Identification of Potential Probiotics Producing Bacteriocins Active against Listeria monocytogenes by a Combination of Screening Tools
Abstract
:1. Introduction
2. Results
2.1. Isolation and Typing of LAB from Raw Cow Milk
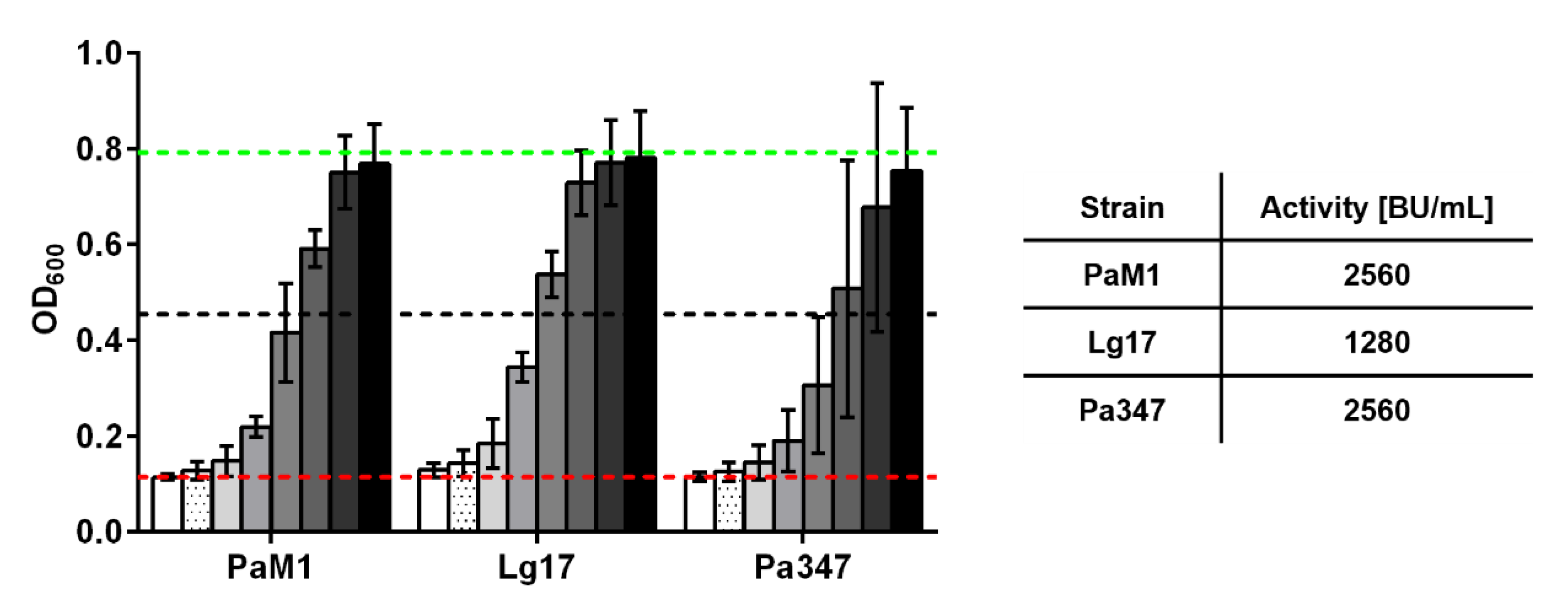
2.2. Screening of Supernatants for Antimicrobial Activity
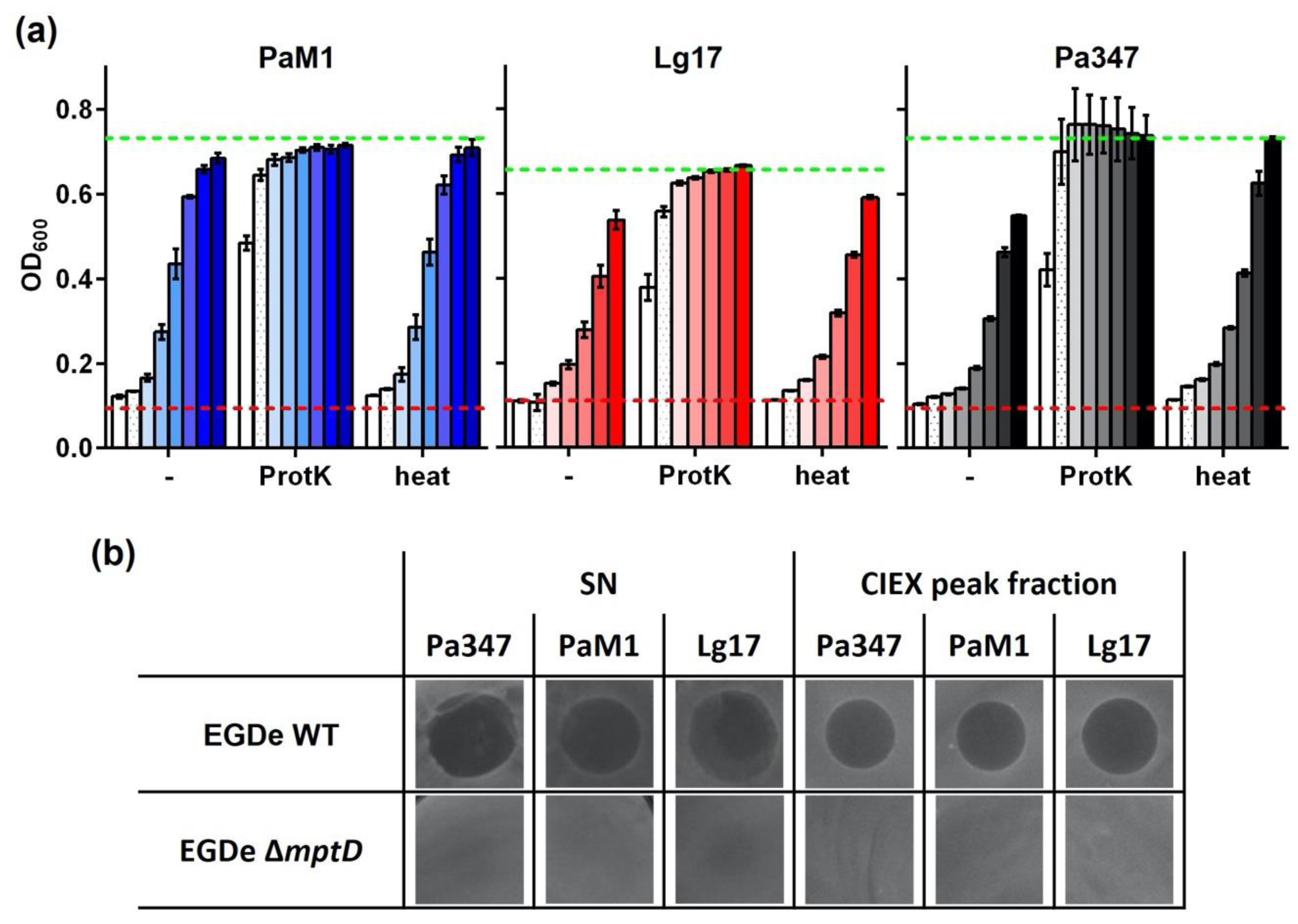
2.3. Characterization of the Antimicrobial Activity of Representative Isolates
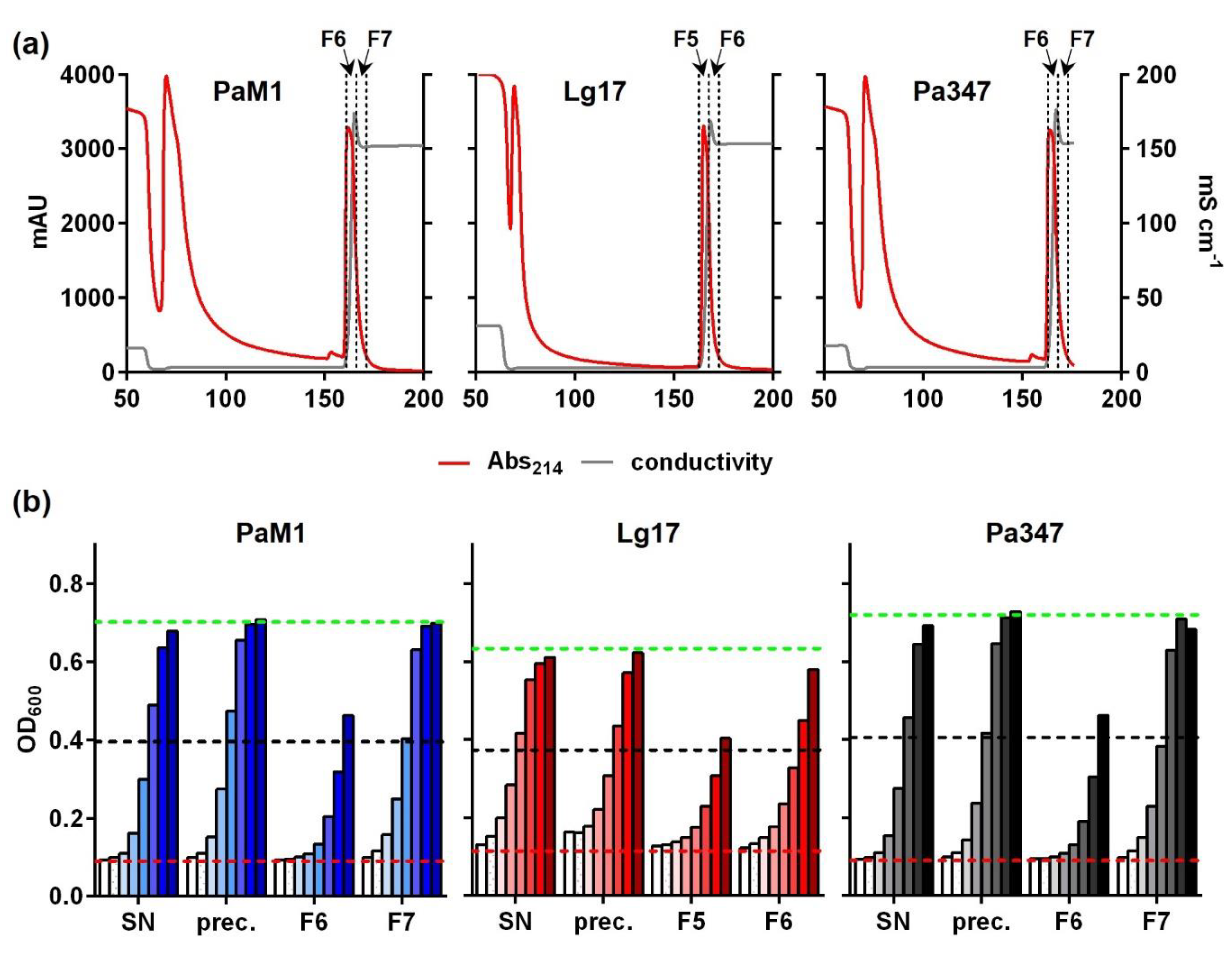
2.4. Partial Purification and Characterization of the Antimicrobial Compounds of P. Acidilacici M1 and L. Garvieae 17
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Identification of Raw Milk Isolates
4.3. Screening Procedures
4.4. Purification and Chromatography of Antimicrobial Compounds
4.5. Antimicrobial Activity Assays
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Freitag, N.E.; Port, G.C.; Miner, M. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat. Rev. Genet. 2009, 7, 623–628. [Google Scholar] [CrossRef]
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gahan, C.G.M.; Hill, C. Listeria monocytogenes: Survival and adaptation in the gastrointestinal tract. Front. Cell. Infect. Microbiol. 2014, 4, 9. [Google Scholar] [CrossRef] [Green Version]
- De Noordhout, C.M.; Devleesschauwer, B.; Angulo, F.J.; Verbeke, G.; Haagsma, J.; Kirk, M.; Havelaar, A.; Speybroeck, N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 1073–1082. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority; European Center for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 2016, 14, 14. [Google Scholar] [CrossRef]
- Mills, S.; Stanton, C.; Hill, C.; Ross, R. New Developments and Applications of Bacteriocins and Peptides in Foods. Annu. Rev. Food Sci. Technol. 2011, 2, 299–329. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Hill, C.; Ross, R.P. Food microbiology: Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Balciunas, E.M.; Martinez, F.C.; Todorov, S.; Franco, B.D.G.D.M.; Converti, A.; Oliveira, R.P.D.S. Novel biotechnological applications of bacteriocins: A review. Food Control. 2013, 32, 134–142. [Google Scholar] [CrossRef]
