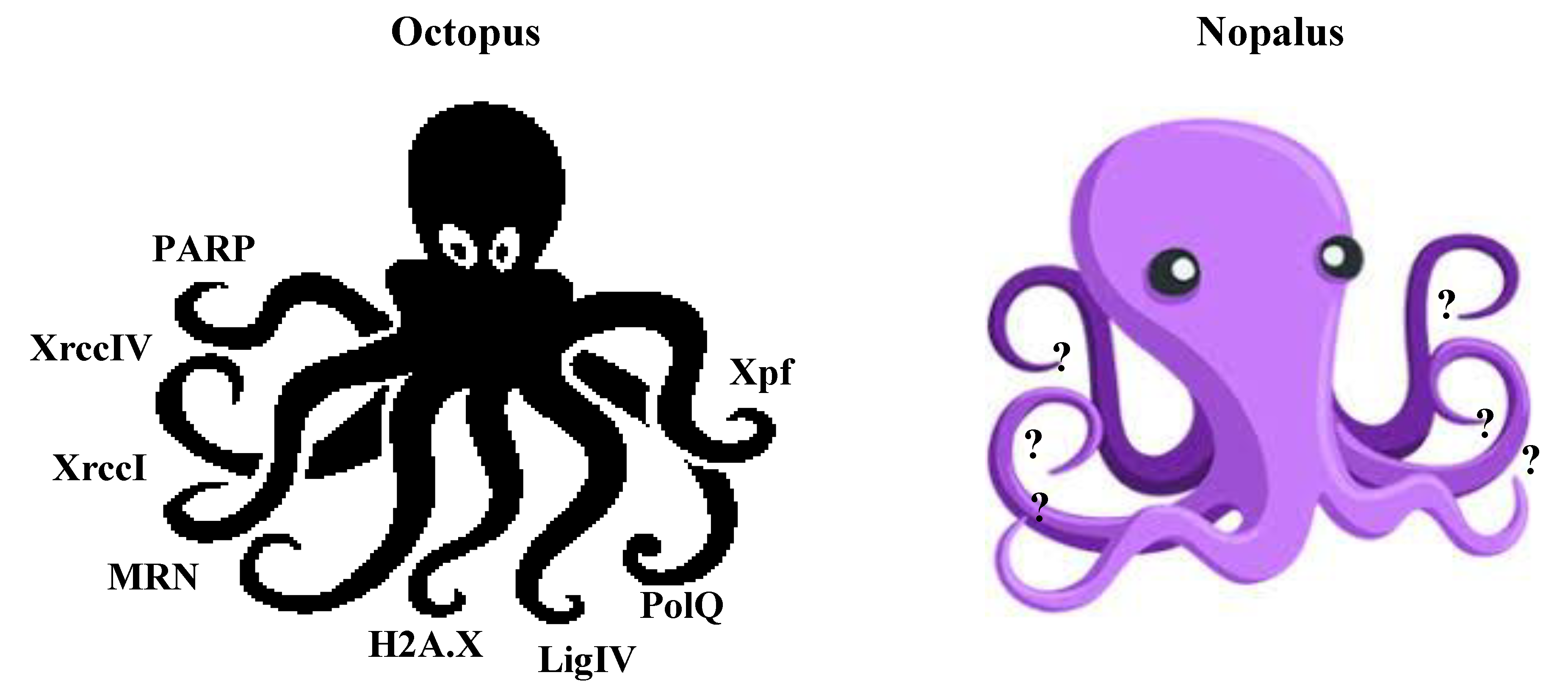
7. T-DNA Integration: An Octopus or a Nopalus?
8. Where Do We Go from Here?
9. Important Remaining Questions to Answer Regarding T-DNA Integration
- Are plant single-strand nicks, double-strand breaks, neither, or both required for T-DNA integration?
- Are there particular aspects of plant chromatin that are more conducive for integration to occur?
- What are the roles, if any, of Agrobacterium virulence effector proteins (such as VirD2, VirE2, and possibly others) in T-DNA integration?
- What plant proteins are important for integration? Related to this question, what (if any) plant DNA repair pathway(s) is/are used for integration?
- Transient transformation of most plant species and tissues is considerably more efficient than is stable transformation. Why is this? If many T-strands enter the nucleus and can initially be converted to double-strand transcription-competent forms (either linear or circular), why is T-DNA integration relatively rare in these nuclei? Could greater expression of plant DNA repair genes important for T-DNA integration increase the percentage of stably transformed cells?
