Pyranoanthocyanins Interfering with the Quorum Sensing of Pseudomonas aeruginosa and Staphylococcus aureus
Abstract
:1. Introduction
2. Results
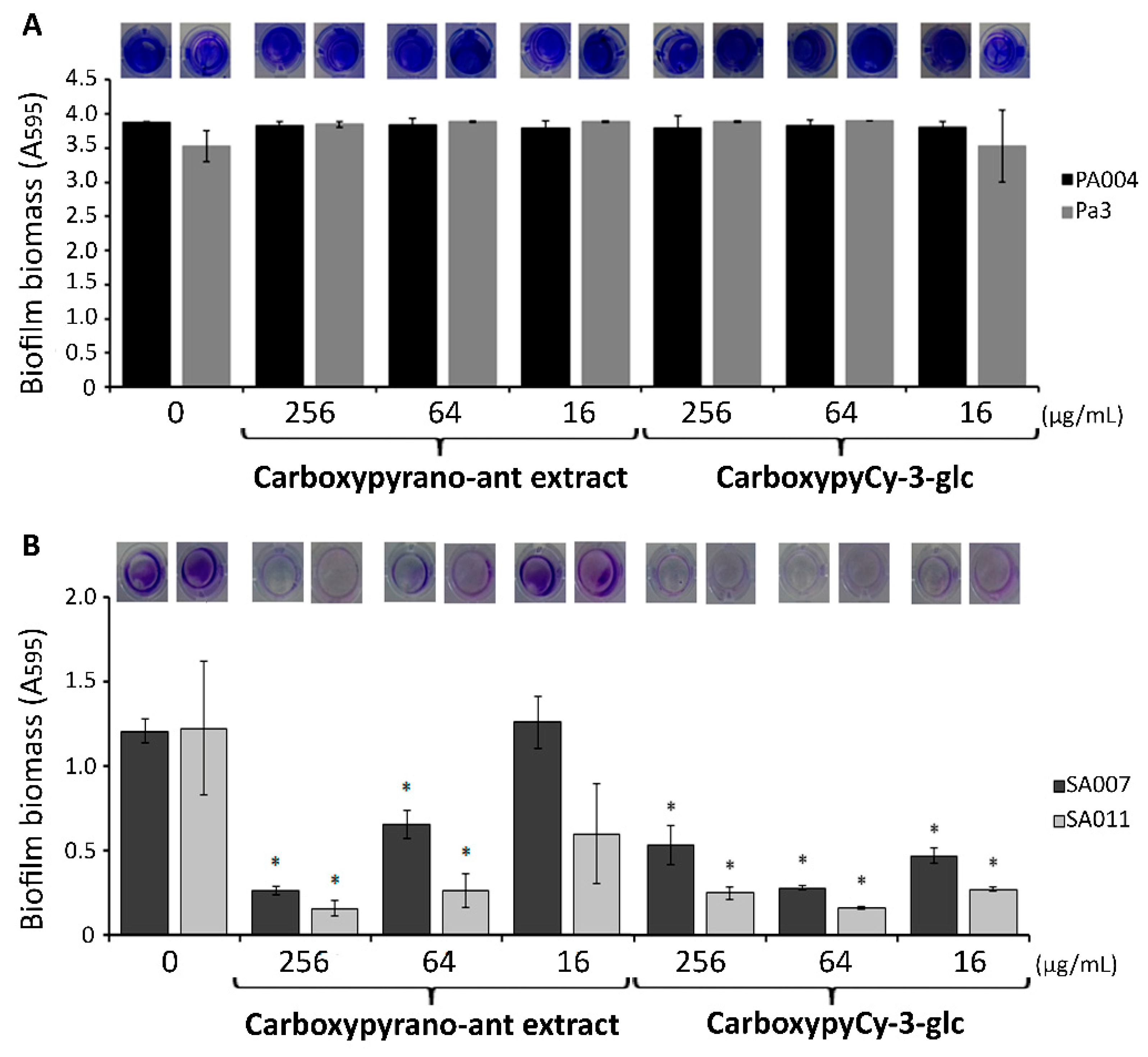
2.1. Antimicrobial and Biofilm Inhibition Activities of Carboxypyrano-ant Extract and CarboxypyCy-3-glc against P. aeruginosa and S. aureus MDR Isolates
2.2. Impact of Carboxypyrano-ant Extract and CarboxypyCy-3-glc on the Biofilm Formation—Qualitative Microscopic Analysis
2.3. Interference of Carboxypyrano-ant Extract and CarboxypyCy-3-glc with the QS-Mediated Production of Violacein by C. violaceum
2.4. Interference of Carboxypyrano-ant Extract and CarboxypyCy-3-glc with the Expression of QS-Regulated Genes in P. aeruginosa and S. aureus Biofilms
2.5. Toxicity and Assessment of Carboxypyrano-ant Extract and CarboxypyCy-3-glc in G. mellonella
3. Discussion
4. Materials and Methods
4.1. Synthesis of Carboxypyranocyanidin-3-O-glucoside and Carboxypyranoanthocyanins Extract
4.2. Bacterial Strains and Growth Conditions
4.3. Minimum Inhibitory Concentration (MIC) Assay and Biofilm Formation Assay against MDR Isolates
4.4. Microscopic Visualization of Biofilm Formation
4.5. Effects on Violacein Production by C. violaceum
4.6. Biofilm Growth for RNA Extraction
4.7. RNA Extraction
4.7.1. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR) for P. aeruginosa
4.7.2. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR) for S. aureus
4.8. Galleria Mellonella Larvae Toxicity Assay
4.9. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations The Review on Antimicrobial Resistance. 2016. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 15 April 2020).
- WHO. Global Action Plan on Antimicrobial Resistance. Microbe 2015, 10, 354–355. [Google Scholar] [CrossRef]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 20 October 2020).
- Rossiter, S.E.; Fletcher, M.H.; Wuest, W.M. Natural Products as Platforms to Overcome Antibiotic Resistance. Chem. Rev. 2017, 117, 12415–12474. [Google Scholar] [CrossRef]
- Dodds, D.R. Antibiotic Resistance: A Current Epilogue. Biochem. Pharmacol. 2017, 134, 139–146. [Google Scholar] [CrossRef]
- Allegretta, G.; Maurer, C.K.; Eberhard, J.; Maura, D.; Hartmann, R.W.; Rahme, L.; Empting, M. In-Depth Profiling of MvfR-Regulated Small Molecules in Pseudomonas aeruginosa after Quorum Sensing Inhibitor Treatment. Front. Microbiol. 2017, 8, 924. [Google Scholar] [CrossRef]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9, e02331-17. [Google Scholar] [CrossRef] [Green Version]
- Bäuerle, T.; Fischer, A.; Speck, T.; Bechinger, C. Self-Organization of Active Particles by Quorum Sensing Rules. Nat. Commun. 2018, 9, 3232. [Google Scholar] [CrossRef] [PubMed]
