Specific and Global RNA Regulators in Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Base-Pairing RNAs in Pae
2.1. ErsA
2.2. Sr0161
2.3. Sr006
2.4. PrrF1/2
2.5. PrrH
2.6. ReaL
2.7. RgsA
2.8. PaiI
2.9. NrsZ
2.10. PhrS
3. Translational Repressors and Their RNA Decoys
3.1. The Hfq Regulon
3.2. CrcZ, a Decoy for Hfq
3.3. The RsmA and RsmN/F Regulons
3.4. RsmV/W/Y/Z, Decoys for RsmA/N/F
4. Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Toledo-Arana, A.; Repoila, F.; Cossart, P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007, 10, 182–188. [Google Scholar] [CrossRef]
- Waters, L.S.; Storz, G. Regulatory RNAs in bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef] [Green Version]
- Holmqvist, E.; Wagner, E.G.H. Impact of bacterial sRNAs in stress responses. Biochem. Soc. Trans. 2017, 45, 1203–1212. [Google Scholar] [CrossRef] [Green Version]
- Hör, J.; Vogel, J. Global snapshots of bacterial RNA networks. EMBO J. 2017, 36, 245–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durica-Mitic, S.; Göpel, Y.; Görke, B. Carbohydrate utilization in bacteria: Making the most out of sugars with the help of small regulatory RNAs. Microbiol. Spectr. 2018, 6, 1–19. [Google Scholar] [CrossRef]
- Chakravarty, S.; Massé, E. RNA-dependent regulation of virulence in pathogenic bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 337. [Google Scholar] [CrossRef]
- Diallo, I.; Provost, P. RNA-sequencing analyses of small bacterial RNAs and their emergence as virulence factors in host-pathogen interactions. Int. J. Mol. Sci. 2020, 21, 1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González Plaza, J.J. Small RNAs as fundamental players in the transference of information during bacterial infectious diseases. Front. Mol. Biosci. 2020, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Iosub, I.A.; van Nues, R.W.; McKellar, S.W.; Nieken, K.J.; Marchioretto, M.; Sy, B.; Tree, J.J.; Viero, G.; Granneman, S. Hfq CLASH uncovers sRNA-target interaction networks linked to nutrient availability adaptation. Elife 2020, 9, e54655. [Google Scholar] [CrossRef]
- Mediati, D.G.; Wu, S.; Wu, W.; Tree, J.J. Networks of resistance: Small RNA control of antibiotic resistance. Trends Genet. 2021, 37, 35–45. [Google Scholar] [CrossRef]
- Sorek, R.; Cossart, P. Prokaryotic transcriptomics: A new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 2010, 11, 9–16. [Google Scholar] [CrossRef]
- Dar, D.; Shamir, M.; Mellin, J.R.; Koutero, M.; Stern-Ginossar, N.; Cossart, P.; Sorek, R. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 2016, 352, aad9822. [Google Scholar] [CrossRef] [Green Version]
- Wagner, E.G.H.; Romby, P. Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv. Genet. 2015, 90, 133–208. [Google Scholar] [CrossRef] [PubMed]
- Massé, E.; Escorcia, F.E.; Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003, 17, 2374–2383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, T.; Tomizawa, J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. USA 1980, 77, 2450–2454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kittle, J.D.; Simons, R.W.; Lee, J.; Kleckner, N. Insertion sequence IS10 anti-sense pairing initiates by an interaction between the 5’ end of the target RNA and a loop in the anti-sense RNA. J. Mol. Biol. 1989, 210, 561–572. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 2008, 322, 1843–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wassarman, K.M. 6S RNA: A regulator of transcription. Mol. Microbiol. 2007, 65, 1425–1431. [Google Scholar] [CrossRef]
- Wassarman, K.M. 6S RNA: A small RNA regulator of transcription. Curr. Opin. Microbiol. 2007, 10, 164–168. [Google Scholar] [CrossRef]
- Sedlyarova, N.; Shamovsky, I.; Bharati, B.K.; Epshtein, V.; Chen, J.; Gottesman, S.; Schroeder, R.; Nudler, E. sRNA-mediated control of transcription termination in E. coli. Cell 2016, 167, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Wassarman, K.M. 6S RNA, a global regulator of transcription. Microbiol. Spectr. 2018, 6, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Morita, T.; Gottesman, S. Regulation of transcription termination of small RNAs and by small RNAs: Molecular mechanisms and biological functions. Front. Cell. Infect. Microbiol. 2019, 9, 201. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papenfort, K.; Vanderpool, C.K. Target activation by regulatory RNAs in bacteria. FEMS Microbiol. Rev. 2015, 39, 362–378. [Google Scholar] [CrossRef] [Green Version]
- Jørgensen, M.G.; Pettersen, J.S.; Kallipolitis, B.H. sRNA-mediated control in bacteria: An increasing diversity of regulatory mechanisms. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194504. [Google Scholar] [CrossRef] [PubMed]
- Babitzke, P.; Romeo, T. CsrB sRNA family: Sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007, 10, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Lapouge, K.; Schubert, M.; Allain, F.H.-T.; Haas, D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: From RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008, 67, 241–253. [Google Scholar] [CrossRef]
- Romeo, T.; Babitzke, P. Global regulation by CsrA and its RNA antagonists. Microbiol. Spectr. 2018, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sonnleitner, E.; Bläsi, U. Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet. 2014, 10, e1004440. [Google Scholar] [CrossRef]
- Azam, M.S.; Vanderpool, C.K. Talk among yourselves: RNA sponges mediate cross talk between functionally related messenger RNAs. EMBO J. 2015, 34, 1436–1438. [Google Scholar] [CrossRef] [Green Version]
- Miyakoshi, M.; Chao, Y.; Vogel, J. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 2015, 34, 1478–1492. [Google Scholar] [CrossRef]
- Denham, E.L. The Sponge RNAs of bacteria—How to find them and their role in regulating the post-transcriptional network. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194565. [Google Scholar] [CrossRef]