- What form of T-DNA (single-strand, double-strand linear, double-strand circular) is the substrate or template for integration?
- What is the molecular basis for differences in the frequency of T-DNA integration among plant species, or even among varieties/cultivars of the same species?
- During transformation, many plant cells are exposed to Agrobacterium, but only a few may be transformed (either transiently or stably). What is the basis for plant cell transformation competency?
Funding
Conflicts of Interest
References
- Mayerhofer, R.; Koncz-Kalman, Z.; Nawrath, C.; Bakkeren, G.; Cramer, A.; Angelis, K.; Redei, G.P.; Schell, J.; Hohn, B.; Koncz, C. T-DNA integration: A mode of illegitimate recombination in plants. EMBO J. 1991, 10, 697–704. [Google Scholar] [CrossRef]
- Tinland, B.; Hohn, B. Recombination between prokaryotic and eukaryotic DNA: Integration of Agrobacterium tumefaciens T-DNA into the plant genome. In Genetic Engineering; Setlow, J.K., Ed.; Plenum Press: New York, NY, USA, 1995; pp. 209–229. [Google Scholar]
- Tinland, B. The integration of T-DNA into plant genomes. Trends Plant Sci. 1996, 1, 178–184. [Google Scholar] [CrossRef]
- Tzfira, T.; Li, J.; Lacroix, B.; Citovsky, V. Agrobacterium T-DNA integration: Molecules and models. Trends Genet. 2004, 20, 375–383. [Google Scholar] [CrossRef]
- Bilichak, A.; Yao, Y.; Kovalchuk, I. Transient down-regulation of the RNA silencing machinery increases efficiency of Agrobacterium-mediated transformation of Arabidopsis. Plant Biotechnol. J. 2014, 12, 590–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saika, H.; Nishizawa-Yokoi, A.; Toki, S. The non-homologous end-joining pathway is involved in stable transformation in rice. Front. Plant Sci. 2014, 5, 560. [Google Scholar] [CrossRef] [Green Version]
- van Kregten, M.; de Pater, S.; Romeijn, R.; van Schendel, R.; Hooykaas, P.J.J.; Tijsterman, M. T-DNA integration in plants results from polymerase-theta-mediated DNA repair. Nature Plants 2016, 2, 16164. [Google Scholar] [CrossRef] [PubMed]
- Gelvin, S.B. Integration of Agrobacterium T-DNA into the plant genome. Annu. Rev. Genet. 2017, 51, 195–217. [Google Scholar] [CrossRef]
- Lacroix, B.; Citovsky, V. Pathways of DNA transfer to plants from Agrobacterium tumefaciens and related bacterial species. Annu. Rev. Phytopathol. 2019, 57, 231–251. [Google Scholar] [CrossRef]
- Dong, O.X.; Ronald, P.C. Targeted DNA insertion in plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2004834117. [Google Scholar] [CrossRef] [PubMed]
- Yanofsky, M.F.; Porter, S.G.; Young, C.; Albright, L.M.; Gordon, M.P.; Nester, E.W. The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell 1986, 47, 471–477. [Google Scholar] [CrossRef]
- Stachel, S.E.; Timmerman, B.; Zambryski, P. Generation of single-stranded T-DNA molecules during the initial stages of T-DNA transfer from Agrobacterium tumefaciens to plant cells. Nature 1986, 322, 706–712. [Google Scholar] [CrossRef]
- Jayaswal, R.K.; Veluthambi, K.; Gelvin, S.B.; Slightom, J.L. Double-stranded cleavage of T-DNA and generation of single-stranded T-DNA molecules in Escherichia coli by a virD-encoded border-specific endonuclease from Agrobacterium tumefaciens. J. Bacteriol. 1987, 169, 5035–5045. [Google Scholar] [CrossRef] [Green Version]
- Veluthambi, K.; Jayaswal, R.K.; Gelvin, S.B. Virulence genes A, G, and D mediate the double-stranded border cleavage of T-DNA from the Agrobacterium Ti plasmid. Proc. Natl. Acad. Sci. USA 1987, 84, 1881–1885. [Google Scholar] [CrossRef] [Green Version]