- Turan, N.B.; Chormey, D.S.; Büyükpınar, Ç.; Engin, G.O.; Bakirdere, S. Quorum Sensing: Little Talks for an Effective Bacterial Coordination. TrAC Trend Anal. Chem. 2017, 91, 1–11. [Google Scholar] [CrossRef]
- Butrico, C.E.; Cassat, J.E. Quorum Sensing and Toxin Production in Staphylococcus aureus Osteomyelitis: Pathogenesis and Paradox. Toxins 2020, 12, 516. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum Sensing Signal-Response Systems in Gram-Negative Bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Geng, M.; Bai, L. Targeting Biofilms Therapy: Current Research Strategies and Development Hurdles. Microorganisms 2020, 8, 1222. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Chen, J.; Yang, C.; Yin, Y.; Yao, K.; Song, D. Quorum Sensing: A Prospective Therapeutic Target for Bacterial Diseases. Biomed. Res. Int. 2019, 2019, 2015978. [Google Scholar] [CrossRef] [Green Version]
- Paczkowski, J.E.; Mukherjee, S.; McCready, A.R.; Cong, J.P.; Aquino, C.J.; Kim, H.; Henke, B.R.; Smith, C.D.; Bassler, B.L. Flavonoids Suppress Pseudomonas aeruginosa Virulence through Allosteric Inhibition of Quorum-Sensing Receptors. Int. J. Biol. Chem. 2017, 292, 4064–4076. [Google Scholar] [CrossRef] [Green Version]
- Palliyil, S.; Downham, C.; Broadbent, I.; Charlton, K.; Porter, A.J. High-Sensitivity Monoclonal Antibodies Specific for Homoserine Lactones Protect Mice from Lethal Pseudomonas aeruginosa Infections. Appl Environ. Microbiol. 2014, 80, 462–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soh, E.Y.C.; Chhabra, S.R.; Halliday, N.; Heeb, S.; Müller, C.; Birmes, F.S.; Fetzner, S.; Cámara, M.; Chan, K.G.; Williams, P. Biotic Inactivation of the Pseudomonas aeruginosa Quinolone Signal Molecule. Environ. Microbiol. 2015, 17, 4352–4365. [Google Scholar] [CrossRef] [PubMed]
- Truchado, P.; Larrosa, M.; Castro-Ibáñez, I.; Allende, A. Plant Food Extracts and Phytochemicals: Their Role as Quorum Sensing Inhibitors. Trends Food Sci. Technol. 2015, 43, 189–204. [Google Scholar] [CrossRef]
- Gopu, V.; Kothandapani, S.; Shetty, P.H. Quorum Quenching Activity of Syzygium cumini (L.) Skeels and Its Anthocyanin Malvidin against Klebsiella pneumoniae. Microb. Pathog. 2015, 79, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Deryabin, D.; Galadzhieva, A.; Kosyan, D.; Duskaev, G. Plant-Derived Inhibitors of AHL-Mediated Quorum Sensing in Bacteria: Modes of Action. Int. J. Mol. Sci. 2019, 20, 5588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopu, V.; Shetty, P.H. Cyanidin Inhibits Quorum Signalling Pathway of a Food Borne Opportunistic Pathogen. J. Food Sci. Technol. 2016, 53, 968–976. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, F.; Bao, X.; Feng, J.; Fu, L. Inhibition of Biogenic Amines in Shewanella baltica by Anthocyanins Involving a Quorum Sensing System. J. Food Prot. 2019, 82, 589–596. [Google Scholar] [CrossRef]
