Hepatic Proteomic Analysis of Selenoprotein T Knockout Mice by TMT: Implications for the Role of Selenoprotein T in Glucose and Lipid Metabolism
Abstract
:1. Introduction
2. Results
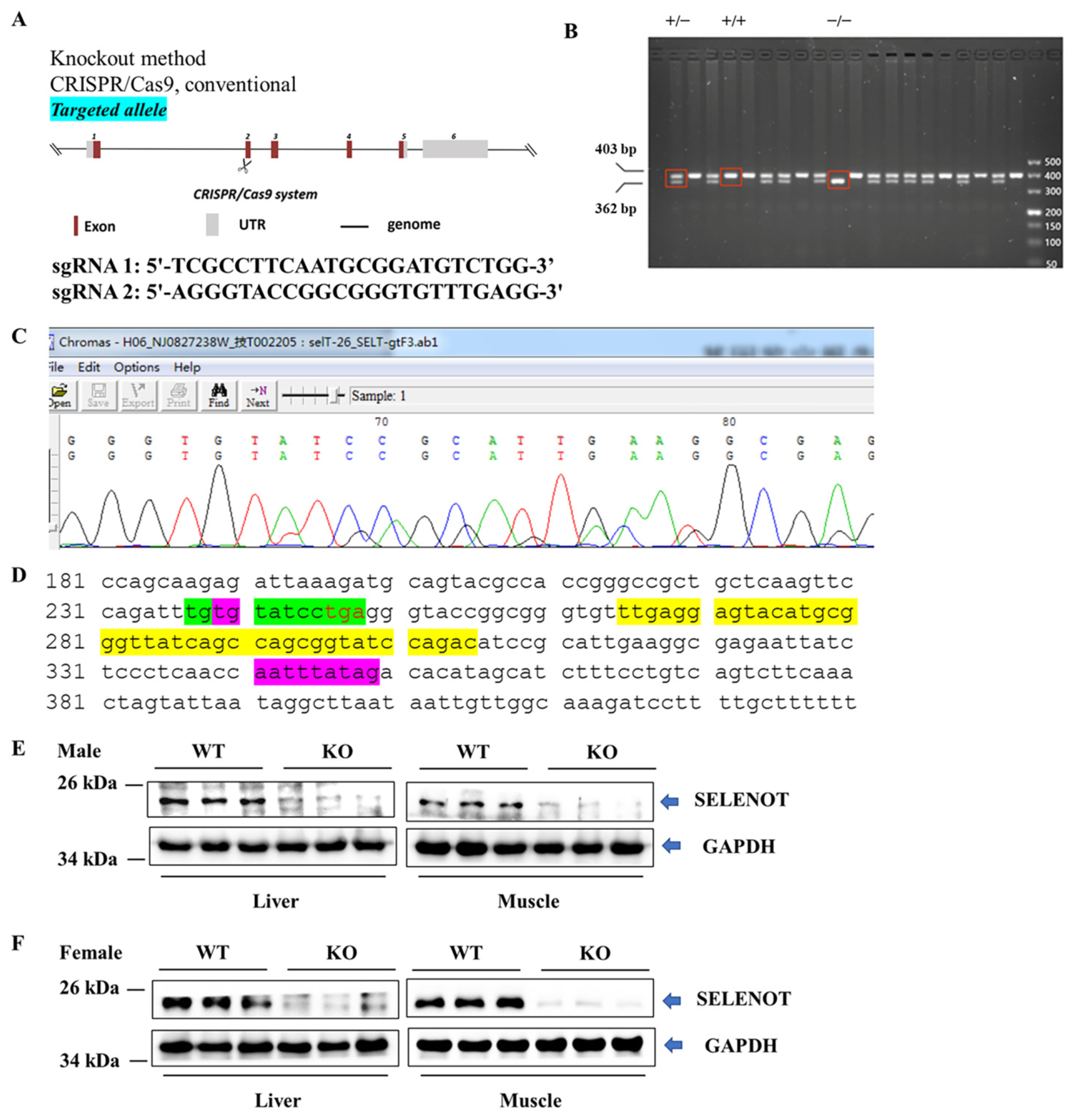
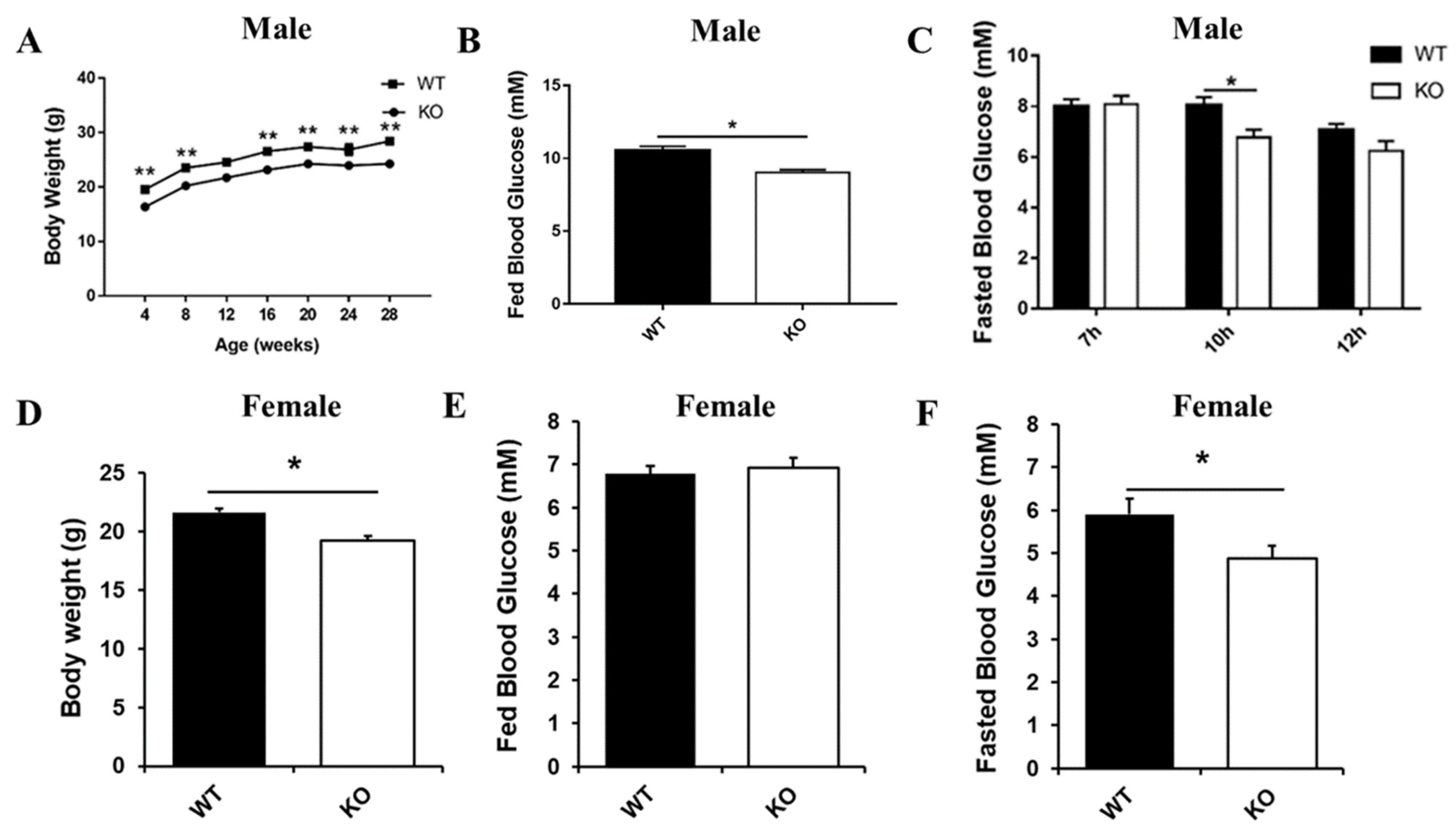
2.1. Generation and Metabolic Phenotype Analysis of Selenot KO Mice
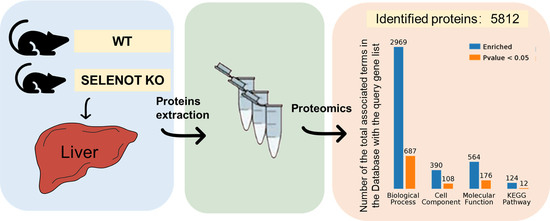
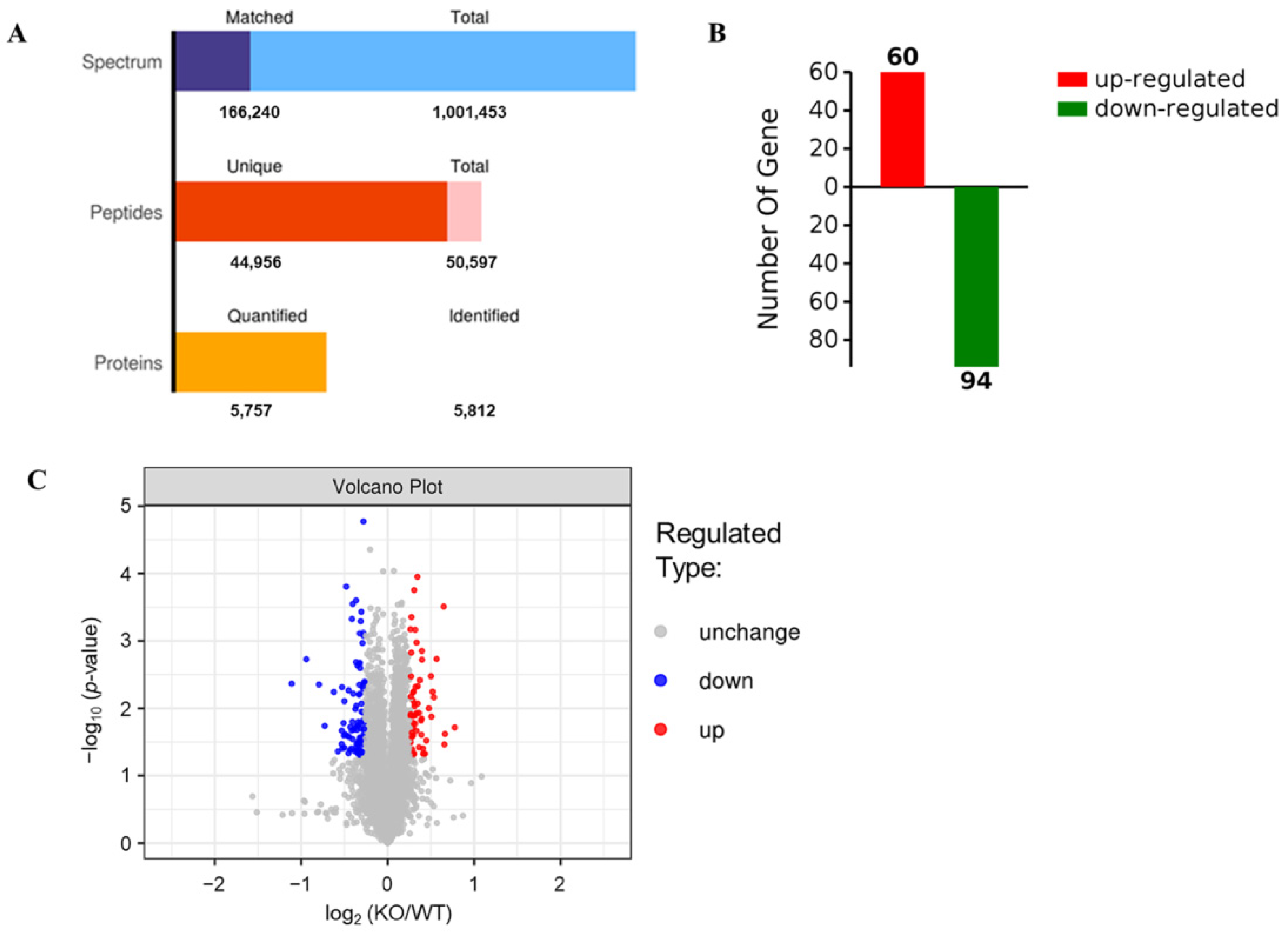
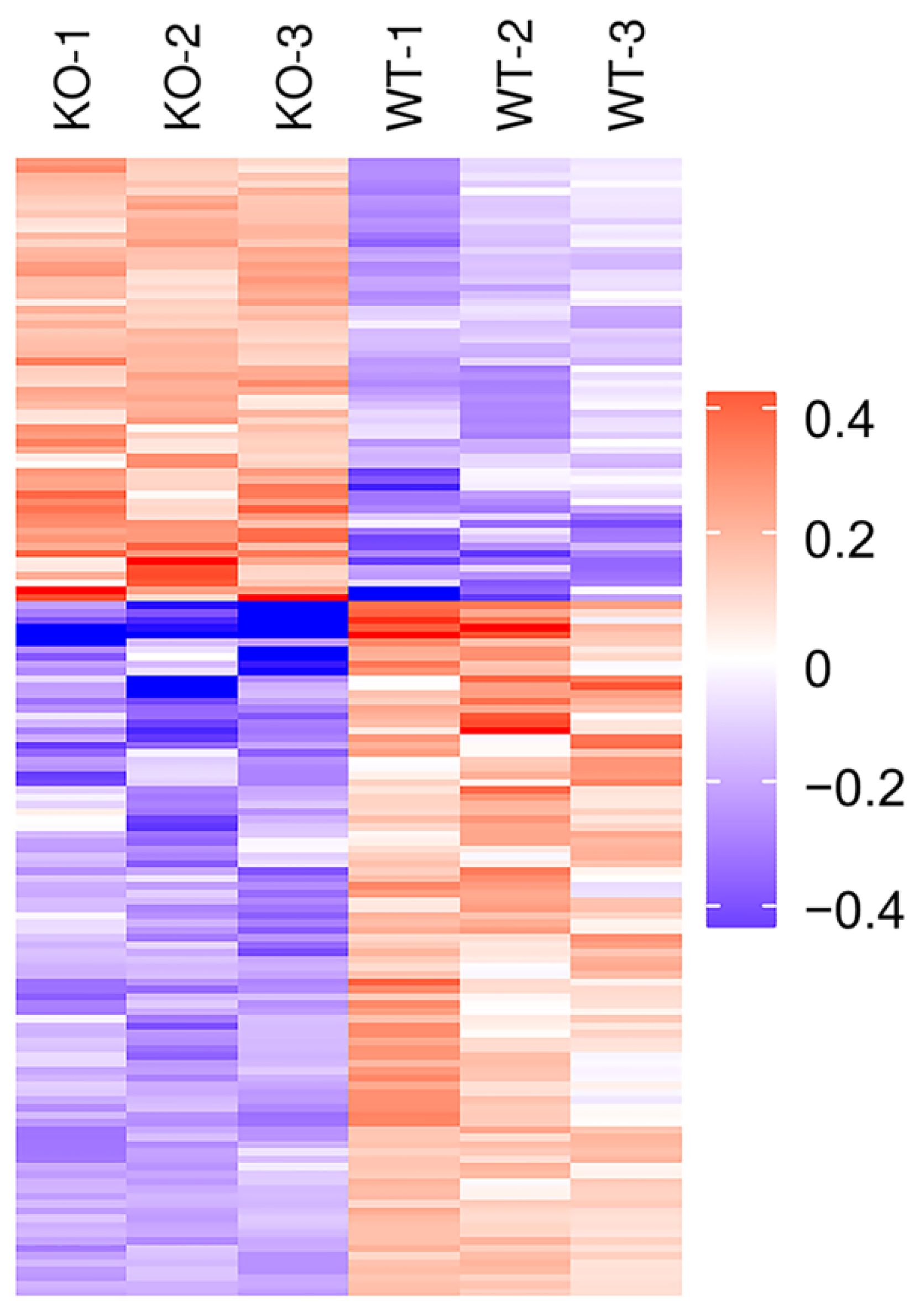
2.2. Identification of Differentially Expressed Proteins (DEPs) by TMT Proteomic Analysis
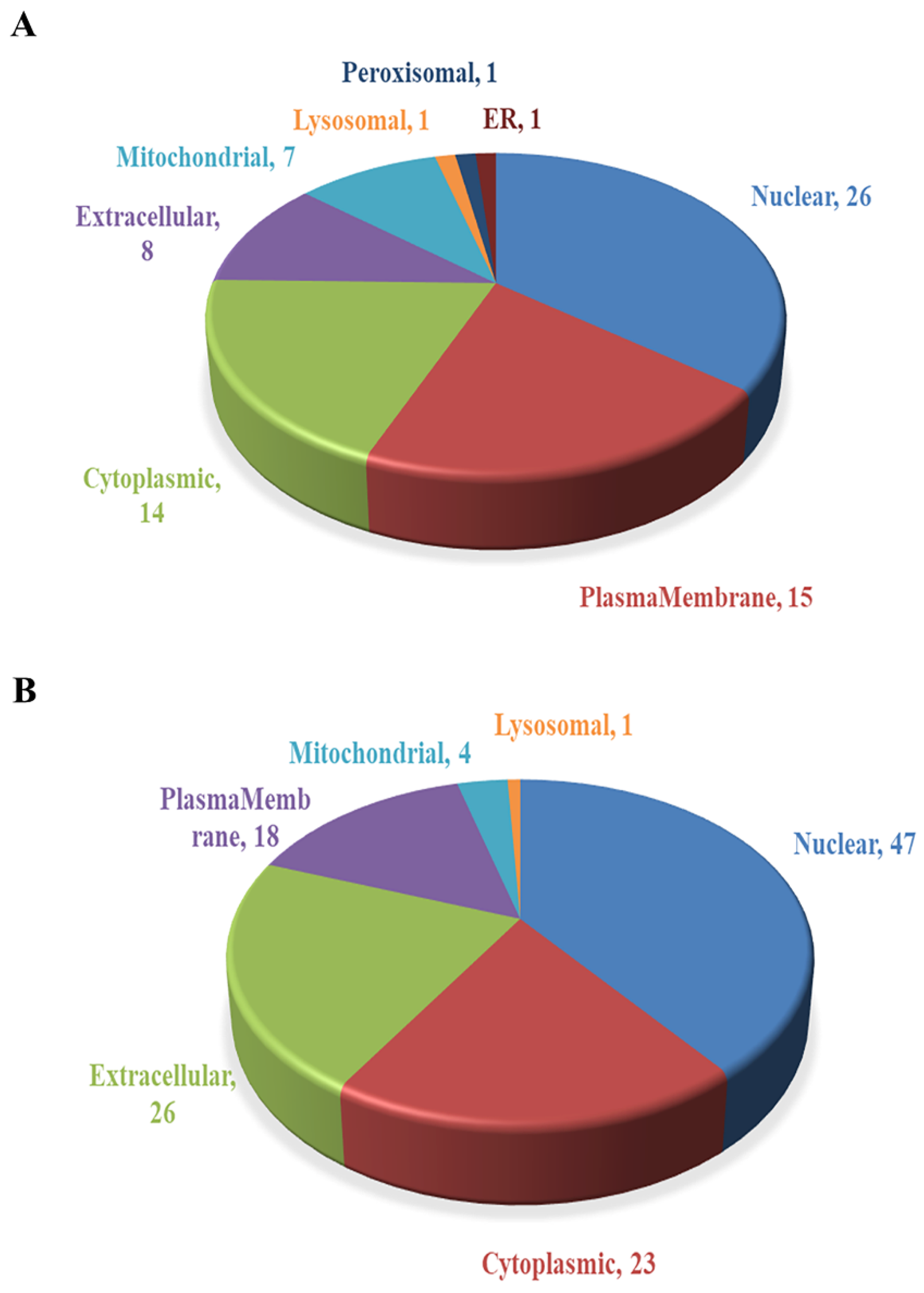
2.3. Subcellular Localization Analysis of DEPs
2.4. Domain Analysis of DEPs
2.5. Gene Ontology (GO) Categorization of DEPs
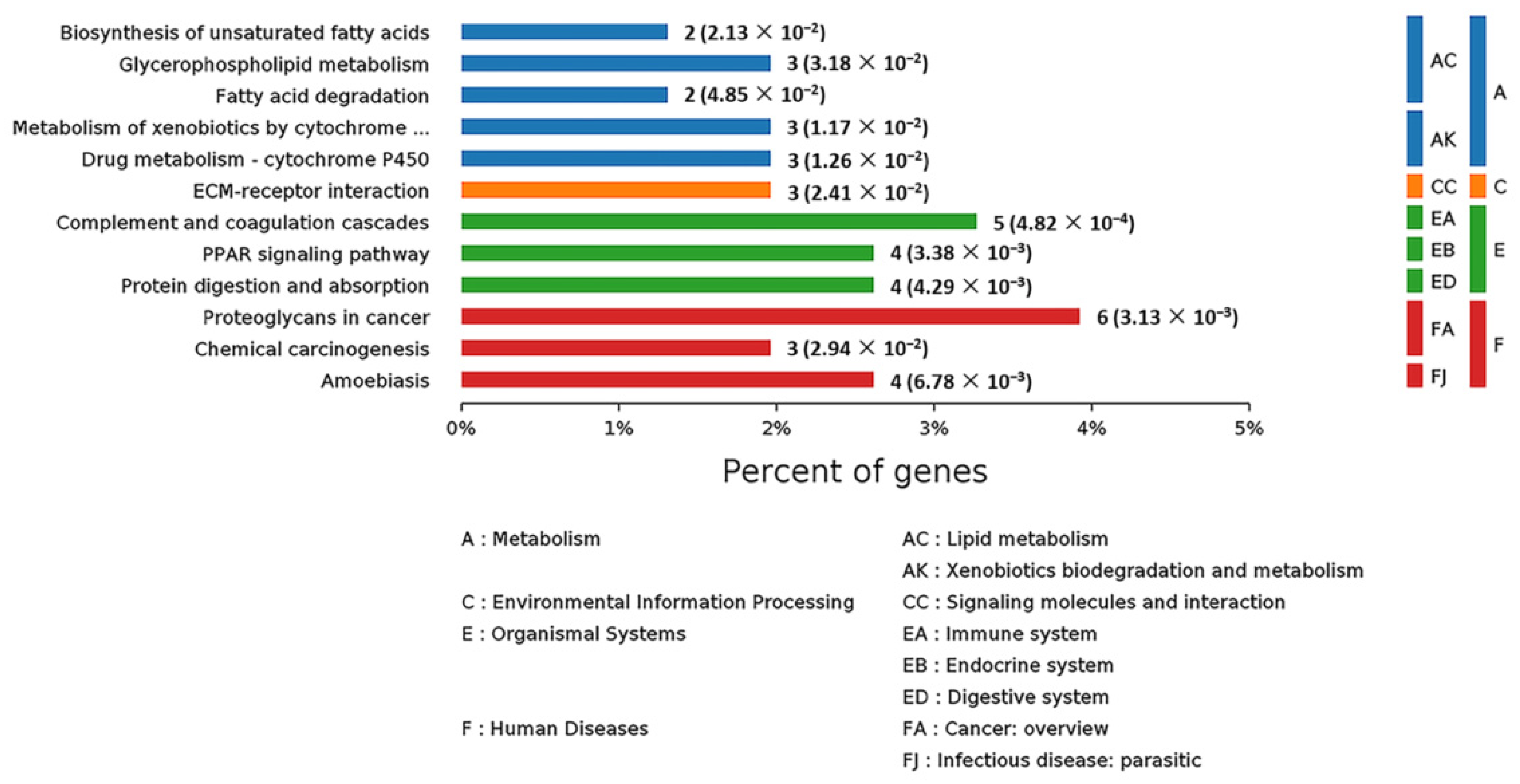
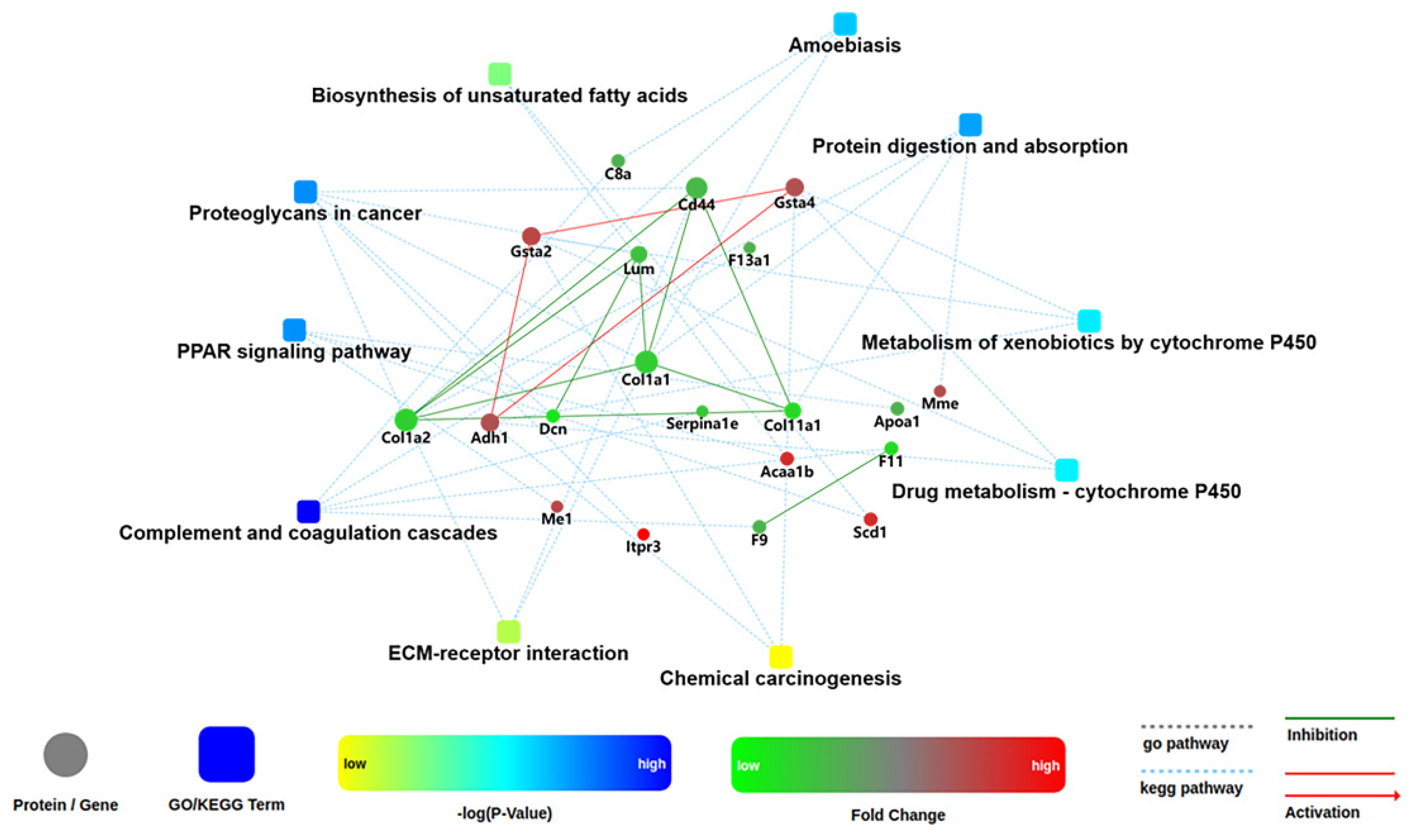
2.6. Analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways and Protein–Protein Interaction (PPI) among the DEPs
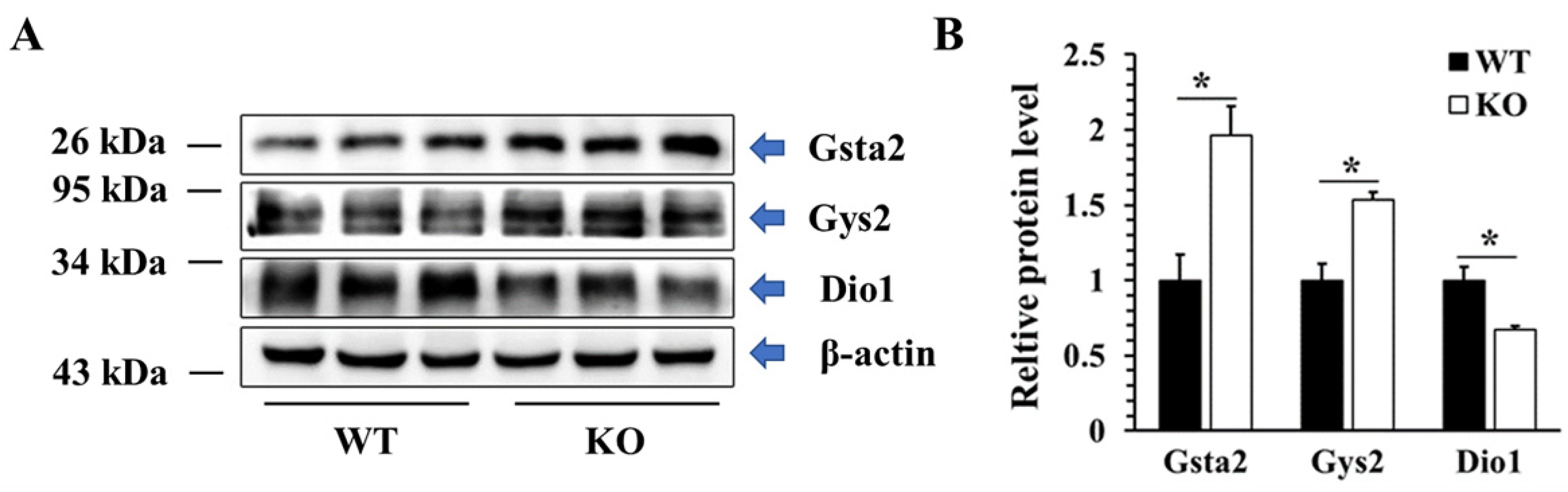
2.7. Validation of DEPs by Western Blotting
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Proteomic Analysis
4.2.1. Protein Extraction and Digestion
4.2.2. TMT Labeling
4.2.3. High pH Reversed-Phase Fractionation
4.2.4. LC-MS/MS Analysis
4.2.5. Identification and Quantitation of Proteins
4.3. Bioinformatics Analysis
4.3.1. Protein Cluster Analysis
4.3.2. Subcellular Location Analysis
4.3.3. Protein Structure Domain Analysis
4.3.4. GO Analysis, KEGG Pathway Analysis and PPI Network Analysis
4.4. Western Blot
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Combs, G.F., Jr.; Clark, L.C.; Turnbull, B.W. An analysis of cancer prevention by selenium. Biofactors 2001, 14, 153–159. [Google Scholar] [CrossRef]
- Rayman, M.P. Selenium in cancer prevention: A review of the evidence and mechanism of action. Proc. Nutr. Soc. 2005, 64, 527–542. [Google Scholar] [CrossRef] [Green Version]
- Whanger, P.D. Selenium and its relationship to cancer: An update. Br. J. Nutr. 2004, 91, 11–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Romero, F.J.; Kryukov, G.V.; Lobanov, A.V.; Carlson, B.A.; Lee, B.J.; Gladyshev, V.N.; Hatfield, D.L. Selenium metabolism in Drosophila: Selenoproteins, selenoprotein mRNA expression, fertility, and mortality. J. Biol. Chem. 2001, 276, 29798–29804. [Google Scholar] [CrossRef] [Green Version]
- Ursini, F.; Heim, S.; Kiess, M.; Maiorino, M.; Roveri, A.; Wissing, J.; Flohé, L. Dual function of the selenoprotein PHGPx during sperm maturation. Science 1999, 285, 1393–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef]