- Sassone-Corsi, M.; Nuccio, S.-P.; Liu, H.; Hernandez, D.; Vu, C.T.; Takahashi, A.A.; Edwards, R.A.; Raffatellu, M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nat. Cell Biol. 2016, 540, 280–283. [Google Scholar] [CrossRef]
- Kim, S.; Becattini, S.; Moody, T.U.; Shliaha, P.V.; Littmann, E.R.; Seok, R.; Gjonbalaj, M.; Eaton, V.; Fontana, E.; Amoretti, L.; et al. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nat. Cell Biol. 2019, 572, 665–669. [Google Scholar] [CrossRef]
- Claesen, J.; Spagnolo, J.B.; Ramos, S.F.; Kurita, K.L.; Byrd, A.L.; Aksenov, A.A.; Melnik, A.V.; Wong, W.R.; Wang, S.; Hernandez, R.D.; et al. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl. Med. 2020, 12, eaay5445. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef] [Green Version]
- Corr, S.; Li, Y.; Riedel, C.U.; O’Toole, P.; Hill, C.; Gahan, C. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. USA 2007, 104, 7617–7621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobson, A.; Cotter, P.; Ross, R.; Hill, C. Bacteriocin Production: A Probiotic Trait? Appl. Environ. Microbiol. 2011, 78, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadimitriou, K.; Zoumpopoulou, G.; Foligne, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering probiotic microorganisms: In vitro, in vivo, genetic and omics approaches. Front. Microbiol. 2015, 6, 58. [Google Scholar] [CrossRef] [Green Version]
- Cotter, P.D.; Ross, R.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Genet. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.; Dicks, L.M. Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of lactic acid bacteria: Extending the family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [Green Version]
- Wiedemann, I.; Breukink, E.; van Kraaij, C.; Kuipers, O.P.; Bierbaum, G.; de Kruijff, B.; Sahl, H.-G. Specific Binding of Nisin to the Peptidoglycan Precursor Lipid II Combines Pore Formation and Inhibition of Cell Wall Biosynthesis for Potent Antibiotic Activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef] [Green Version]