- Pejin, B.; Ciric, A.; Dimitric Markovic, J.; Glamoclija, J.; Nikolic, M.; Sokovic, M. An Insight into Anti-Biofilm and Anti-Quorum Sensing Activities of the Selected Anthocyanidins: The Case Study of Pseudomonas aeruginosa PAO1. Nat. Prod. Res. 2017, 31, 1177–1180. [Google Scholar] [CrossRef]
- Kordbacheh, H.; Eftekhar, F.; Ebrahimi, S.N. Anti-Quorum Sensing Activity of Pistacia atlantica against Pseudomonas aeruginosa PAO1 and Identification of Its Bioactive Compounds. Microb. Pathog. 2017, 110, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Correia, P.; Araújo, P.; Ribeiro, C.; Oliveira, H.; Pereira, A.R.; Mateus, N.; de Freitas, V.; Brás, N.F.; Gameiro, P.; Coelho, P.; et al. Anthocyanin-Related Pigments: Natural Allies for Skin Health Maintenance and Protection. Antioxidants 2021, 10, 1038. [Google Scholar] [CrossRef] [PubMed]
- Monnet, V.; Gardan, R. Quorum-Sensing Regulators in Gram-Positive Bacteria: “Cherchez Le Peptide. ” Mol. Microbiol. 2015, 97, 181–184. [Google Scholar] [CrossRef]
- Hemmati, F.; Salehi, R.; Ghotaslou, R.; Kafil, H.S.; Hasani, A.; Gholizadeh, P.; Nouri, R.; Rezaee, M.A. Quorum Quenching: A Potential Target for Antipseudomonal Therapy. Infect. Drug Resist. 2020, 13, 2989–3005. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Li, S.; Chen, S.; Fan, J.; Cao, Z.; Ouyang, W.; Tong, N.; Hu, X.; Hu, J.; Li, P.; Feng, Z.; et al. Anti-Biofilm Effect of Novel Thiazole Acid Analogs against Pseudomonas aeruginosa through IQS Pathways. Eur. J. Med. Chem. 2018, 145, 64–73. [Google Scholar] [CrossRef]
- Fong, J.; Yuan, M.; Jakobsen, T.H.; Mortensen, K.T.; Delos Santos, M.M.S.; Chua, S.L.; Yang, L.; Tan, C.H.; Nielsen, T.E.; Givskov, M. Disulfide Bond-Containing Ajoene Analogues As Novel Quorum Sensing Inhibitors of Pseudomonas aeruginosa. J. Med. Chem. 2017, 60, 215–227. [Google Scholar] [CrossRef]
- Pérez-Pérez, M.; Jorge, P.; Pérez Rodríguez, G.; Pereira, M.O.; Lourenço, A. Quorum Sensing Inhibition in Pseudomonas aeruginosa Biofilms: New Insights through Network Mining. Biofouling 2017, 33, 128–142. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Zhou, L.; Jin, K.; Jiang, H.; He, Y.W. Quorum Sensing Systems Differentially Regulate the Production of Phenazine-1-Carboxylic Acid in the Rhizobacterium Pseudomonas aeruginosa PA1201. Sci. Rep. 2016, 6, 30352. [Google Scholar] [CrossRef] [PubMed]
- Feltner, J.B.; Wolter, D.J.; Pope, C.E.; Groleau, M.C.; Smalley, N.E.; Greenberg, E.P.; Mayer-Hamblett, N.; Burns, J.; Déziel, E.; Hoffman, L.R.; et al. LasR Variant Cystic Fibrosis Isolates Reveal an Adaptable Quorum-Sensing Hierarchy in Pseudomonas aeruginosa. mBio 2016, 7, e01513-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Zhang, L. The Hierarchy Quorum Sensing Network in Pseudomonas aeruginosa. Protein Cell 2014, 6, 26–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus Virulence. Microbiol. Spectr. 2019, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Lin, Y.Y.; Tu, C.C.; Lan, C.Y. Distribution of Different Efflux Pump Genes in Clinical Isolates of Multidrug-Resistant Acinetobacter baumannii and Their Correlation with Antimicrobial Resistance. J. Microbiol. Immunol. Infect. 2017, 50, 224–231. [Google Scholar] [CrossRef] [Green Version]