- Wilderman, P.J.; Sowa, N.A.; FitzGerald, D.J.; FitzGerald, P.C.; Gottesman, S.; Ochsner, U.A.; Vasil, M.L. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 2004, 101, 9792–9797. [Google Scholar] [CrossRef] [Green Version]
- Livny, J.; Brencic, A.; Lory, S.; Waldor, M.K. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006, 34, 3484–3493. [Google Scholar] [CrossRef]
- Sonnleitner, E.; Sorger-Domenigg, T.; Madej, M.J.; Findeiss, S.; Hackermuller, J.; Huttenhofer, A.; Stadler, P.F.; Blasi, U.; Moll, I. Detection of small RNAs in Pseudomonas aeruginosa by RNomics and structure-based bioinformatic tools. Microbiology 2008, 154, 3175–3187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dötsch, A.; Eckweiler, D.; Schniederjans, M.; Zimmermann, A.; Jensen, V.; Scharfe, M.; Geffers, R.; Häussler, S. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS ONE 2012, 7, e31092. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, S.; Brugnoli, M.; De Bonis, A.; Righetti, F.; Delvillani, F.; Deho, G.; Horner, D.; Briani, F.; Bertoni, G. Comparative profiling of Pseudomonas aeruginosa strains reveals differential expression of novel unique and conserved small RNAs. PLoS ONE 2012, 7, e36553. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Lozano, M.; Marvig, R.L.; Molin, S.; Long, K.S. Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ. Microbiol. 2012, 14, 2006–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wurtzel, O.; Yoder-Himes, D.R.; Han, K.; Dandekar, A.A.; Edelheit, S.; Greenberg, E.P.; Sorek, R.; Lory, S. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 2012, 8, e1002945. [Google Scholar] [CrossRef]
- Gomez-Lozano, M.; Marvig, R.L.; Molina-Santiago, C.; Tribelli, P.M.; Ramos, J.-L.; Molin, S. Diversity of small RNAs expressed in Pseudomonas species. Environ. Microbiol. Rep. 2015, 7, 227–236. [Google Scholar] [CrossRef]
- Pita, T.; Feliciano, J.R.; Leitão, J.H. Small noncoding regulatory RNAs from Pseudomonas aeruginosa and Burkholderia cepacia complex. Int. J. Mol. Sci. 2018, 19, 3759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Huang, D.; Cheung, M.K.; Nong, W.; Huang, Q.; Kwan, H.S. BSRD: A repository for bacterial small regulatory RNA. Nucleic Acids Res. 2013, 41, D233–D238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Assche, E.; Van Puyvelde, S.; Vanderleyden, J.; Steenackers, H.P. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 2015, 6, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodson, S.A.; Panja, S.; Santiago-Frangos, A. Proteins that chaperone RNA regulation. Microbiol. Spectr. 2018, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, E.; Vogel, J. RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 2018, 16, 601–615. [Google Scholar] [CrossRef]
- Quendera, A.P.; Seixas, A.F.; dos Santos, R.F.; Santos, I.; Silva, J.P.N.; Arraiano, C.M.; Andrade, J.M. RNA-binding proteins driving the regulatory activity of small non-coding RNAs in bacteria. Front. Mol. Biosci. 2020, 7, 78. [Google Scholar] [CrossRef]
- Sonnleitner, E.; Prindl, K.; Bläsi, U. The Pseudomonas aeruginosa CrcZ RNA interferes with Hfq-mediated riboregulation. PLoS ONE 2017, 12, e0180887. [Google Scholar] [CrossRef] [Green Version]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef] [Green Version]
- Santiago-Frangos, A.; Woodson, S.A. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA 2018, 9, e1475. [Google Scholar] [CrossRef]
- Gottesman, S.; McCullen, C.A.; Guillier, M.; Vanderpool, C.K.; Majdalani, N.; Benhammou, J.; Thompson, K.M.; FitzGerald, P.C.; Sowa, N.A.; FitzGerald, D.J. Small RNA regulators and the bacterial response to stress. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, S.; Carloni, S.; Fulco, R.; Falcone, M.; Macchi, R.; Bertoni, G. Post-transcriptional regulation of the virulence-associated enzyme AlgC by the sigma(22) -dependent small RNA ErsA of Pseudomonas aeruginosa. Environ. Microbiol. 2015, 17, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.F.; Han, K.; Chandler, C.E.; Tjaden, B.; Ernst, R.K.; Lory, S. Probing the sRNA regulatory landscape of P. aeruginosa: Post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility. Mol. Microbiol. 2017, 106, 919–937. [Google Scholar] [CrossRef] [Green Version]
- Padgett, P.J.; Phibbs, P.V. Phosphomannomutase activity in wild-type and alginate-producing strains of Pseudomonas aeruginosa. Curr. Microbiol. 1986, 14, 187–192. [Google Scholar] [CrossRef]
- Sá-Correia, I.; Darzins, A.; Wang, S.K.; Berry, A.; Chakrabarty, A.M. Alginate biosynthetic enzymes in mucoid and nonmucoid Pseudomonas aeruginosa: Overproduction of phosphomannose isomerase, phosphomannomutase, and GDP-mannose pyrophosphorylase by overexpression of the phosphomannose isomerase (pmi) gene. J. Bacteriol. 1987, 169, 3224–3231. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Wang, J.; Wang, S.; Anderson, E.M.; Lam, J.S.; Parsek, M.R.; Wozniak, D.J. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ. Microbiol. 2012, 14, 1995–2005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg, J.B.; Hatano, K.; Pier, G.B. Synthesis of lipopolysaccharide O side chains by Pseudomonas aeruginosa PAO1 requires the enzyme phosphomannomutase. J. Bacteriol. 1993, 175, 1605–1611. [Google Scholar] [CrossRef] [Green Version]
- Coyne, M.J.J.; Russell, K.S.; Coyle, C.L.; Goldberg, J.B. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 1994, 176, 3500–3507. [Google Scholar] [CrossRef] [Green Version]
- Olvera, C.; Goldberg, J.B.; Sánchez, R.; Soberón-Chávez, G. The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol. Lett. 1999, 179, 85–90. [Google Scholar] [CrossRef] [Green Version]
- Deretic, V.; Schurr, M.J.; Yu, H. Pseudomonas aeruginosa, mucoidy and the chronic infection phenotype in cystic fibrosis. Trends Microbiol. 1995, 3, 351–356. [Google Scholar] [CrossRef]