- Breukink, E.; De Kruijff, B. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 2006, 5, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Tymoszewska, A.; Diep, D.B.; Aleksandrzak-Piekarczyk, T. The extracellular loop of Man-PTS subunit IID is responsible for the sensitivity of Lactococcus garvieae to garvicins A., B and C. Sci. Rep. 2018, 8, 15790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tymoszewska, A.; Diep, D.B.; Wirtek, P.; Aleksandrzak-Piekarczyk, T. The Non-Lantibiotic Bacteriocin Garvicin Q Targets Man-PTS in a Broad Spectrum of Sensitive Bacterial Genera. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Diep, D.B.; Skaugen, M.; Salehian, Z.; Holo, H.; Nes, I.F. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. USA 2007, 104, 2384–2389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kjos, M.; Salehian, Z.; Nes, I.F.; Diep, D.B. An Extracellular Loop of the Mannose Phosphotransferase System Component IIC Is Responsible for Specific Targeting by Class IIa Bacteriocins. J. Bacteriol. 2010, 192, 5906–5913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabrielsen, C.; Brede, D.A.; Hernandez, P.E.; Nes, I.F.; Diep, D.B. The Maltose ABC Transporter in Lactococcus lactis Facilitates High-Level Sensitivity to the Circular Bacteriocin Garvicin ML. Antimicrob. Agents Chemother. 2012, 56, 2908–2915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, C.; Silva, S.P.M.; Ribeiro, S.C. Application of Bacteriocins and Protective Cultures in Dairy Food Preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef]
- Zou, J.; Jiang, H.; Cheng, H.; Fang, J.; Huang, G. Strategies for screening, purification and characterization of bacteriocins. Int. J. Biol. Macromol. 2018, 117, 781–789. [Google Scholar] [CrossRef]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Crauwels, P.; Schäfer, L.; Weixler, D.; Bar, N.S.; Diep, D.B.; Riedel, C.U.; Seibold, G.M. Intracellular pHluorin as Sensor for Easy Assessment of Bacteriocin-Induced Membrane-Damage in Listeria monocytogenes. Front. Microbiol. 2018, 9, 3038. [Google Scholar] [CrossRef] [PubMed]
- Miesenboeck, G.; De Angelis, D.A.; Rothman, J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nat. Cell Biol. 1998, 394, 192–195. [Google Scholar] [CrossRef]