- Martinez, J.L. General Principles of Antibiotic Resistance in Bacteria. Drug Discov. Today Technol. 2014, 11, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Pontes, D.S.; de Araujo, R.S.A.; Dantas, N.; Scotti, L.; Scotti, M.T.; de Moura, R.O.; Mendonca-Junior, F.J.B. Genetic Mechanisms of Antibiotic Resistance and the Role of Antibiotic Adjuvants. Curr. Top. Med. Chem. 2018, 18, 42–74. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.S.; Lee, J.H.; Cho, M.H.; Lee, J. Red Wines and Flavonoids Diminish Staphylococcus aureus Virulence with Anti-Biofilm and Anti-Hemolytic Activities. Biofouling 2015, 31, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Lee, J.H.; Beyenal, H.; Lee, J. Fatty Acids as Antibiofilm and Antivirulence Agents. Trends Microbiol. 2020, 28, 753–768. [Google Scholar] [CrossRef] [PubMed]
- Saidi, N.; Owlia, P.; Marashi, S.M.A.; Saderi, H. Inhibitory Effect of Probiotic Yeast Saccharomyces cerevisiae on Biofilm Formation and Expression of α-Hemolysin and Enterotoxin A Genes of Staphylococcus aureus. Iran. J. Microbiol. 2019, 11, 246–254. [Google Scholar] [CrossRef] [Green Version]
- Kothari, V.; Sharma, S.; Padia, D. Recent Research Advances on Chromobacterium violaceum. Asian Pac. J. Trop. Med. 2017, 10, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Devescovi, G.; Kojic, M.; Covaceuszach, S.; Cámara, M.; Williams, P.; Bertani, I.; Subramoni, S.; Venturi, V. Negative Regulation of Violacein Biosynthesis in Chromobacterium violaceum. Front. Microbiol. 2017, 8, 349. [Google Scholar] [CrossRef] [Green Version]
- Mok, N.; Chan, S.Y.; Liu, S.Y.; Chua, S.L. Vanillin Inhibits PqsR-Mediated Virulence in Pseudomonas aeruginosa. Food Funct. 2020, 11, 6496–6508. [Google Scholar] [CrossRef]
- Groleau, M.-C.; de Oliveira Pereira, T.; Dekimpe, V.; Déziel, E. PqsE Is Essential for RhlR-Dependent Quorum Sensing Regulation in Pseudomonas aeruginosa. mSystems 2020, 5, e00194-20. [Google Scholar] [CrossRef]
- Mukherjee, S.; Moustafa, D.A.; Stergioula, V.; Smith, C.D.; Goldberg, J.B.; Bassler, B.L. The PqsE and RhlR Proteins Are an Autoinducer Synthase–Receptor Pair That Control Virulence and Biofilm Development in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2018, 115, 9411–9418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostylev, M.; Kim, D.Y.; Smalley, N.E.; Salukhe, I.; Peter Greenberg, E.; Dandekar, A.A. Evolution of the Pseudomonas aeruginosa Quorum-Sensing Hierarchy. Proc. Natl. Acad. Sci. USA 2019, 116, 7027–7032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okkotsu, Y.; Tieku, P.; Fitzsimmons, L.F.; Churchill, M.E.; Schurr, M.J. Pseudomonas aeruginosa AlgR Phosphorylation Modulates Rhamnolipid Production and Motility. J. Bacteriol. 