- Kieliszek, M.; Błażejak, S. Selenium: Significance, and outlook for supplementation. Nutrition 2013, 29, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigó, R.; Gladyshev, V.N. Characterization of Mammalian Selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef] [Green Version]
- Boukhzar, L.; Hamieh, A.; Cartier, D.; Tanguy, Y.; Alsharif, I.; Castex, M.; Arabo, A.; El Hajji, S.; Bonnet, J.J.; Errami, M.; et al. Selenoprotein T exerts an essential oxidoreductase activity that protects dopaminergic neurons in mouse models of Parkinson’s Disease. Antioxid. Redox Signal. 2016, 24, 557–574. [Google Scholar] [CrossRef] [PubMed]
- Anouar, Y.; Lihrmann, I.; Falluel-Morel, A.; Boukhzar, L. Selenoprotein T is a key player in ER proteostasis, endocrine homeostasis and neuroprotection. Free Radic. Biol. Med. 2018, 127, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Pothion, H.; Jehan, C.; Tostivint, H.; Cartier, D.; Bucharles, C.; Falluel-Morel, A.; Boukhzar, L.; Anouar, Y.; Lihrmann, I. Selenoprotein T: An Essential Oxidoreductase Serving as a Guardian of Endoplasmic Reticulum Homeostasis. Antioxid. Redox Signal. 2020, 33, 1257–1275. [Google Scholar] [CrossRef]
- Prevost, G.; Arabo, A.; Jian, L.; Quelennec, E.; Cartier, D.; Hassan, S.; Falluel-Morel, A.; Tanguy, Y.; Gargani, S.; Lihrmann, I.; et al. The PACAP-regulated gene selenoprotein T is abundantly expressed in mouse and human β-cells and its targeted inactivation impairs glucose tolerance. Endocrinology 2013, 154, 3796–3806. [Google Scholar] [CrossRef] [Green Version]
- Tanguy, Y.; Falluel-Morel, A.; Arthaud, S.; Boukhzar, L.; Manecka, D.L.; Chagraoui, A.; Prevost, G.; Elias, S.; Dorval-Coiffec, I.; Lesage, J.; et al. The PACAP-regulated gene selenoprotein T is highly induced in nervous, endocrine, and metabolic tissues during ontogenetic and regenerative processes. Endocrinology 2011, 152, 4322–4335. [Google Scholar] [CrossRef] [Green Version]
- Bian, Y.; Qin, C.; Xin, Y.; Yu, Y.; Chen, H.; Wang, G.; Xie, K.; Yu, Y. iTRAQ-based quantitative proteomic analysis of lungs in murine polymicrobial sepsis with hydrogen gas treatment. Shock 2018, 49, 187–195. [Google Scholar] [CrossRef] [PubMed]
- McAlister, G.C.; Huttlin, E.L.; Haas, W.; Ting, L.; Jedrychowski, M.P.; Rogers, J.C.; Kuhn, K.; Pike, I.; Grothe, R.A.; Blethrow, J.D.; et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 2012, 84, 7469–7478. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davi, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef] [Green Version]