- Wang, S.; Yu, S.; Zhang, Z.; Wei, Q.; Yan, L.; Ai, G.; Liu, H.; Ma, L.Z. Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2014, 80, 6724–6732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zielinski, N.A.; Chakrabarty, A.M.; Berry, A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 1991, 266, 9754–9763. [Google Scholar] [CrossRef]
- Zielinski, N.A.; Maharaj, R.; Roychoudhury, S.; Danganan, C.E.; Hendrickson, W.; Chakrabarty, A.M. Alginate synthesis in Pseudomonas aeruginosa: Environmental regulation of the algC promoter. J. Bacteriol. 1992, 174, 7680–7688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lizewski, S.E.; Lundberg, D.S.; Schurr, M.J. The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis. Infect. Immun. 2002, 70, 6083–6093. [Google Scholar] [CrossRef] [Green Version]
- Falcone, M.; Ferrara, S.; Rossi, E.; Johansen, H.K.; Molin, S.; Bertoni, G. The small RNA ErsA of Pseudomonas aeruginosa contributes to biofilm development and motility through post-transcriptional modulation of AmrZ. Front. Microbiol. 2018, 9, 238. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, S.; Rossi, A.; Ranucci, S.; De Fino, I.; Bragonzi, A.; Cigana, C.; Bertoni, G. The small RNA ErsA plays a role in the regulatory network of Pseudomonas aeruginosa pathogenicity in airway infections. mSphere 2020, 5, e00909-20. [Google Scholar] [CrossRef] [PubMed]
- Trias, J.; Nikaido, H. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J. Biol. Chem. 1990, 265, 15680–15684. [Google Scholar] [CrossRef]
- Trias, J.; Nikaido, H. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1990, 34, 52–57. [Google Scholar] [CrossRef] [Green Version]
- Henrichfreise, B.; Wiegand, I.; Pfister, W.; Wiedemann, B. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 2007, 51, 4062–4070. [Google Scholar] [CrossRef] [Green Version]
- Diene, S.M.; L’homme, T.; Bellulo, S.; Stremler, N.; Dubus, J.-C.; Mely, L.; Leroy, S.; Degand, N.; Rolain, J.-M. ISPa46, a novel insertion sequence in the oprD porin gene of an imipenem-resistant Pseudomonas aeruginosa isolate from a cystic fibrosis patient in Marseille, France. Int. J. Antimicrob. Agents 2013, 42, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Llanes, C.; Pourcel, C.; Richardot, C.; Plésiat, P.; Fichant, G.; Cavallo, J.-D.; Mérens, A. Diversity of β-lactam resistance mechanisms in cystic fibrosis isolates of Pseudomonas aeruginosa: A French multicentre study. J. Antimicrob. Chemother. 2013, 68, 1763–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardot, C.; Plésiat, P.; Fournier, D.; Monlezun, L.; Broutin, I.; Llanes, C. Carbapenem resistance in cystic fibrosis strains of Pseudomonas aeruginosa as a result of amino acid substitutions in porin OprD. Int. J. Antimicrob. Agents 2015, 45, 529–532. [Google Scholar] [CrossRef]
- Chalhoub, H.; Sáenz, Y.; Rodriguez-Villalobos, H.; Denis, O.; Kahl, B.C.; Tulkens, P.M.; Van Bambeke, F. High-level resistance to meropenem in clinical isolates of Pseudomonas aeruginosa in the absence of carbapenemases: Role of active efflux and porin alterations. Int. J. Antimicrob. Agents 2016, 48, 740–743. [Google Scholar] [CrossRef]
- Courtois, N.; Caspar, Y.; Maurin, M. Phenotypic and genetic resistance traits of Pseudomonas aeruginosa strains infecting cystic fibrosis patients: A French cohort study. Int. J. Antimicrob. Agents 2018, 52, 358–364. [Google Scholar] [CrossRef]
- Sherrard, L.J.; Wee, B.A.; Duplancic, C.; Ramsay, K.A.; Dave, K.A.; Ballard, E.; Wainwright, C.E.; Grimwood, K.; Sidjabat, H.E.; Whiley, D.M.; et al. Emergence and impact of oprD mutations in Pseudomonas aeruginosa strains in cystic fibrosis. J. Cyst. Fibros. Off. J. Eur. Cyst. Fibros. Soc. 2021. [Google Scholar] [CrossRef] [PubMed]
- Sonnleitner, E.; Pusic, P.; Wolfinger, M.T.; Bläsi, U. Distinctive regulation of carbapenem susceptibility in Pseudomonas aeruginosa by Hfq. Front. Microbiol. 2020, 11, 1001. [Google Scholar] [CrossRef] [PubMed]
- Ernst, R.K.; Yi, E.C.; Guo, L.; Lim, K.B.; Burns, J.L.; Hackett, M.; Miller, S.I. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 1999, 286, 1561–1565. [Google Scholar] [CrossRef] [PubMed]
- Ernst, R.K.; Hajjar, A.M.; Tsai, J.H.; Moskowitz, S.M.; Wilson, C.B.; Miller, S.I. Pseudomonas aeruginosa lipid A diversity and its recognition by Toll-like receptor 4. J. Endotoxin Res. 2003, 9, 395–400. [Google Scholar] [CrossRef]
- Ernst, R.K.; Adams, K.N.; Moskowitz, S.M.; Kraig, G.M.; Kawasaki, K.; Stead, C.M.; Trent, M.S.; Miller, S.I. The Pseudomonas aeruginosa lipid A deacylase: Selection for expression and loss within the cystic fibrosis airway. J. Bacteriol. 2006, 188, 191–201. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, K.; China, K.; Nishijima, M. Release of the lipopolysaccharide deacylase PagL from latency compensates for a lack of lipopolysaccharide aminoarabinose modification-dependent resistance to the antimicrobial peptide polymyxin B in Salmonella enterica. J. Bacteriol. 2007, 189, 4911–4919. [Google Scholar] [CrossRef] [Green Version]
- Han, M.-L.; Velkov, T.; Zhu, Y.; Roberts, K.D.; Le Brun, A.P.; Chow, S.H.; Gutu, A.D.; Moskowitz, S.M.; Shen, H.-H.; Li, J. Polymyxin-induced lipid A deacylation in Pseudomonas aeruginosa perturbs polymyxin penetration and confers high-level resistance. ACS Chem. Biol. 2018, 13, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, S.M.; Ernst, R.K. The role of Pseudomonas lipopolysaccharide in cystic fibrosis airway infection. Subcell. Biochem. 2010, 53, 241–253. [Google Scholar] [CrossRef] [Green Version]
- Massé, E.; Gottesman, S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 4620–4625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaballa, A.; Antelmann, H.; Aguilar, C.; Khakh, S.K.; Song, K.-B.; Smaldone, G.T.; Helmann, J.D. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 11927–11932. [Google Scholar] [CrossRef] [Green Version]