2013, 195, 5499–5515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Déziel, E.; Groleau, M.-C.; Schaefer, A.L.; Greenberg, E.P. Social Cheating in a Pseudomonas aeruginosa Quorum-Sensing Variant. Proc. Natl. Acad. Sci. USA 2019, 116, 7021–7026. [Google Scholar] [CrossRef] [Green Version]
- Dastgheyb, S.S.; Villaruz, A.E.; Le, K.Y.; Tan, V.Y.; Duong, A.C.; Chatterjee, S.S.; Cheung, G.Y.C.; Joo, H.S.; Hickok, N.J.; Otto, M. Role of Phenol-Soluble Modulins in Formation of Staphylococcus aureus Biofilms in Synovial Fluid. Infect. Immun. 2015, 83, 2966–2975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Khan, B.A.; Cheung, G.Y.C.; Bach, T.H.L.; Jameson-Lee, M.; Kong, K.F.; Queck, S.Y.; Otto, M. Staphylococcus epidermidis Surfactant Peptides Promote Biofilm Maturation and Dissemination of Biofilm-Associated Infection in Mice. J. Clin. Investig. 2011, 121, 238–248. [Google Scholar] [CrossRef]
- Zielinska, A.K.; Beenken, K.E.; Joo, H.S.; Mrak, L.N.; Griffin, L.M.; Luong, T.T.; Lee, C.Y.; Otto, M.; Shaw, L.N.; Smeltzer, M.S. Defining the Strain-Dependent Impact of the Staphylococcal accessory regulator (SarA) on the Alpha-Toxin Phenotype of Staphylococcus aureus. J. Bacteriol. 2011, 193, 2948–2958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omidi, M.; Firoozeh, F.; Saffari, M.; Sedaghat, H.; Zibaei, M.; Khaledi, A. Ability of Biofilm Production and Molecular Analysis of spa and ica Genes among Clinical Isolates of Methicillin-Resistant Staphylococcus aureus. BMC Res. Notes 2020, 13, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montgomery, C.P.; Boyle-Vavra, S.; Roux, A.; Ebine, K.; Sonenshein, A.L.; Daumb, R.S. CodY Deletion Enhances in vivo Virulence of Community-Associated Methicillin-Resistant Staphylococcus aureus Clone USA300. Infect. Immun. 2012, 80, 2382–2389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassat, J.E.; Hammer, N.D.; Campbell, J.P.; Benson, M.A.; Perrien, D.S.; Mrak, L.N.; Smeltzer, M.S.; Torres, V.J.; Skaar, E.P. A Secreted Bacterial Protease Tailors the Staphylococcus aureus Virulence Repertoire to Modulate Bone Remodeling during Osteomyelitis. Cell Host Microbe 2013, 13, 759–772. [Google Scholar] [CrossRef] [Green Version]
- Kolar, S.L.; Antonio Ibarra, J.; Rivera, F.E.; Mootz, J.M.; Davenport, J.E.; Stevens, S.M.; Horswill, A.R.; Shaw, L.N. Extracellular Proteases Are Key Mediators of Staphylococcus aureus Virulence via the Global Modulation of Virulence-Determinant Stability. Microbiol. 2013, 2, 18–34. [Google Scholar] [CrossRef]