- Dikiy, A.; Novoselov, S.V.; Fomenko, D.E.; Sengupta, A.; Carlson, B.A.; Cerny, R.L.; Ginalski, K.; Grishin, N.V.; Hatfield, D.L.; Gladyshev, V.N. SelT, SelW, SelH, and Rdx12: Genomics and molecular insights into the functions of selenoproteins of a novel thioredoxin-like family. Biochemistry 2007, 46, 6871–6882. [Google Scholar] [CrossRef] [PubMed]
- Grumolato, L.; Ghzili, H.; Montero-Hadjadje, M.; Gasman, S.; Lesage, J.; Tanguy, Y.; Galas, L.; Ait-Ali, D.; Leprince, J.; Guérineau, N.C.; et al. Selenoprotein T is a PACAP-regulated gene involved in intracellular Ca2+ mobilization and neuroendocrine secretion. FASEB J. 2008, 22, 1756–1768. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Bao, D.; Lei, C.L.; Tang, H.; Zhang, C.Y.; Su, H.; Zhang, C. Selenoprotein T protects against cisplatin-induced acute kidney injury through suppression of oxidative stress and apoptosis. FASEB J. 2020, 34, 11983–11996. [Google Scholar] [CrossRef]
- Merry, T.L.; Tran, M.; Stathopoulos, M.; Wiede, F.; Fam, B.C.; Dodd, G.T.; Clarke, I.; Watt, M.J.; Andrikopoulos, S.; Tiganis, T. High-Fat-Fed Obese Glutathione Peroxidase 1-Deficient Mice Exhibit Defective Insulin Secretion but Protection from Hepatic Steatosis and Liver Damage. Antioxid. Redox Signal. 2014, 20, 2114–2129. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Ren, B.; Wang, H.; Huang, R.; Zhou, J.; Liu, H.; Tian, J.; Huang, K. Hepatic proteomic analysis of selenoprotein F knockout mice by iTRAQ: An implication for the roles of selenoprotein F in metabolism and diseases. J. Proteom. 2020, 215, 103653. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Ren, B.; Li, X.; Yan, H.; Xie, Q.; Liu, H.; Zhou, J.; Tian, J.; Huang, K. Selenoprotein F Knockout Leads to Glucose and Lipid Metabolism Disorders in Mice. J. Biol. Inorg. Chem. 2020, 25, 1009–1022. [Google Scholar] [CrossRef] [PubMed]
- Raman, A.V.; Pitts, M.W.; Seyedali, A.; Hashimoto, A.C.; Seale, L.A.; Bellinger, F.P.; Berry, M.J. Absence of selenoprotein P but not selenocysteine lyase results in severe neurological dysfunction. Genes Brain Behav. 2012, 11, 601–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasaikina, M.V.; Hatfield, D.L.; Gladyshev, V.N. Understanding selenoprotein function and regulation through the use of rodent models. Biochim. Biophys. Acta 2012, 1823, 1633–1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castex, M.T.; Arabo, A.; Bénard, M.; Roy, V.; Le Joncour, V.; Prévost, G.; Bonnet, J.J.; Anouar, Y.; Falluel-Morel, A. Selenoprotein T Deficiency Leads to Neurodevelopmental Abnormalities and Hyperactive Behavior in Mice. Mol. Neurobiol. 2016, 53, 5818–5832. [Google Scholar] [CrossRef] [PubMed]