- Vasil, M.L. How we learnt about iron acquisition in Pseudomonas aeruginosa: A series of very fortunate events. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 2007, 20, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Oglesby, A.G.; Farrow, J.M., 3rd; Lee, J.-H.; Tomaras, A.P.; Greenberg, E.P.; Pesci, E.C.; Vasil, M.L. The influence of iron on Pseudomonas aeruginosa physiology: A regulatory link between iron and quorum sensing. J. Biol. Chem. 2008, 283, 15558–15567. [Google Scholar] [CrossRef] [Green Version]
- Han, K.; Tjaden, B.; Lory, S. GRIL-seq provides a method for identifying direct targets of bacterial small regulatory RNA by in vivo proximity ligation. Nat. Microbiol. 2016, 2, 16239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrieta-Ortiz, M.L.; Hafemeister, C.; Shuster, B.; Baliga, N.S.; Bonneau, R.; Eichenberger, P. Inference of bacterial small RNA regulatory networks and integration with transcription factor-driven regulatory networks. mSystems 2020, 5, e00057-20. [Google Scholar] [CrossRef]
- Chihara, K.; Bischler, T.; Barquist, L.; Monzon, V.A.; Noda, N.; Vogel, J.; Tsuneda, S. Conditional Hfq association with small noncoding RNAs in Pseudomonas aeruginosa revealed through comparative UV cross-linking immunoprecipitation followed by high-throughput sequencing. mSystems 2019, 4, e00590-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesci, E.C.; Milbank, J.B.J.; Pearson, J.P.; McKnight, S.; Kende, A.S.; Greenberg, E.P.; Iglewski, B.H. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1999, 96, 11229–11234. [Google Scholar] [CrossRef] [Green Version]
- McKnight, S.L.; Iglewski, B.H.; Pesci, E.C. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 2702–2708. [Google Scholar] [CrossRef] [Green Version]
- Dubern, J.-F.; Diggle, S.P. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol. BioSyst. 2008, 4, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Hänsch, G.M.; Prior, B.; Brenner-Weiss, G.; Obst, U.; Overhage, J. The Pseudomonas quinolone signal (PQS) stimulates chemotaxis of polymorphonuclear neutrophils. J. Appl. Biomater. Funct. Mater. 2014, 12, 21–26. [Google Scholar] [CrossRef]
- Reinhart, A.A.; Nguyen, A.T.; Brewer, L.K.; Bevere, J.; Jones, J.W.; Kane, M.A.; Damron, F.H.; Barbier, M.; Oglesby-Sherrouse, A.G. The Pseudomonas aeruginosa PrrF small RNAs regulate iron homeostasis during acute murine lung infection. Infect. Immun. 2017, 85, e00764-16. [Google Scholar] [CrossRef] [Green Version]
- Djapgne, L.; Panja, S.; Brewer, L.K.; Gans, J.H.; Kane, M.A.; Woodson, S.A.; Oglesby-Sherrouse, A.G. The Pseudomonas aeruginosa PrrF1 and PrrF2 small regulatory RNAs promote 2-alkyl-4- quinolone production through redundant regulation of the antR mRNA. J. Bacteriol. 2018, 200, e00704-17. [Google Scholar] [CrossRef] [Green Version]
- Nelson, C.E.; Huang, W.; Brewer, L.K.; Nguyen, A.T.; Kane, M.A.; Wilks, A.; Oglesby-Sherrouse, A.G. Proteomic analysis of the Pseudomonas aeruginosa iron starvation response reveals PrrF small regulatory RNA-dependent iron regulation of twitching motility, amino acid metabolism, and zinc homeostasis proteins. J. Bacteriol. 2019, 201, e00754-1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oglesby-Sherrouse, A.G.; Vasil, M.L. Characterization of a heme-regulated non-coding RNA encoded by the prrF locus of Pseudomonas aeruginosa. PLoS ONE 2010, 5, e9930. [Google Scholar] [CrossRef] [Green Version]
- Osborne, J.; Djapgne, L.; Tran, B.Q.; Goo, Y.A.; Oglesby-Sherrouse, A.G. A method for in vivo identification of bacterial small RNA-binding proteins. Microbiologyopen 2014, 3, 950–960. [Google Scholar] [CrossRef]
- Kawasaki, S.; Arai, H.; Kodama, T.; Igarashi, Y. Gene cluster for dissimilatory nitrite reductase (nir) from Pseudomonas aeruginosa: Sequencing and identification of a locus for heme d1 biosynthesis. J. Bacteriol. 1997, 179, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Reinhart, A.A.; Powell, D.A.; Nguyen, A.T.; O’Neill, M.; Djapgne, L.; Wilks, A.; Ernst, R.K.; Oglesby-Sherrouse, A.G. The prrF-encoded small regulatory RNAs are required for iron homeostasis and virulence of Pseudomonas aeruginosa. Infect. Immun. 2015, 83, 863–875. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, M.J.; Wilks, A. The P. aeruginosa heme binding protein PhuS is a heme oxygenase titratable regulator of heme uptake. ACS Chem. Biol. 2013, 8, 1794–1802. [Google Scholar] [CrossRef] [Green Version]
- Llamas, M.A.; van der Sar, A.; Chu, B.C.H.; Sparrius, M.; Vogel, H.J.; Bitter, W. A Novel extracytoplasmic function (ECF) sigma factor regulates virulence in Pseudomonas aeruginosa. PLoS Pathog. 2009, 5, e1000572. [Google Scholar] [CrossRef]
- Quesada, J.M.; Otero-Asman, J.R.; Bastiaansen, K.C.; Civantos, C.; Llamas, M.A. The activity of the Pseudomonas aeruginosa virulence regulator σ(VreI) is modulated by the anti-σ factor VreR and the transcription factor PhoB. Front. Microbiol. 2016, 7, 1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otero-Asman, J.R.; Quesada, J.M.; Jim, K.K.; Ocampo-Sosa, A.; Civantos, C.; Bitter, W.; Llamas, M.A. The extracytoplasmic function sigma factor σVreI is active during infection and contributes to phosphate starvation-induced virulence of Pseudomonas aeruginosa. Sci. Rep. 2020, 10, 3139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorger-Domenigg, T. Novel Pseudomonas aeruginosa Small Regulatory RNAs. Ph.D. Thesis, University of Vienna, Vienna, Austria, 2010. [Google Scholar]
- Carloni, S.; Macchi, R.; Sattin, S.; Ferrara, S.; Bertoni, G. The small RNA ReaL: A novel regulatory element embedded in the Pseudomonas aeruginosa quorum sensing networks. Environ. Microbiol. 2017, 19, 4220–4237. [Google Scholar] [CrossRef]
- Thi Bach Nguyen, H.; Romero A., D.; Amman, F.; Sorger-Domenigg, T.; Tata, M.; Sonnleitner, E.; Bläsi, U. Negative control of RpoS synthesis by the sRNA ReaL in Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 2488. [Google Scholar] [CrossRef]
- Xia, Y.; Weng, Y.; Xu, C.; Wang, D.; Pan, X.; Tian, Z.; Xia, B.; Li, H.; Chen, R.; Liu, C.; et al. Endoribonuclease YbeY is essential for RNA processing and virulence in Pseudomonas aeruginosa. MBio 2020, 11, e00659-20. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [Green Version]