- Veluthambi, K.; Ream, W.; Gelvin, S.B. Virulence genes, borders, and overdrive generate single-stranded T-DNA molecules from the A6 Ti plasmid of Agrobacterium tumefaciens. J. Bacteriol. 1988, 170, 1523–1532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, N.S.; Van der Leyden, J.; Bennett, D.R.; Barnes, W.M.; Chilton, M.-D. Short direct repeats flank the T-DNA on a nopaline Ti plasmid. Proc. Natl. Acad. Sci. USA 1982, 79, 6322–6326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Herrera-Estrella, L.; Van Montagu, M.; Zambryski, P. Right 25 bp terminus sequence of the nopaline T-DNA is essential for and determines direction of DNA transfer from Agrobacterium to the plant genome. Cell 1984, 38, 455–462. [Google Scholar] [CrossRef]
- Herrera-Estrella, A.; Chen, Z.-M.; Van Montagu, M.; Wang, K. VirD proteins of Agrobacterium tumefaciens are required for the formation of a covalent DNA-protein complex at the 5′ terminus of T-strand molecules. EMBO J. 1988, 7, 4055–4062. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.R.; Barnes, W.M. VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science 1988, 242, 927–930. [Google Scholar] [CrossRef]
- Young, C.; Nester, E.W. Association of the VirD2 protein with the 5′ end of T strands in Agrobacterium tumefaciens. J. Bacteriol. 1988, 170, 3367–3374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durrenberger, F.; Crameri, A.; Hohn, B.; Koukolikova-Nicola, Z. Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc. Natl. Acad. Sci. USA 1989, 86, 9154–9158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard, E.A.; Winsor, B.A.; De Vos, G.; Zambryski, P. Activation of the T-DNA transfer process in Agrobacterium results in the generation of a T-strand-protein complex: Tight association of VirD2 with the 5′ ends of T-strands. Proc. Natl. Acad. Sci. USA 1989, 86, 4017–4021. [Google Scholar] [CrossRef] [Green Version]
- Tinland, B.; Hohn, B.; Puchta, H. Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc. Natl. Acad. Sci. USA 1994, 91, 8000–8004. [Google Scholar] [CrossRef] [Green Version]
- Yusibov, V.M.; Steck, T.R.; Gupta, V.; Gelvin, S.B. Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl. Acad. Sci. USA 1994, 91, 2994–2998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pansegrau, W.; Schoumacher, F.; Hohn, B.; Lanka, E. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: Analogy to bacterial conjugation. Proc. Natl. Acad. Sci. USA 1993, 90, 11538–11542. [Google Scholar] [CrossRef] [Green Version]
- Ziemienowicz, A.; Tinland, B.; Bryant, J.; Gloeckler, V.; Hohn, B. Plant enzymes but not Agrobacterium VirD2 mediate T-DNA ligation in vitro. Mol. Cell. Biol. 2000, 20, 6317–6322. [Google Scholar] [CrossRef] [PubMed]
- Shurvinton, C.E.; Hodges, L.; Ream, W. A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc. Natl. Acad. Sci. USA 1992, 89, 11837–11841. [Google Scholar] [CrossRef] [Green Version]
- Narasimhulu, S.B.; Deng, X.-B.; Sarria, R.; Gelvin, S.B. Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 1996, 8, 873–886. [Google Scholar] [PubMed] [Green Version]
- Mysore, K.S.; Bassuner, B.; Deng, X.-b.; Darbinian, N.S.; Motchoulski, A.; Ream, W.; Gelvin, S.B. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant.-Microbe Interact. 1998, 11, 668–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tinland, B.; Schoumacher, F.; Gloeckler, V.; Bravo-Angel, A.M.; Hohn, B. The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 1995, 14, 3585–3595. [Google Scholar] [CrossRef]
- Bravo-Angel, A.M.; Hohn, B.; Tinland, B. The omega sequence of VirD2 is important but not essential for efficient transfer of T-DNA by Agrobacterium tumefaciens. Mol. Plant.-Microbe Interact. 1997, 11, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Nuti, M.P.; Ledeboer, A.M.; Durante, M.; Nuti-Ronchi, V.; Schilperoort, R.A. Detection of Ti-plasmid sequences in infected tissues by in situ hybridization. Plant Sci. Lett. 1980, 18, 1–6. [Google Scholar] [CrossRef]
- Ambros, P.F.; Matzke, A.J.M.; Matzke, M.A. Localization of Agrobacterium rhizogenes T-DNA in plant chromosomes by in situ hybridization. EMBO J. 1986, 5, 2073–2078. [Google Scholar] [CrossRef]