- Rocca, C.; Boukhzar, L.; Granieri, M.C.; Alsharif, I.; Mazza, R.; Lefranc, B.; Tota, B.; Leprince, J.; Cerra, M.C.; Anouar, Y.; et al. A selenoprotein T-derived peptide protects the heart against ischaemia/reperfusion injury through inhibition of apoptosis and oxidative stress. Acta Physiol. 2018, 223, e13067. [Google Scholar] [CrossRef]
- Moustafa, M.E.; Antar, H.A. A bioinformatics approach to characterize mammalian selenoprotein T. Biochem. Genet. 2012, 50, 736–747. [Google Scholar] [CrossRef]
- Liu, J.; Rozovsky, S. Membrane-bound selenoproteins. Antioxid. Redox Signal. 2015, 23, 795–813. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Förster, H.; Boersma, A.; Seiler, A.; Wehnes, H.; Sinowatz, F.; Neumüller, C.; Deutsch, M.J.; Walch, A.; Hrabé de Angelis, M.; et al. Mitochondrial glutathione peroxidase 4 disruption causes male infertility. FASEB J. 2009, 23, 3233–3242. [Google Scholar] [CrossRef] [Green Version]
- Braakman, I.; Bulleid, N.J. Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 2011, 80, 71–99. [Google Scholar] [CrossRef]
- Chen, S.L.; Zhang, C.Z.; Liu, L.L.; Lu, S.X.; Pan, Y.H.; Wang, C.H.; He, Y.F.; Lin, C.S.; Yang, X.; Xie, D.; et al. A GYS2/p53 Negative Feedback Loop Restricts Tumor Growth in HBV-Related Hepatocellular Carcinoma. Cancer Res. 2018, 79, 534–545. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Kim, M.S. The role of GSK3 in glucose homeostasis and the development of insulin resistance. Diabetes Res. Clin. Pract. 2007, 77, S49–S57. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhou, Y.; Liu, Y.; Ping, J.; Shou, Q.; Chen, F.; Ruo, R. Metformin improves hepatic IRS2/PI3K/Akt signaling in insulin-resistant rats of NASH and cirrhosis. J. Endocrinol. 2016, 229, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.Y.; Shi, C.X.; Gao, R.; Sun, H.J.; Xiong, X.Q.; Ding, L.; Chen, Q.; Li, Y.H.; Wang, J.J.; Kang, Y.M.; et al. Irisin inhibits hepatic gluconeogenesis and increases glycogen synthesis via the PI3K/Akt pathway in type 2 diabetic mice and hepatocytes. Clin. Sci. 2015, 129, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Koenig, R.J. Regulation of Type 1 Iodothyronine Deiodinase in Health and Disease. Thyroid 2005, 15, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Koenig, R.J. Induction of Type 1 Iodothyronine Deiodinase to Prevent the Nonthyroidal Illness Syndrome in Mice. Endocrinology 2006, 147, 3580–3585. [Google Scholar] [CrossRef] [Green Version]