- Häussler, S.; Becker, T. The Pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 2008, 4, e1000166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas quinolone signal (PQS): Not just for quorum sensing anymore. Front. Cell. Infect. Microbiol. 2018, 8, 230. [Google Scholar] [CrossRef]
- Diggle, S.P.; Cornelis, P.; Williams, P.; Cámara, M. 4-quinolone signalling in Pseudomonas aeruginosa: Old molecules, new perspectives. Int. J. Med. Microbiol. 2006, 296, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Nilsson, M.; Gjermansen, M.; Givskov, M.; Tolker-Nielsen, T. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol. Microbiol. 2009, 74, 1380–1392. [Google Scholar] [CrossRef]
- Guo, Q.; Kong, W.; Jin, S.; Chen, L.; Xu, Y.; Duan, K. PqsR-dependent and PqsR-independent regulation of motility and biofilm formation by PQS in Pseudomonas aeruginosa PAO1. J. Basic Microbiol. 2014, 54, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.M.; Neely, A.; Stintzi, A.; Georges, C.; Holder, I.A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 1996, 64, 518–523. [Google Scholar] [CrossRef] [Green Version]
- Lau, G.W.; Ran, H.; Kong, F.; Hassett, D.J.; Mavrodi, D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 2004, 72, 4275–4278. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, L.A.; McKnight, S.L.; Kuznetsova, M.S.; Pesci, E.C.; Manoil, C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 2002, 184, 6472–6480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lépine, F.; Déziel, E.; Milot, S.; Rahme, L.G. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim. Biophys. Acta 2003, 1622, 36–41. [Google Scholar] [CrossRef]
- Déziel, E.; Lépine, F.; Milot, S.; He, J.; Mindrinos, M.N.; Tompkins, R.G.; Rahme, L.G. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 2004, 101, 1339–1344. [Google Scholar] [CrossRef] [Green Version]
- D’Argenio, D.A.; Wu, M.; Hoffman, L.R.; Kulasekara, H.D.; Déziel, E.; Smith, E.E.; Nguyen, H.; Ernst, R.K.; Larson Freeman, T.J.; Spencer, D.H.; et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 2007, 64, 512–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schertzer, J.W.; Boulette, M.L.; Whiteley, M. More than a signal: Non-signaling properties of quorum sensing molecules. Trends Microbiol. 2009, 17, 189–195. [Google Scholar] [CrossRef]
- Williams, P.; Cámara, M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef]
- Wade, D.S.; Calfee, M.W.; Rocha, E.R.; Ling, E.A.; Engstrom, E.; Coleman, J.P.; Pesci, E.C. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 4372–4380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, N.; Heeb, S.; Valverde, C.; Kay, E.; Reimmann, C.; Junier, T.; Haas, D. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genom. 2008, 9, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, P.; Wang, Y.; Zhang, Y.; Hu, Y.; Thompson, K.M.; Chen, S. RpoS-dependent sRNA RgsA regulates Fis and AcpP in Pseudomonas aeruginosa. Mol. Microbiol. 2016, 102, 244–259. [Google Scholar] [CrossRef] [PubMed]
- Sonnleitner, E.; González, N.; Haas, D. Small RNAs of Pseudomonas spp. In Pseudomonas; Ramos, J., Filloux, A., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 3–28. ISBN 978-90-481-3909-5. [Google Scholar]
- Park, S.H.; Butcher, B.G.; Anderson, Z.; Pellegrini, N.; Bao, Z.; D’Amico, K.; Filiatrault, M.J. Analysis of the small RNA P16/RgsA in the plant pathogen Pseudomonas syringae pv. tomato strain DC3000. Microbiology 2013, 159, 296–306. [Google Scholar] [CrossRef]
- Koch, C.; Vandekerckhove, J.; Kahmann, R. Escherichia coli host factor for site-specific DNA inversion: Cloning and characterization of the fis gene. Proc. Natl. Acad. Sci. USA 1988, 85, 4237–4241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, R.; Lurz, R.; Lüder, G.; Tolksdorf, C.; Travers, A.; Muskhelishvili, G. An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res. 2001, 29, 5107–5114. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.J.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef]
- Long, Y.; Fu, W.; Wang, S.; Deng, X.; Jin, Y.; Bai, F.; Cheng, Z.; Wu, W. Fis contributes to resistance of Pseudomonas aeruginosa to ciprofloxacin by regulating pyocin synthesis. J. Bacteriol. 2020, 202, e00064-20. [Google Scholar] [CrossRef]
- Tata, M.; Amman, F.; Pawar, V.; Wolfinger, M.T.; Weiss, S.; Häussler, S.; Bläsi, U. The anaerobically induced sRNA PaiI affects denitrification in Pseudomonas aeruginosa PA14. Front. Microbiol. 2017, 8, 2312. [Google Scholar] [CrossRef]
- Schreiber, K.; Krieger, R.; Benkert, B.; Eschbach, M.; Arai, H.; Schobert, M.; Jahn, D. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol. 2007, 189, 4310–4314. [Google Scholar] [CrossRef] [Green Version]
- Wenner, N.; Maes, A.; Cotado-Sampayo, M.; Lapouge, K. NrsZ: A novel, processed, nitrogen-dependent, small non-coding RNA that regulates Pseudomonas aeruginosa PAO1 virulence. Environ. Microbiol. 2014, 16, 1053–1068. [Google Scholar] [CrossRef] [Green Version]
- Papenfort, K.; Said, N.; Welsink, T.; Lucchini, S.; Hinton, J.C.D.; Vogel, J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 2009, 74, 139–158. [Google Scholar] [CrossRef]
- Davis, B.M.; Waldor, M.K. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol. Microbiol. 2007, 65, 373–385. [Google Scholar] [CrossRef] [Green Version]