- Shabala, L.; Budde, B.; Ross, T.; Siegumfeldt, H.; McMeekin, T. Responses of Listeria monocytogenes to acid stress and glucose availability monitored by measurements of intracellular pH and viable counts. Int. J. Food Microbiol. 2002, 75, 89–97. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Tosukhowong, A.; Zendo, T.; Visessanguan, W.; Roytrakul, S.; Pumpuang, L.; Jaresitthikunchai, J.; Sonomoto, K. Garvieacin Q, a Novel Class II Bacteriocin from Lactococcus garvieae BCC 43578. Appl. Environ. Microbiol. 2012, 78, 1619–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porto, M.C.W.; Kuniyoshi, T.M.; Azevedo, P.O.D.S.D.; Vitolo, M.; Oliveira, R. Pediococcus spp.: An important genus of lactic acid bacteria and pediocin producers. Biotechnol. Adv. 2017, 35, 361–374. [Google Scholar] [CrossRef]
- Dalet, K.; Cossart, P.; Cenatiempo, Y.; Héchard, Y. A σ54-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology 2001, 147, 3263–3269. [Google Scholar] [CrossRef] [Green Version]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellmann, A.; Cloud, J.; Maier, T.; Keckevoet, U.; Ramminger, I.; Iwen, P.; Dunn, J.; Hall, G.; Wilson, D.; LaSala, P.; et al. Evaluation of Matrix-Assisted Laser Desorption Ionization-Time-of-Flight Mass Spectrometry in Comparison to 16S rRNA Gene Sequencing for Species Identification of Nonfermenting Bacteria. J. Clin. Microbiol. 2008, 46, 1946–1954. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 41, D36–D42. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Yang, Q.; Kudinha, T.; Zhang, L.; Xiao, M.; Kong, F.; Zhao, Y.; Xu, Y.-C. Using Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) Complemented with Selected 16S rRNA and gyrB Genes Sequencing to Practically Identify Clinical Important Viridans Group Streptococci (VGS). Front. Microbiol. 2016, 7, 1328. [Google Scholar] [CrossRef] [Green Version]
- Patel, R. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry in Clinical Microbiology. Clin. Infect. Dis. 2013, 57, 564–572. [Google Scholar] [CrossRef] [Green Version]
- Eigner, U.; Holfelder, M.; Oberdorfer, K.; Betz-Wild, U.; Bertsch, D.; Fahr, A.-M. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin. Lab. 2009, 55, 289–296. [Google Scholar] [PubMed]
- Timperio, A.M.; Gorrasi, S.; Zolla, L.; Fenice, M. Evaluation of MALDI-TOF mass spectrometry and MALDI BioTyper in comparison to 16S rDNA sequencing for the identification of bacteria isolated from Arctic sea water. PLoS ONE 2017, 12, e0181860. [Google Scholar] [CrossRef] [PubMed]