- Wang, J.; Lewis, M.E.; Whallon, J.H.; Sink, K.C. Chromosomal mapping of T-DNA inserts in transgenic Petunia by in situ hybridization. Transgen. Res. 1995, 4, 241–246. [Google Scholar] [CrossRef]
- Koncz, C.; Martini, N.; Mayerhofer, R.; Koncz-Kalman, Z.; Korber, H.; Redei, G.P.; Schell, J. High-frequency T-DNA-mediated gene tagging in plants. Proc. Natl. Acad. Sci. USA 1989, 86, 8467–8471. [Google Scholar] [CrossRef] [Green Version]
- Brunaud, V.; Balzergue, S.; Dubreucq, B.; Aubourg, S.; Samson, F.; Chauvin, S.; Bechtold, N.; Cruaud, C.; DeRose, R.; Pelletier, G.; et al. T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep. 2002, 3, 1152–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szabados, L.; Kovacs, I.; Oberschall, A.; Abraham, E.; Kerekes, I.; Zsigmond, L.; Nagy, R.; Alvarado, M.; Krasovskaja, I.; Gal, M.; et al. Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J. 2002, 32, 233–242. [Google Scholar] [CrossRef]
- Chen, S.; Jin, W.; Wang, M.; Zhang, F.; Zhou, J.; Jia, Q.; Wu, Y.; Liu, F.; Wu, P. Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J. 2003, 36, 105–113. [Google Scholar] [CrossRef]
- Sallaud, C.; Gay, C.; Larmande, P.; Bes, M.; Piffanelli, P.; Piegu, B.; Droc, G.; Regad, F.; Bourgeois, E.; Meynard, D.; et al. High throughput T-DNA insertion mutagenesis in rice: A first step towards in silico reverse genetics. Plant J. 2004, 39, 450–464. [Google Scholar] [CrossRef]
- Schneeberger, R.G.; Zhang, K.; Tatarinova, T.; Troukhan, M.; Kwok, S.F.; Drais, J.; Klinger, K.; Orejudos, F.; Macy, K.; Bhakta, A.; et al. Agrobacterium T-DNA integration in Arabidopsis is correlated with DNA sequence compositions that occur frequently in gene promoter regions. Funct. Integr. Genom. 2005, 5, 240–253. [Google Scholar] [CrossRef]
- Kim, S.I.; Veena; Gelvin, S.B. Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 2007, 51, 779–791. [Google Scholar] [CrossRef]
- Shilo, S.; Tripathi, P.; Melamed-Bessudo, C.; Tzfadia, O.; Muth, T.R.; Levy, A.A. T-DNA-genome junctions form early after infection and are influenced by the chromatin state of the host genome. PLoS Genet. 2017, 13, e1006875. [Google Scholar] [CrossRef] [Green Version]
- Windels, P.; De Buck, S.; Van Bockstaele, E.; De Loose, M.; Depicker, A. T-DNA integration in Arabidopsis chromosomes: Presence and origin of filler DNA sequences. Plant Physiol. 2003, 133, 2061–2068. [Google Scholar] [CrossRef] [Green Version]
- Muller, A.E.; Atkinson, R.G.; Sandoval, R.B.; Jorgensen, R.A. Microhomologies between T-DNA ends and target sites often occur in inverted orientation and may be responsible for the high frequency of T-DNA-associated inversions. Plant Cell Rep. 2007, 26, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Kleinboelting, N.; Huep, G.; Appelhagen, I.; Viehoever, P.; Li, Y.; Weisshaar, B. The structural features of thousands of T-DNA insertion sites are consistent with a double-strand break repair based insertion mechanism. Mol. Plant 2015, 8, 1651–1664. [Google Scholar] [CrossRef]
- Nam, J.; Mysore, K.S.; Zheng, C.; Knue, M.K.; Matthysse, A.G.; Gelvin, S.B. Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 1999, 261, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Mysore, K.S.; Nam, J.; Gelvin, S.B. An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl. Acad. Sci. USA 2000, 97, 948–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; He, X.-W.; Ying, Y.-H.; Lu, J.-F.; Gelvin, S.B.; Shou, H.-X. Expression of the Arabidopsis thaliana histone gene AtHTA1 enhances rice transformation efficiency. Mol. Plant 2009, 2, 832–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, H.; Mysore, K.S.; Gelvin, S. Expression of the Arabidopsis histone H2A-1 gene correlates with susceptibility to Agrobacterium transformation. Plant J. 2002, 32, 285–298. [Google Scholar] [CrossRef]