- Caiazza, N.C.; Shanks, R.M.Q.; O’Toole, G.A. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 7351–7361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnleitner, E.; Gonzalez, N.; Sorger-Domenigg, T.; Heeb, S.; Richter, A.S.; Backofen, R.; Williams, P.; Huttenhofer, A.; Haas, D.; Blasi, U. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol. Microbiol. 2011, 80, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Déziel, E.; Gopalan, S.; Tampakaki, A.P.; Lépine, F.; Padfield, K.E.; Saucier, M.; Xiao, G.; Rahme, L.G. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: Multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 2005, 55, 998–1014. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Silistre, H.; Lovelock, L.; Wright, V.J.; Chan, K.-G.; Hong, K.-W.; Williams, P.; Camara, M.; Heeb, S. Genome-wide mapping of the RNA targets of the Pseudomonas aeruginosa riboregulatory protein RsmN. Nucleic Acids Res. 2018, 46, 6823–6840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, L.; Breidenstein, E.B.M.; Taylor, P.K.; Bains, M.; De La Fuente-Nunez, C.; Fang, Y.; Foster, L.J.; Hancock, R.E.W. Interconnection of post-transcriptional regulation: The RNA-binding protein Hfq is a novel target of the Lon protease in Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26811. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.; Pu, Q.; Wu, Q.; Zhou, C.; Wang, B.; Schettler, J.; Wang, Z.; Qin, S.; Gao, P.; Li, R.; et al. High-throughput screen reveals sRNAs regulating crRNA biogenesis by targeting CRISPR leader to repress Rho termination. Nat. Commun. 2019, 10, 3728. [Google Scholar] [CrossRef]
- Kambara, T.K.; Ramsey, K.M.; Dove, S.L. Pervasive targeting of nascent transcripts by Hfq. Cell Rep. 2018, 23, 1543–1552. [Google Scholar] [CrossRef]
- Gebhardt, M.J.; Kambara, T.K.; Ramsey, K.M.; Dove, S.L. Widespread targeting of nascent transcripts by RsmA in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2020, 117, 10520–10529. [Google Scholar] [CrossRef]
- Sun, X.; Zhulin, I.; Wartell, R.M. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002, 30, 3662–3671. [Google Scholar] [CrossRef] [Green Version]
- Scofield, D.G.; Lynch, M. Evolutionary diversification of the Sm family of RNA-associated proteins. Mol. Biol. Evol. 2008, 25, 2255–2267. [Google Scholar] [CrossRef]
- MacGregor, C.H.; Wolff, J.A.; Arora, S.K.; Phibbs, P.V.J. Cloning of a catabolite repression control (crc) gene from Pseudomonas aeruginosa, expression of the gene in Escherichia coli, and identification of the gene product in Pseudomonas aeruginosa. J. Bacteriol. 1991, 173, 7204–7212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnleitner, E.; Wulf, A.; Campagne, S.; Pei, X.-Y.; Wolfinger, M.T.; Forlani, G.; Prindl, K.; Abdou, L.; Resch, A.; Allain, F.H.-T.; et al. Interplay between the catabolite repression control protein Crc, Hfq and RNA in Hfq-dependent translational regulation in Pseudomonas aeruginosa. Nucleic Acids Res. 2018, 46, 1470–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, X.Y.; Dendooven, T.; Sonnleitner, E.; Chen, S.; Bläsi, U.; Luisi, B.F. Architectural principles for Hfq/Crc-mediated regulation of gene expression. Elife 2019, 8, e43158. [Google Scholar] [CrossRef] [PubMed]
- Sonnleitner, E.; Hagens, S.; Rosenau, F.; Wilhelm, S.; Habel, A.; Jäger, K.-E.; Bläsi, U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 2003, 35, 217–228. [Google Scholar] [CrossRef]
- Sonnleitner, E.; Schuster, M.; Sorger-Domenigg, T.; Greenberg, E.P.; Blasi, U.; Bläsi, U. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 2006, 59, 1542–1558. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Ding, S.; Chen, F.; Zhang, X.; Xia, Y.; Di, H.; Cao, Q.; Deng, X.; Wu, M.; Wong, C.C.L.; et al. The Crc protein participates in down-regulation of the Lon gene to promote rhamnolipid production and rhl quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 2015, 96, 526–547. [Google Scholar] [CrossRef]
- Pusic, P.; Sonnleitner, E.; Krennmayr, B.; Heitzinger, D.A.; Wolfinger, M.T.; Resch, A.; Bläsi, U. Harnessing metabolic regulation to increase Hfq-dependent antibiotic susceptibility in Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 2709. [Google Scholar] [CrossRef] [Green Version]
- Zheng, A.; Panja, S.; Woodson, S.A. Arginine patch predicts the RNA annealing activity of Hfq from Gram-negative and Gram-positive bacteria. J. Mol. Biol. 2016, 428, 2259–2264. [Google Scholar] [CrossRef] [Green Version]
- Møller, T.; Franch, T.; Højrup, P.; Keene, D.R.; Bächinger, H.P.; Brennan, R.G.; Valentin-Hansen, P. Hfq: A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 2002, 9, 23–30. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Pearson, R.F.; Møller, T.; Valentin-Hansen, P.; Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 2002, 21, 3546–3556. [Google Scholar] [CrossRef] [Green Version]
- Mikulecky, P.J.; Kaw, M.K.; Brescia, C.C.; Takach, J.C.; Sledjeski, D.D.; Feig, A.L. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004, 11, 1206–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Ribeiro, E.A.J.; Beich-Frandsen, M.; Konarev, P.V.; Shang, W.; Vecerek, B.; Kontaxis, G.; Hämmerle, H.; Peterlik, H.; Svergun, D.I.; Bläsi, U.; et al. Structural flexibility of RNA as molecular basis for Hfq chaperone function. Nucleic Acids Res. 2012, 40, 8072–8084. [Google Scholar] [CrossRef] [PubMed]
- Otaka, H.; Ishikawa, H.; Morita, T.; Aiba, H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc. Natl. Acad. Sci. USA 2011, 108, 13059–13064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauer, E.; Weichenrieder, O. Structural basis for RNA 3’-end recognition by Hfq. Proc. Natl. Acad. Sci. USA 2011, 108, 13065–13070. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, H.; Otaka, H.; Maki, K.; Morita, T.; Aiba, H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3’ poly(U) tail. RNA 2012, 18, 1062–1074. [Google Scholar] [CrossRef] [Green Version]
- Sauer, E.; Schmidt, S.; Weichenrieder, O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. USA 2012, 109, 9396–9401. [Google Scholar] [CrossRef] [Green Version]
- Panja, S.; Schu, D.J.; Woodson, S.A. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013, 41, 7536–7546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Curtis, J.E.; Fang, X.; Woodson, S.A. Structural model of an mRNA in complex with the bacterial chaperone Hfq. Proc. Natl. Acad. Sci. USA 2014, 111, 17134–17139. [Google Scholar] [CrossRef] [Green Version]