- Beckett, G.J.; Beddows, S.E.; Morrice, P.C.; Nicol, F.; Arthur, J.R. Inhibition of hepatic deiodination of thyroxine is caused by selenium deficiency in rats. Biochem. J. 1987, 248, 443–447. [Google Scholar] [CrossRef] [Green Version]
- Beckett, G.J.; MacDougall, D.A.; Nicol, F.; Arthur, R. Inhibition of type I and type II iodothyronine deiodinase activity in rat liver, kidney and brain produced by selenium deficiency. Biochem. J. 1989, 259, 887–892. [Google Scholar] [CrossRef] [Green Version]
- Behne, D.; Kyriakopoulos, A.; Meinhold, H.; Köhrle, J. Identification of type I iodothyronine 5-deiodinase as a selenoenzyme. Biochem. Biophys. Res. Commun. 1990, 173, 1143–1149. [Google Scholar] [CrossRef]
- Baur, A.; Buchfelder, M.; Köhrle, J. Expression of 5-deiodinase enzymes in normal pituitaries and in various human pituitary adenomas. Eur. J. Endocrinol. 2002, 147, 263–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Short, S.P.; Whitten-Barrett, C.; Williams, C.S. Selenoprotein P in colitis-associated carcinoma. Mol. Cell. Oncol. 2015, 3, e1075094. [Google Scholar] [CrossRef]
- Hatfield, D.L.; Yoo, M.H.; Carlson, B.A.; Gladyshev, V.N. Selenoproteins that function in cancer prevention and promotion. Biochim. Biophys. Acta 2009, 1790, 1541–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, M.H.; Xu, X.M.; Carlson, B.A.; Patterson, A.D.; Gladyshev, V.N.; Hatfield, D.L. Targeting thioredoxin reductase 1 reduction in cancer cells inhibits self-sufficient growth and DNA replication. PLoS ONE 2007, 2, e1112. [Google Scholar] [CrossRef] [PubMed]
- Rundlöf, A.K.; Arnér, E.S. Regulation of the mammalian selenoprotein thioredoxin reductase 1 in relation to cellular phenotype, growth, and signaling events. Antioxid. Redox Signal. 2004, 6, 41–52. [Google Scholar] [CrossRef]
- Irons, R.; Tsuji, P.A.; Carlson, B.A.; Ouyang, P.; Yoo, M.H.; Xu, X.M.; Hatfield, D.L.; Gladyshev, V.N.; Davis, C.D. Deficiency in the 15-kDa selenoprotein inhibits tumorigenicity and metastasis of colon cancer cells. Cancer Prev. Res. 2010, 3, 630–639. [Google Scholar] [CrossRef] [Green Version]