- Yi, H.; Sardesai, N.; Fujinuma, T.; Chan, C.W.; Veena; Gelvin, S.B. Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell 2006, 18, 1575–1589. [Google Scholar] [CrossRef] [Green Version]
- Tenea, G.N.; Spantzel, J.; Lee, L.-Y.; Zhu, Y.; Lin, K.; Johnson, S.J.; Gelvin, S.B. Overexpression of several Arabidopsis histone genes increases Agrobacterium-mediated transformation and transgene expression in plants. Plant Cell 2009, 21, 3350–3367. [Google Scholar] [CrossRef] [Green Version]
- Gelvin, S.B.; Kim, S.-I. Effect of chromatin upon Agrobacterium T-DNA integration and transgene expression. Biophys. Biochem. Acta 2007, 1769, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Wolterink-van Loo, S.; Escamilla Ayala, A.A.; Hooykaas, P.J.J.; van Heudsden, G.P.H. Interaction of the Agrobacterium tumefaciens virulence protein VirD2 with histones. Microbiology 2015, 161, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Howard, E.; Citovsky, V. The emerging structure of the Agrobacterium T-DNA transfer complex. BioEssays 1990, 12, 103–108. [Google Scholar] [CrossRef]
- Tzfira, T.; Vaidya, M.; Citovsky, V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001, 20, 3596–3607. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Krichevsky, A.; Vaidya, M.; Tzfira, T.; Citovsky, V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. USA 2005, 102, 5733–5738. [Google Scholar] [CrossRef] [Green Version]
- Loyter, A.; Rosenbluh, J.; Zakai, N.; Li, J.; Kozlovsky, S.V.; Tzfira, T.; Citovsky, V. The plant VirE2 interacting protein 1. A molecular link between the Agrobacterium T-complex and the host cell chromatin? Plant Physiol. 2005, 138, 1318–1321. [Google Scholar] [CrossRef] [Green Version]
- Lacriox, B.; Loyter, A.; Citovsky, V. Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc. Natl. Acad. Sci. USA 2008, 105, 15429–15434. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Lee, L.-Y.; Gelvin, S.B. Is VIP1 important for Agrobacterium-mediated transformation? Plant J. 2014, 79, 848–860. [Google Scholar] [CrossRef]
- Lapham, R.; Lee, L.-Y.; Tsugama, D.; Lee, S.; Mengiste, T.; Gelvin, S.B. VIP1 and its homologs are not required for Agrobacterium-mediated transformation, but play a role in Botrytis and salt stress responses. Front. Plant Sci. 2018, 9, 749. [Google Scholar] [CrossRef]
- Crane, Y.M.; Gelvin, S.B. RNAi-mediated gene silencing reveals involvement of Arabidopsis chromatin-related genes in Agrobacterium-mediated root transformation. Proc. Natl. Acad. Sci. USA 2007, 104, 15156–15161. [Google Scholar] [CrossRef] [Green Version]
- Soltani, J.; van Heudsden, G.P.H.; Hooykaas, P.J.J. Deletion of host histone acetyltransferases and deacetylases strongly affects Agrobacterium-mediated transformation of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2009, 298, 228–233. [Google Scholar] [CrossRef] [Green Version]
- Köhler, F.; Cardon, G.; Pöhlman, M.; Gill, R.; Schieder, O. Enhancement of transformation rates in higher plants by low-dose irradiation: Are DNA repair systems involved in the incorporation of exogenous DNA into the plant genome? Plant Mol. Biol. 1989, 12, 189–199. [Google Scholar] [CrossRef]
- Gorbunova, V.; Levy, A.A. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucl. Acids Res. 1997, 25, 4650–4657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirley, B.W.; Hanley, S.; Goodman, H.M. Effects of ionizing radiation on a plant genome: Analysis of two Arabidopsis transparent testa mutations. Plant Cell 1992, 4, 333–347. [Google Scholar] [PubMed]
- Somers, D.A.; Makarevitch, I. Transgene integration in plants: Poking or patching holes in promiscuous genomes? Curr. Opin. Biotechnol. 2004, 15, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Salomon, S.; Puchta, H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998, 17, 6086–6095. [Google Scholar] [CrossRef]