- Lozano, C.; Azcona-Gutiérrez, J.M.; van Bambeke, F.; Sáenz, Y. Great Phenotypic and Genetic Variation among Successive Chronic Pseudomonas aeruginosa from a Cystic Fibrosis Patient. PLoS ONE 2018, 13, e0204167. [Google Scholar] [CrossRef]
- Tan, L.; Li, S.R.; Jiang, B.; Hu, X.M.; Li, S. Therapeutic Targeting of the Staphylococcus aureus Accessory Gene Regulator (Agr) System. Front. Microbiol. 2018, 9, 55. [Google Scholar] [CrossRef]
- Oliveira, J.; Fernandes, V.; Miranda, C.; Santos-Buelga, C.; Silva, A.; de Freitas, V.; Mateus, N. Color Properties of Four Cyanidin-Pyruvic Acid Adducts. J. Agric. Food Chem. 2006, 54, 6894–6903. [Google Scholar] [CrossRef] [PubMed]
- Bessa, L.J.; Eaton, P.; Dematei, A.; Plácido, A.; Vale, N.; Gomes, P.; Delerue-Matos, C.; Leite, J.R.S.A.; Gameiro, P. Synergistic and Antibiofilm Properties of Ocellatin Peptides against Multidrug-Resistant Pseudomonas aeruginosa. Future Microbiol. 2018, 13, 151–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| PA004 | Pa3 | SA007 | SA011 | |
|---|---|---|---|---|
| MIC (MBC) | ||||
| Carboxypyrano-ant extract | >512 (−) | 512 (>512) | >512 (−) | >512 (−) |
| CarboxypyCy-3-glc | >512 (−) | >512 (−) | >512 (−) | >512 (−) |
| Components | (mg/mg ± SD) |
|---|---|
| Pyranoanthocyanins | 0.18 ± 0.002 |
| Low weight polyphenols | 0.010 ± 0.002 |
| Flavonols | 0.056 ± 0.006 |
| Protein content | 0.096 ± 0.002 |
| Total lipids | 0.05 ± 0.01 |
| Sugar content | 0.0184 ± 0.0023 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coelho, P.; Oliveira, J.; Fernandes, I.; Araújo, P.; Pereira, A.R.; Gameiro, P.; Bessa, L.J. Pyranoanthocyanins Interfering with the Quorum Sensing of Pseudomonas aeruginosa and Staphylococcus aureus. Int. J. Mol. Sci. 2021, 22, 8559. https://doi.org/10.3390/ijms22168559
Coelho P, Oliveira J, Fernandes I, Araújo P, Pereira AR, Gameiro P, Bessa LJ. Pyranoanthocyanins Interfering with the Quorum Sensing of Pseudomonas aeruginosa and Staphylococcus aureus. International Journal of Molecular Sciences. 2021; 22(16):8559. https://doi.org/10.3390/ijms22168559
Chicago/Turabian StyleCoelho, Patrícia, Joana Oliveira, Iva Fernandes, Paula Araújo, Ana Rita Pereira, Paula Gameiro, and Lucinda J. Bessa. 2021. "Pyranoanthocyanins Interfering with the Quorum Sensing of Pseudomonas aeruginosa and Staphylococcus aureus" International Journal of Molecular Sciences 22, no. 16: 8559. https://doi.org/10.3390/ijms22168559
APA StyleCoelho, P., Oliveira, J., Fernandes, I., Araújo, P., Pereira, A. R., Gameiro, P., & Bessa, L. J. (2021). Pyranoanthocyanins Interfering with the Quorum Sensing of Pseudomonas aeruginosa and Staphylococcus aureus. International Journal of Molecular Sciences, 22(16), 8559. https://doi.org/10.3390/ijms22168559