- Tsuji, P.A.; Carlson, B.A.; Naranjo-Suarez, S.; Yoo, M.H.; Xu, X.M.; Fomenko, D.E.; Gladyshev, V.N.; Hatfield, D.L.; Davis, C.D. Knockout of the 15 kDa selenoprotein protects against chemically-induced aberrant crypt formation in mice. PLoS ONE 2012, 7, e50574. [Google Scholar] [CrossRef]
- Romanelli-Cedrez, L.; Carrera, I.; Otero, L.; Miranda-Vizuete, A.; Mariotti, M.; Alkema, M.J.; Salinas, G. Selenoprotein T is required for pathogenic bacteria avoidance in Caenorhabditis elegans. Free Radic. Biol. Med. 2017, 108, 174–182. [Google Scholar] [CrossRef]
- Kang, K.W.; Lee, S.J.; Kim, S.G. Molecular mechanism of Nrf2 activation by oxidative stress, Antioxid. Redox Signal. 2005, 7, 1664–1673. [Google Scholar] [CrossRef]
- Liu, J.; Liu, H.; Zhao, Z.; Wang, J.; Guo, D.; Liu, Y. Regulation of Actg1 and Gsta2 is possible mechanism by which capsaicin alleviates apoptosis in cell model of 6-OHDA-induced Parkinson’s disease. Biosci. Rep. 2020, 40, BSR20191796. [Google Scholar] [CrossRef]
- Boušová, I.; Košťáková, Š.; Matoušková, P.; Bártíková, H.; Szotáková, B.; Skálová, L. Monosodium glutamate-induced obesity changed the expression and activity of glutathione S-transferases in mouse heart and kidney. Pharmazie 2017, 72, 257–259. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Niu, L.; Li, Q. Proteomics Analysis Reveals an Important Role for the PPAR Signaling Pathway in DBDCT-Induced Hepatotoxicity Mechanisms. Molecules 2017, 22, 1113. [Google Scholar] [CrossRef] [Green Version]
- Vinceti, M.; Filippini, T.; Wise, L.A. Environmental Selenium and Human Health: An Update. Curr. Environ. Health Rep. 2018, 5, 464–485. [Google Scholar] [CrossRef]
- Vinceti, M.; Filippini, T.; Rothman, K.J. Selenium exposure and the risk of type 2 diabetes: A systematic review and meta-analysis. Eur. J. Epidemiol. 2018, 33, 789–810. [Google Scholar] [CrossRef]
- Vinceti, M.; Filippini, T.; Wise, L.A.; Rothman, K.J. A systematic review and dose-response meta-analysis of exposure to environmental selenium and the risk of type 2 diabetes in nonexperimental studies. Environ. Res. 2021, 197, 111210. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Huang, K.; Lei, X.G. Selenium and diabetes—evidence from animal studies. Free Radic. Biol. Med. 2013, 65, 1548–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa-Wong, A.N.; Berry, M.J.; Seale, L.A. Selenium and Metabolic Disorders: An Emphasis on Type 2 Diabetes Risk. Nutrients 2016, 8, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, Y. Selenoprotein P as a significant regulator of pancreatic beta cell function. J. Biochem. 2020, 167, 119–124. [Google Scholar] [CrossRef]