- Pomastowski, P.; Złoch, M.; Rodzik, A.; Ligor, M.; Kostrzewa, M.; Buszewski, B. Analysis of bacteria associated with honeys of different geographical and botanical origin using two different identification approaches: MALDI-TOF MS and 16S rDNA PCR technique. PLoS ONE 2019, 14, e0217078. [Google Scholar] [CrossRef] [Green Version]
- Carrasco, G.; Caballero, J.D.D.; Garrido, N.; Valdezate, S.; Cantón, R.; Sáez-Nieto, J.A. Shortcomings of the Commercial MALDI-TOF MS Database and Use of MLSA as an Arbiter in the Identification of Nocardia Species. Front. Microbiol. 2016, 7, 542. [Google Scholar] [CrossRef]
- Kopcakova, A.; Stramova, Z.; Kvasnova, S.; Godany, A.; Perhacova, Z.; Pristas, P. Need for database extension for reliable identification of bacteria from extreme environments using MALDI TOF mass spectrometry. Chem. Pap. 2014, 68, 1435–1442. [Google Scholar] [CrossRef]
- Yang, E.; Fan, L.; Jiang, Y.; Doucette, C.; Fillmore, S. Antimicrobial activity of bacteriocin-producing lactic acid bacteria isolated from cheeses and yogurts. AMB Express 2012, 2, 48. [Google Scholar] [CrossRef] [Green Version]
- Russell, J.B.; Diez-Gonzalez, F. The Effects of Fermentation Acids on Bacterial Growth. Adv. Microb. Physiol. 1997, 39, 205–234. [Google Scholar] [CrossRef]
- Shelef, L.A. Antimicrobial Effects of Lactates: A Review. J. Food Prot. 1994, 57, 445–450. [Google Scholar] [CrossRef]
- Makhloufi, K.M.; Carré-Mlouka, A.; Peduzzi, J.; Lombard, C.; Van Reenen, C.A.; Dicks, L.; Rebuffat, S. Characterization of Leucocin B-KM432Bz from Leuconostoc pseudomesenteroides Isolated from Boza, and Comparison of its Efficiency to Pediocin PA-1. PLoS ONE 2013, 8, e70484. [Google Scholar] [CrossRef] [Green Version]
- Anastasiadou, S.; Papagianni, M.; Filiousis, G.; Ambrosiadis, I.; Koidis, P. Pediocin SA-1, an antimicrobial peptide from Pediococcus acidilactici NRRL B5627: Production conditions, purification and characterization. Bioresour. Technol. 2008, 99, 5384–5390. [Google Scholar] [CrossRef]
- Telke, A.A.; Ovchinnikov, K.V.; Vuoristo, K.; Mathiesen, G.; Thorstensen, T.; Diep, D.B. Over 2000-Fold Increased Production of the Leaderless Bacteriocin Garvicin KS by Increasing Gene Dose and Optimization of Culture Conditions. Front. Microbiol. 2019, 10, 389. [Google Scholar] [CrossRef]