- Chilton, M.-D.; Que, Q. Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: New insights on the mechanism of T-DNA integration. Plant Physiol. 2003, 133, 956–965. [Google Scholar] [CrossRef] [Green Version]
- Tzfira, T.; Frankman, L.R.; Vaidya, M.; Citovsky, V. Site-specific integration of Agrobacterium tumefaciens T-DNA via double-stranded intermediates. Plant Physiol. 2003, 133, 1011–1023. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Xing, H.-L.; Wang, Z.-P.; Zhang, H.-Y.; Yang, F.; Wang, X.-C.; Chen, Q.-J. Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention. Plant Mol. Biol. 2018, 96, 445–456. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Eggenberer, A.L.; Banakar, R.; McCaw, M.; Zhu, H.; Main, M.; Kang, M.; Gelvin, S.B.; Wang, K. CRISPR/Cas9 targeted T-DNA integration in rice. Plant Mol. Biol. 2019, 99, 317–328. [Google Scholar] [CrossRef]
- Song, J.; Bent, A.F. Microbial pathogens trigger host DNA double-strand breaks whose abundance is reduced by plant defense responses. PLoS Pathog. 2014, 10, e1004030. [Google Scholar] [CrossRef]
- Joseph, J.T.; Chandhini, S.; Das, S.; Mysore, K.S.; Shah, J.M. Methylation status of Arabidopsis DNA repair gene promoters during Agrobacterium infection reveals epigenetic changes in three generations. Plant Mol. Biol. Rep. 2021. [Google Scholar] [CrossRef]
- Wei, F.-J.; Lee, L.-Y.; Hsing, Y.-I.; Gelvin, S.B.; Academia Sinica, Taipei, Taiwan and Purdue University, West Lafayette, IN, USA. Unpublished work. 2021.
- Marton, L.; Hrouda, M.; Pecsvaradi, A.; Czako, M. T-DNA-independent mutations induced in transformed plant cells during Agrobacterium co-cultivation. Transgen. Res. 1994, 3, 317–325. [Google Scholar] [CrossRef]
- Budziszewski, G.J.; Lewis, S.P.; Glover, L.W.; Reineke, J.; Jones, G.; Ziemnik, L.S.; Lonowski, J.; Nyfeler, B.; Aux, G.; Zhou, Q.; et al. Arabidopsis genes essential for seedling viability: Isolation of insertional mutants and molecular cloning. Genetics 2001, 159, 1765–1778. [Google Scholar] [CrossRef]
- McElver, J.; Tzafrir, I.; Aux, G.; Rogers, R.; Ashby, C.; Smith, K.; Thomas, C.; Schetter, A.; Zhou, Q.; Cushman, M.A.; et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 2001, 159, 1751–1763. [Google Scholar] [CrossRef]
- Castle, L.A.; Errampalli, D.; Atherton, T.L.; Franzmann, L.H.; Yoon, E.S.; Meinke, D.W. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol. Gen. Genet. 1993, 241, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Nacry, P.; Camilleri, C.; Courtial, B.; Caboche, M.; Bouchez, D. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 1998, 149, 641–650. [Google Scholar] [CrossRef]
- Tax, F.E.; Vernon, D.M. T-DNA-associated duplication/translocations in Arabidopsis. Implications for mutant analysis and functional genomics. Plant Physiol. 2001, 126, 1527–1538. [Google Scholar] [CrossRef] [Green Version]
- Lafleuriel, J.; Degroote, F.; Depeiges, A.; Picard, G. A reciprocal translocation, induced by a canonical integration of a single T-DNA, interrupts the HMG-I/Y Arabidopsis thaliana gene. Plant Physiol. Biochem. 2004, 42, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.J.; Belcram, K.; Bollmann, S.R.; Tominey, C.M.; Hoffman, P.D.; Mercier, R.; Hays, J.B. Reciprocal chromosome translocation associated with T-DNA-insertion mutation in Arabidopsis: Genetic and cytological analyses of consequences for gametophyte development and for construction of doubly mutant lines. Planta 2009, 229, 731–745. [Google Scholar] [CrossRef] [Green Version]