- Saito, Y. Selenoprotein P as an in vivo redox regulator: Disorders related to its deficiency and excess. J. Clin. Biochem. Nutr. 2020, 66, 19–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsutsumi, R.; Saito, Y. Selenoprotein P; P for Plasma, Prognosis, Prophylaxis, and More. Biol. Pharm. Bull. 2020, 43, 366–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takamura, T. Hepatokine Selenoprotein P-mediated reductive stress causes resistance to intracellular signal transduction. Antioxid. Redox Signal. 2020, 33, 517–524. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]




| Uniprot ID | Gene Name | Fold Change KO/WT | Protein Name | p Value |
|---|---|---|---|---|
| P70227 | Itpr3 | 1.7142 | Inositol 1,4,5-trisphosphate receptor type 3 | 0.0193 |
| Q810C0 | Slitrk2 | 1.5838 | SLIT and NTRK-like protein 2 | 0.0240 |
| Q99JH8 | Kdelr1 | 1.5758 | ER lumen protein-retaining receptor 1 | 0.0344 |
| P19426 | Nelfe | 1.5667 | Negative elongation factor E | 0.0003 |
| Q80V94 | Ap4e1 | 1.4803 | AP-4 complex subunit epsilon-1 | 0.0019 |
| Q8C4X7 | Minar2 | 1.4486 | Major intrinsically disordered NOTCH2-binding receptor 1-like homolog | 0.0069 |
| Q8VCH0 | Acaa1b | 1.4334 | 3-ketoacyl-CoA thiolase B, peroxisomal | 0.0057 |
| Q80X71 | Tmem106b | 1.4200 | Transmembrane protein 106B | 0.0133 |
| P13516 | Scd1 | 1.4148 | Acyl-CoA desaturase 1 | 0.0033 |
| Q9CR30 | Josd2 | 1.3915 | Josephin-2 | 0.0100 |
| Q6P1E7 | Primpol | 1.3635 | DNA-directed primase/polymerase protein | 0.0301 |
| Q9EQQ2 | Yipf5 | 1.3481 | Protein YIPF5 | 0.0472 |
| Q80ZJ8 | Cracr2b | 1.3299 | EF-hand calcium-binding domain-containing protein 4A | 0.0473 |
| E9Q2M9 | Wdfy4 | 1.3271 | WD repeat- and FYVE domain-containing protein 4 | 0.0395 |
| Q9JIA7 | Sphk2 | 1.3167 | Sphingosine kinase 2 | 0.0019 |
| Q8BGS7 | Cept1 | 1.3154 | Choline/ethanolaminephosphotransferase 1 | 0.0014 |
| Q9JMI0 | Ecel1 | 1.3130 | Endothelin-converting enzyme-like 1 | 0.0143 |
| Q6P2L6 | Nsd3 | 1.3102 | Histone-lysine N-methyltransferase NSD3 | 0.0247 |
| Q02242 | Pdcd1 | 1.3066 | Programmed cell death protein 1 | 0.0151 |
| Q920M5 | Coro6 | 1.2939 | Coronin-6 | 0.0038 |
| Uniprot ID | Gene Name | Fold Change KO/WT | Protein Name | p Value |
|---|---|---|---|---|
| Q61646 | Hp | 0.7401 | Haptoglobin | 0.0418 |
| Q71KU9 | Fgl1 | 0.7342 | Fibrinogen-like protein 1 | 0.0267 |