- Schu, D.J.; Zhang, A.; Gottesman, S.; Storz, G. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 2015, 34, 2557–2573. [Google Scholar] [CrossRef] [Green Version]
- Link, T.M.; Valentin-Hansen, P.; Brennan, R.G. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. USA 2009, 106, 19292–19297. [Google Scholar] [CrossRef] [Green Version]
- Murina, V.; Lekontseva, N.; Nikulin, A. Hfq binds ribonucleotides in three different RNA-binding sites. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1504–1513. [Google Scholar] [CrossRef]
- Robinson, K.E.; Orans, J.; Kovach, A.R.; Link, T.M.; Brennan, R.G. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 2014, 42, 2736–2749. [Google Scholar] [CrossRef]
- Zhang, A.; Schu, D.J.; Tjaden, B.C.; Storz, G.; Gottesman, S. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J. Mol. Biol. 2013, 425, 3678–3697. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, W.; Li, F.; Zhang, J.; Wu, J.; Gong, Q.; Shi, Y. Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq. Nucleic Acids Res. 2015, 43, 2400–2411. [Google Scholar] [CrossRef] [Green Version]
- Sonnleitner, E.; Valentini, M.; Wenner, N.; Haichar, F.E.Z.; Haas, D.; Lapouge, K. Novel targets of the CbrAB/Crc carbon catabolite control system revealed by transcript abundance in Pseudomonas aeruginosa. PLoS ONE 2012, 7, e44637. [Google Scholar] [CrossRef] [Green Version]
- Linares, J.F.; Moreno, R.; Fajardo, A.; Martínez-Solano, L.; Escalante, R.; Rojo, F.; Martínez, J.L. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ. Microbiol. 2010, 12, 3196–3212. [Google Scholar] [CrossRef]
- Corona, F.; Reales-Calderón, J.A.; Gil, C.; Martínez, J.L. The development of a new parameter for tracking post-transcriptional regulation allows the detailed map of the Pseudomonas aeruginosa Crc regulon. Sci. Rep. 2018, 8, 16793. [Google Scholar] [CrossRef]
- Romeo, T.; Vakulskas, C.A.; Babitzke, P. Post-transcriptional regulation on a global scale: Form and function of Csr/Rsm systems. Environ. Microbiol. 2013, 15, 313–324. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Potts, A.H.; Babitzke, P.; Ahmer, B.M.M.; Romeo, T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 2015, 79, 193–224. [Google Scholar] [CrossRef] [Green Version]
- Holmqvist, E.; Wright, P.R.; Li, L.; Bischler, T.; Barquist, L.; Reinhardt, R.; Backofen, R.; Vogel, J. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016, 35, 991–1011. [Google Scholar] [CrossRef]
- Kusmierek, M.; Dersch, P. Regulation of host-pathogen interactions via the post-transcriptional Csr/Rsm system. Curr. Opin. Microbiol. 2018, 41, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Farrow, J.M., 3rd; Wells, G.; Palethorpe, S.; Adams, M.D.; Pesci, E.C. CsrA supports both environmental persistence and host-associated growth of Acinetobacter baumannii. Infect. Immun. 2020, 88, e00259-20. [Google Scholar] [CrossRef]
- Butz, H.A.; Mey, A.R.; Ciosek, A.L.; Crofts, A.A.; Davies, B.W.; Payne, S.M. Regulatory effects of CsrA in Vibrio cholerae. MBio 2021, 12, e03380-20. [Google Scholar] [CrossRef] [PubMed]
- Burrowes, E.; Baysse, C.; Adams, C.; O’Gara, F. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 2006, 152, 405–418. [Google Scholar] [CrossRef] [Green Version]
- Dubey, A.K.; Baker, C.S.; Romeo, T.; Babitzke, P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 2005, 11, 1579–1587. [Google Scholar] [CrossRef] [Green Version]
- Schubert, M.; Lapouge, K.; Duss, O.; Oberstrass, F.C.; Jelesarov, I.; Haas, D.; Allain, F.H.-T. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 2007, 14, 807–813. [Google Scholar] [CrossRef]
- Marden, J.N.; Diaz, M.R.; Walton, W.G.; Gode, C.J.; Betts, L.; Urbanowski, M.L.; Redinbo, M.R.; Yahr, T.L.; Wolfgang, M.C. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2013, 110, 15055–15060. [Google Scholar] [CrossRef] [Green Version]
- Schulmeyer, K.H.; Diaz, M.R.; Bair, T.B.; Sanders, W.; Gode, C.J.; Laederach, A.; Wolfgang, M.C.; Yahr, T.L. Primary and secondary sequence structure requirements for recognition and discrimination of target RNAs by Pseudomonas aeruginosa RsmA and RsmF. J. Bacteriol. 2016, 198, 2458–2469. [Google Scholar] [CrossRef] [Green Version]
- Chihara, K.; Barquist, L.; Takasugi, K.; Noda, N.; Tsuneda, S. Global identification of RsmA/N binding sites in Pseudomonas aeruginosa by in vivo UV CLIP-seq. RNA Biol. 2021, 1–16. [Google Scholar] [CrossRef]
- Brencic, A.; Lory, S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 2009, 72, 612–632. [Google Scholar] [CrossRef] [Green Version]
- Heurlier, K.; Williams, F.; Heeb, S.; Dormond, C.; Pessi, G.; Singer, D.; Cámara, M.; Williams, P.; Haas, D. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2004, 186, 2936–2945. [Google Scholar] [CrossRef] [Green Version]
- Frangipani, E.; Visaggio, D.; Heeb, S.; Kaever, V.; Cámara, M.; Visca, P.; Imperi, F. The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ. Microbiol. 2014, 16, 676–688. [Google Scholar] [CrossRef]
- Sonnleitner, E.; Romeo, A.; Bläsi, U. Small regulatory RNAs in Pseudomonas aeruginosa. RNA Biol. 2012, 9, 364–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Rosa, R.; Behrends, V.; Williams, H.D.; Bundy, J.G.; Rojo, F. Influence of the Crc regulator on the hierarchical use of carbon sources from a complete medium in Pseudomonas. Environ. Microbiol. 2016, 18, 807–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina, L.; La Rosa, R.; Nogales, J.; Rojo, F. Influence of the Crc global regulator on substrate uptake rates and the distribution of metabolic fluxes in Pseudomonas putida KT2440 growing in a complete medium. Environ. Microbiol. 2019, 21, 4446–4459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hester, K.L.; Lehman, J.; Najar, F.; Song, L.; Roe, B.A.; MacGregor, C.H.; Hager, P.W.; Phibbs, P.V.J.; Sokatch, J.R. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 1144–1149. [Google Scholar] [CrossRef] [Green Version]