- Ruprecht, C.; Carroll, A.; Persson, S. T-DNA-induced chromosomal translocation in feronia and anxur2 mutants reveal implications for the mechanism of collapsed pollen due to chromosomal rearrangements. Mol. Plant 2014, 7, 1591–1594. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Chen, Z.; Zhung, C.; Huang, J. Cascade of chromosomal rearrangements caused by a heterogeneous T-DNA integration supports the double-strand break repair model for T-DNA integration. Plant J. 2017, 90, 954–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jupe, F.; Rivkin, A.C.; Michael, T.P.; Zander, M.; Motley, S.T.; Sandoval, J.P.; Slotkin, R.K.; Chien, H.; Castanon, R.; Nery, J.R.; et al. The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genet. 2019, 15, e1007819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majhi, B.B.; Shah, J.M.; Veluthambi, K. A novel T-DNA integration in rice involving two interchromosomal translocations. Plant Cell Rep. 2014, 33, 929–944. [Google Scholar] [CrossRef]
- Clark, D.A.; Krysan, P.J. Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant J. 2010, 64, 990–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leibowitz, M.L.; Papathanasiou, S.; Doerfler, P.A.; Blaine, L.J.; Sun, L.; Yao, Y.; Zhang, C.-Z.; Weiss, M.J.; Pellman, D. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat. Genet. 2021, 53, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Offringa, R.; van den Elzen, P.J.M.; Hooykaas, P.J.J. Gene targeting in plants using the Agrobacterium vector system. Transgen. Res. 1992, 1, 114–123. [Google Scholar] [CrossRef]
- Bundock, P.; Hooykaas, P.J.J. Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc. Natl. Acad. Sci. USA 1996, 93, 15272–15275. [Google Scholar] [CrossRef] [Green Version]
- van Attikum, H.; Hooykaas, P.J.J. Genetic requirements for the targeted integration of Agrobacterium T-DNA in Saccharomyces cerevisiae. Nucl. Acids Res. 2003, 31, 826–832. [Google Scholar] [CrossRef] [Green Version]
- Mladenov, E.; Iliakis, G. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2011, 711, 61–72. [Google Scholar] [CrossRef]
- Decottignies, A. Alternative end-joining mechanisms: A historical perspective. Front. Genet. 2013, 4, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Xu, X. Microhomology-mediated end joining: New players join the team. Cell. Biosci. 2017, 7, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friesner, J.; Britt, A.B. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 2003, 34, 427–440. [Google Scholar] [CrossRef] [Green Version]
- Gallego, M.E.; Bleuyard, J.-Y.; Daoudal-Cotterell, S.; Jallut, N.; White, C.I. Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 2003, 35, 557–565. [Google Scholar] [CrossRef]
- van Attikum, H.; Bundock, P.; Lee, L.-Y.; Gelvin, S.B.; Hooykaas, P.J.J. The Arabidopsis AtLIG4 gene is involved in the repair of DNA damage, but not in the integration of Agrobacterium T-DNA. Nucl. Acids Res. 2003, 31, 4247–4255. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Vaidya, M.; White, C.; Vainstein, A.; Citovsky, V.; Tzfira, T. Involvement of Ku80 in T-DNA integration in plant cells. Proc. Natl. Acad. Sci. USA 2005, 102, 19231–19236. [Google Scholar] [CrossRef] [Green Version]
- Jia, Q.; Bundock, P.; Hooykaas, P.J.J.; de Pater, S. Agrobacterium tumefaciens T-DNA integration and gene targeting in Arabidopsis thaliana non-homologous end-joining mutants. J. Bot. 2012. [Google Scholar] [CrossRef] [Green Version]
- Nishizawa-Yokoi, A.; Nonaka, S.; Saika, H.; Kwon, Y.-I.; Osakabe, K.; Toki, S. Suppression of Ku70/80 or Lig4 leads to decreased stable transformation and enhanced homologous recombination in rice. New Phytol. 2012, 196, 1048–1059. [Google Scholar] [CrossRef] [Green Version]
- Vaghchhipawala, Z.E.; Vasudevan, B.; Lee, S.; Morsy, M.R.; Mysore, K.S. Agrobacterium may delay plant nonhomologous end-joining DNA repair via XRCC4 to favor T-DNA integration. Plant Cell 2012, 24, 4110–4123. [Google Scholar] [CrossRef] [Green Version]