| Q01149 | Col1a2 | 0.7313 | Collagen alpha-2(I) chain | 0.0054 |
| P11588 | Mup1 | 0.7303 | Major urinary protein 1 | 0.0464 |
| A2AAE1 | Kiaa1109 | 0.7279 | Uncharacterized protein KIAA1109 | 0.0259 |
| C0HKD8 | Mfap1a | 0.7176 | Microfibrillar-associated protein 1A | 0.0002 |
| Q8BL00 | Cdhr3 | 0.7118 | Cadherin-related family member 3 | 0.0243 |
| Q1ERP8 | Cd300lg | 0.7068 | CMRF35-like molecule 9 | 0.0384 |
| Q61245 | Col11a1 | 0.7067 | Collagen alpha-1(XI) chain | 0.0078 |
| Q91Y47 | F11 | 0.7040 | Coagulation factor XI | 0.0253 |
| Q91WR5 | Akr1c21 | 0.7019 | Aldo-keto reductase family 1 member C21 | 0.0166 |
| P97315 | Csrp1 | 0.6954 | Cysteine and glycine-rich protein 1 | 0.0391 |
| Q9D304 | Rnf128 | 0.6938 | E3 ubiquitin-protein ligase RNF128 | 0.0049 |
| P09602 | Hmgn2 | 0.6922 | Non-histone chromosomal protein HMG-17 | 0.0214 |
| P28654 | Dcn | 0.6919 | Decorin | 0.0339 |
| Q9QY24 | Zbp1 | 0.6706 | Z-DNA-binding protein 1 | 0.0435 |
| P14602 | Hspb1 | 0.6493 | Heat shock protein beta-1 | 0.0057 |
| A2ASS6 | Ttn | 0.6033 | Titin | 0.0182 |
| Q63836 | Selenbp2 | 0.5763 | Selenium-binding protein 2 | 0.0045 |
| P02802 | Mt1 | 0.5208 | Metallothionein-1 | 0.0019 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Feng, T.; Liu, L.; Liu, H.; Huang, K.; Zhou, J. Hepatic Proteomic Analysis of Selenoprotein T Knockout Mice by TMT: Implications for the Role of Selenoprotein T in Glucose and Lipid Metabolism. Int. J. Mol. Sci. 2021, 22, 8515. https://doi.org/10.3390/ijms22168515
Li K, Feng T, Liu L, Liu H, Huang K, Zhou J. Hepatic Proteomic Analysis of Selenoprotein T Knockout Mice by TMT: Implications for the Role of Selenoprotein T in Glucose and Lipid Metabolism. International Journal of Molecular Sciences. 2021; 22(16):8515. https://doi.org/10.3390/ijms22168515
Chicago/Turabian StyleLi, Ke, Tiejun Feng, Leyan Liu, Hongmei Liu, Kaixun Huang, and Jun Zhou. 2021. "Hepatic Proteomic Analysis of Selenoprotein T Knockout Mice by TMT: Implications for the Role of Selenoprotein T in Glucose and Lipid Metabolism" International Journal of Molecular Sciences 22, no. 16: 8515. https://doi.org/10.3390/ijms22168515
APA StyleLi, K., Feng, T., Liu, L., Liu, H., Huang, K., & Zhou, J. (2021). Hepatic Proteomic Analysis of Selenoprotein T Knockout Mice by TMT: Implications for the Role of Selenoprotein T in Glucose and Lipid Metabolism. International Journal of Molecular Sciences, 22(16), 8515. https://doi.org/10.3390/ijms22168515