- Kjos, M.; Nes, I.F.; Diep, D.B. Class II one-peptide bacteriocins target a phylogenetically defined subgroup of mannose phosphotransferase systems on sensitive cells. Microbiology 2009, 155, 2949–2961. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, J.; Cintas, L.; Casaus, P.; Martínez, M.; Suárez, A.; Hernandez, P.E. Detection of pediocin PA-1-producing pediococci by rapid molecular biology techniques. Food Microbiol. 1997, 14, 363–371. [Google Scholar] [CrossRef]
- NBSP; Glaser, P.; Frangeul, L.; Buchrieser, C.; Rusniok, C.; Amend, A.; Baquero, F.; Berche, P.; Bloecker, H.; Brandt, P.; et al. Comparative genomics of Listeria species. Science 2001, 294, 849–852. [Google Scholar] [CrossRef] [Green Version]
- Kostrzewa, M. Application of the MALDI Biotyper to clinical microbiology: Progress and potential. Expert Rev. Proteom. 2018, 15, 193–202. [Google Scholar] [CrossRef]
- Sauget, M.; Valot, B.; Bertrand, X.; Hocquet, D. Can MALDI-TOF Mass Spectrometry Reasonably Type Bacteria? Trends Microbiol. 2017, 25, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, H.; Cai, K.; Yu, P.; Liu, Y.; Zhao, G.; Chen, R.; Xu, R.; Yu, M. Evaluation of three sample preparation methods for the identification of clinical strains by using two MALDI-TOF MS systems. J. Mass Spectrom. 2021, 56, e4696. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holo, H.; Nilssen, O.; Nes, I.F. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: Isolation and characterization of the protein and its gene. J. Bacteriol. 1991, 173, 3879–3887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FAO; WHO. Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food; World Health Organization: London, ON, Canada, 2002. [Google Scholar]


| Sample | Volume [mL] | PaM1 | Lg17 | Pa347 | |||
|---|---|---|---|---|---|---|---|
| BU/mL | Total BU (Recovery) | BU/mL | Total BU (Recovery) | BU/mL | Total BU (Recovery) | ||
| SN | 500 | 5120 | 2,560,000 (100%) | 2560 | 1,280,000 (100%) | 5120 | 2,560,000 (100%) |
| Prec. | 50 | 2560 | 128,000 (5%) | 2560 | 256,000 (20%) | 2560 | 128,000 (5%) |
| F6 a | 5 | 20,480 | 102,400 (4%) | 20,480 | 102,400 (8%) | 20,480 | 102,400 (4%) |