- Mestiri, I.; Norre, F.; Gallego, M.E.; White, C.I. Multiple host-cell recombination pathways act in Agrobacterium-mediated transformation of plant cells. Plant J. 2014, 77, 511–520. [Google Scholar] [CrossRef]
- Park, S.-Y.; Vaghchhipawala, Z.; Vasudevan, B.; Lee, L.-Y.; Shen, Y.; Singer, K.; Waterworth, W.M.; Zhang, Z.; West, C.E.; Mysore, K.S.; et al. Agrobacterium T-DNA integration into the plant genome can occur without the activity of key non-homologous end-joining proteins. Plant J. 2015, 81, 934–946. [Google Scholar] [CrossRef]
- Yoshihara, R.; Mitomi, Y.; Okada, M.; Shibata, H.; Tanokami, M.; Nakajima, Y.; Inui, H.; Oono, Y.; Furudate, H.; Tanaka, S. Effects of Arabidopsis Ku80 deletion on the integration of the left border of T-DNA into plant chromosomal DNA via Agrobacterium tumefaciens. Genes Genet. Syst. 2020, 95, 1–10. [Google Scholar] [CrossRef]
- Nishizawa-Yokoi, A.; Saika, H.; Hara, N.; Lee, L.-Y.; Toki, S.; Gelvin, S.B. Agrobacterium T-DNA integration is somatic cells does not require the activity of DNA polymerase theta. New Phytol. 2021, 229, 2859–2872. [Google Scholar] [CrossRef]
- Francis, K.E.; Spiker, S. Identification of Arabidopsis thaliana transformants without selection reveals a high occurrence of silenced T-DNA integrations. Plant J. 2005, 41, 464–477. [Google Scholar] [CrossRef]
- Mysore, K.S.; Kumar, C.T.R.; Gelvin, S.B. Arabidopsis ecotypes and mutants that are recalcitrant to Agrobacterium root transformation are susceptible to germ-line transformation. Plant J. 2000, 21, 9–16. [Google Scholar] [CrossRef]
- Faure, D. Is there a unique integration mechanism of Agrobacterium T-DNA into a plant genome? New Phytol. 2021, 229, 2386–2388. [Google Scholar] [CrossRef] [PubMed]
- Nisa, M.; Bergis, C.; Pedroza-Garcia, J.-A.; Drouin-Wahbi, J.; Mazubert, C.; Bergounioux, C.; Benhamed, M.; Raynaud, C. The plant DNA polymerase theta is essential for the repair of replication-associated DNA damage. Plant J. 2021, 106, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Verhage, L. Solving the theta enigma: Polymerase θ deficiency causes developmental defects. Plant J. 2021, 106, 1195–1196. [Google Scholar] [CrossRef] [PubMed]
- Saika, H.; Shen, Y.; Kim, Y.; Lee, L.-Y.; Gelvin, S.B.; NARO, Tsukuba, Japan and Purdue University, West Lafayette, IN, USA. Unpublished work. 2021.
- Waterworth, W.M.; Kozak, J.; Provost, C.M.; Bray, C.M.; Angelis, K.J.; West, C.E. DNA ligase I deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 2009, 9, 79. [Google Scholar] [CrossRef] [Green Version]
- Bakkeren, G.; Koukolikova-Nicola, Z.; Grimsley, N.; Hohn, B. Recovery of Agrobacterium tumefaciens T-DNA molecules from whole plants early after transfer. Cell 1989, 57, 847–857. [Google Scholar] [CrossRef]
- Singer, K.; Shiboleth, Y.M.; Li, J.; Tzfira, T. Formation of complex extrachromosomal T-DNA structures in Agrobacterium-infected plants. Plant Physiol. 2012, 160, 511–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singer, K.; Lee, L.-Y.; Gelvin, S.B.; Purdue University, West Lafayette, IN, USA. Unpublished work. 2021.

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gelvin, S.B. Plant DNA Repair and Agrobacterium T−DNA Integration. Int. J. Mol. Sci. 2021, 22, 8458. https://doi.org/10.3390/ijms22168458
Gelvin SB. Plant DNA Repair and Agrobacterium T−DNA Integration. International Journal of Molecular Sciences. 2021; 22(16):8458. https://doi.org/10.3390/ijms22168458
Chicago/Turabian StyleGelvin, Stanton B. 2021. "Plant DNA Repair and Agrobacterium T−DNA Integration" International Journal of Molecular Sciences 22, no. 16: 8458. https://doi.org/10.3390/ijms22168458
APA StyleGelvin, S. B. (2021). Plant DNA Repair and Agrobacterium T−DNA Integration. International Journal of Molecular Sciences, 22(16), 8458. https://doi.org/10.3390/ijms22168458