| F7 a | 5 | 2560 | 12,800 (0.5%) | 2560 | 51,200 (4%) | 5120 | 12,800 (0.5%) |
| Strain | Relevant Characteristic | Source/ Reference |
|---|---|---|
| Listeria monocytogenes | ||
| EGDe | Wildtype strain, serotype 1/2 a type | [57] |
| EGY2 | EGDe derivative carrying a deletion of 84 bp in the mptD gene | [37] |
| EGDe pNZ-Phelp-pHluorin | EGDe derivative harboring plasmid pNZ-Phelp-pHluorin for constitutive expression of pHluorin | [31] |
| Pediococcus acidilactici 347 | Natural producer of pediocin PA-1; isolated from Spanish dry fermented sausages | [56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desiderato, C.K.; Sachsenmaier, S.; Ovchinnikov, K.V.; Stohr, J.; Jacksch, S.; Desef, D.N.; Crauwels, P.; Egert, M.; Diep, D.B.; Goldbeck, O.; et al. Identification of Potential Probiotics Producing Bacteriocins Active against Listeria monocytogenes by a Combination of Screening Tools. Int. J. Mol. Sci. 2021, 22, 8615. https://doi.org/10.3390/ijms22168615
Desiderato CK, Sachsenmaier S, Ovchinnikov KV, Stohr J, Jacksch S, Desef DN, Crauwels P, Egert M, Diep DB, Goldbeck O, et al. Identification of Potential Probiotics Producing Bacteriocins Active against Listeria monocytogenes by a Combination of Screening Tools. International Journal of Molecular Sciences. 2021; 22(16):8615. https://doi.org/10.3390/ijms22168615
Chicago/Turabian StyleDesiderato, Christian K., Steffen Sachsenmaier, Kirill V. Ovchinnikov, Jonas Stohr, Susanne Jacksch, Dominique N. Desef, Peter Crauwels, Markus Egert, Dzung B. Diep, Oliver Goldbeck, and et al. 2021. "Identification of Potential Probiotics Producing Bacteriocins Active against Listeria monocytogenes by a Combination of Screening Tools" International Journal of Molecular Sciences 22, no. 16: 8615. https://doi.org/10.3390/ijms22168615
APA StyleDesiderato, C. K., Sachsenmaier, S., Ovchinnikov, K. V., Stohr, J., Jacksch, S., Desef, D. N., Crauwels, P., Egert, M., Diep, D. B., Goldbeck, O., & Riedel, C. U. (2021). Identification of Potential Probiotics Producing Bacteriocins Active against Listeria monocytogenes by a Combination of Screening Tools. International Journal of Molecular Sciences, 22(16), 8615. https://doi.org/10.3390/ijms22168615