- Hester, K.L.; Madhusudhan, K.T.; Sokatch, J.R. Catabolite repression control by crc in 2xYT medium is mediated by posttranscriptional regulation of bkdR expression in Pseudomonas putida. J. Bacteriol. 2000, 182, 1150–1153. [Google Scholar] [CrossRef] [Green Version]
- Sonnleitner, E.; Abdou, L.; Haas, D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2009, 106, 21866–21871. [Google Scholar] [CrossRef]
- Huang, J.; Sonnleitner, E.; Ren, B.; Xu, Y.; Haas, D. Catabolite repression control of pyocyanin biosynthesis at an intersection of primary and secondary metabolism in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012, 78, 5016–5020. [Google Scholar] [CrossRef] [Green Version]
- Reales-Calderón, J.A.; Corona, F.; Monteoliva, L.; Gil, C.; Martínez, J.L. Quantitative proteomics unravels that the post-transcriptional regulator Crc modulates the generation of vesicles and secreted virulence determinants of Pseudomonas aeruginosa. J. Proteomics 2015, 127, 352–364. [Google Scholar] [CrossRef] [Green Version]
- La Rosa, R.; Nogales, J.; Rojo, F. The Crc/CrcZ-CrcY global regulatory system helps the integration of gluconeogenic and glycolytic metabolism in Pseudomonas putida. Environ. Microbiol. 2015, 17, 3362–3378. [Google Scholar] [CrossRef]
- Moreno, R.; Hernández-Arranz, S.; La Rosa, R.; Yuste, L.; Madhushani, A.; Shingler, V.; Rojo, F. The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ. Microbiol. 2015, 17, 105–118. [Google Scholar] [CrossRef]
- Malecka, E.M.; Bassani, F.; Dendooven, T.; Sonnleitner, E.; Rozner, M.; Albanese, T.G.; Resch, A.; Luisi, B.; Woodson, S.; Bläsi, U. Stabilization of Hfq-mediated translational repression by the co-repressor Crc in Pseudomonas aeruginosa. Nucleic Acids Res. 2021, 49, 7075–7087. [Google Scholar] [CrossRef]
- Pusic, P.; Tata, M.; Wolfinger, M.T.; Sonnleitner, E.; Häussler, S.; Bläsi, U. Cross-regulation by CrcZ RNA controls anoxic biofilm formation in Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 39621. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, G.A.; Gibbs, K.A.; Hager, P.W.; Phibbs, P.V.J.; Kolter, R. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 425–431. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Gao, Q.; Chen, W.; Qin, H.; Hengzhuang, W.; Chen, Y.; Yang, L.; Zhang, G. Regulation of PQS quorum sensing via catabolite repression control in Pseudomonas aeruginosa. Microbiology 2013, 159, 1931–1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, R.; Fonseca, P.; Rojo, F. Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol. Microbiol. 2012, 83, 24–40. [Google Scholar] [CrossRef]
- Filiatrault, M.J.; Stodghill, P.V.; Bronstein, P.A.; Moll, S.; Lindeberg, M.; Grills, G.; Schweitzer, P.; Wang, W.; Schroth, G.P.; Luo, S.; et al. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J. Bacteriol. 2010, 192, 2359–2372. [Google Scholar] [CrossRef] [Green Version]
- Filiatrault, M.J.; Stodghill, P.V.; Wilson, J.; Butcher, B.G.; Chen, H.; Myers, C.R.; Cartinhour, S.W. CrcZ and CrcX regulate carbon source utilization in Pseudomonas syringae pathovar tomato strain DC3000. RNA Biol. 2013, 10, 245–255. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Gokhale, C.S.; Rainey, P.B.; Zhang, X.-X. Unravelling the complexity and redundancy of carbon catabolic repression in Pseudomonas fluorescens SBW25. Mol. Microbiol. 2017, 105, 589–605. [Google Scholar] [CrossRef] [Green Version]
- Abdou, L.; Chou, H.T.; Haas, D.; Lu, C.D. Promoter recognition and activation by the global response regulator CbrB in Pseudomonas aeruginosa. J. Bacteriol. 2011, 193, 2784–2792. [Google Scholar] [CrossRef] [Green Version]
- Nishijyo, T.; Haas, D.; Itoh, Y. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol. Microbiol. 2001, 40, 917–931. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lu, C.-D. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 5413–5420. [Google Scholar] [CrossRef] [Green Version]
- Valentini, M.; García-Mauriño, S.M.; Pérez-Martínez, I.; Santero, E.; Canosa, I.; Lapouge, K. Hierarchical management of carbon sources is regulated similarly by the CbrA/B systems in Pseudomonas aeruginosa and Pseudomonas putida. Microbiology 2014, 160, 2243–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romeo, T.; Gong, M.; Liu, M.Y.; Brun-Zinkernagel, A.M. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 1993, 175, 4744–4755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Arranz, S.; Sánchez-Hevia, D.; Rojo, F.; Moreno, R. Effect of Crc and Hfq proteins on the transcription, processing, and stability of the Pseudomonas putida CrcZ sRNA. RNA 2016, 22, 1902–1917. [Google Scholar] [CrossRef] [Green Version]
- Pessi, G.; Williams, F.; Hindle, Z.; Heurlier, K.; Holden, M.T.G.; Cámara, M.; Haas, D.; Williams, P. The global posttranscriptional regulator RsmA modulates production of virulence determinants N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 2001, 183, 6676–6683. [Google Scholar] [CrossRef] [Green Version]
- Morris, E.R.; Hall, G.; Li, C.; Heeb, S.; Kulkarni, R.V.; Lovelock, L.; Silistre, H.; Messina, M.; Cámara, M.; Emsley, J.; et al. Structural rearrangement in an RsmA/CsrA ortholog of Pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure 2013, 21, 1659–1671. [Google Scholar] [CrossRef] [Green Version]
- Romeo, T.; Gong, M. Genetic and physical mapping of the regulatory gene csrA on the Escherichia coli K-12 chromosome. J. Bacteriol. 1993, 175, 5740–5741. [Google Scholar] [CrossRef] [Green Version]
- Sabnis, N.A.; Yang, H.; Romeo, T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 1995, 270, 29096–29104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potts, A.H.; Vakulskas, C.A.; Pannuri, A.; Yakhnin, H.; Babitzke, P.; Romeo, T. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat. Commun. 2017, 8, 1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobrero, P.M.; Valverde, C. Comparative genomics and evolutionary analysis of RNA-binding proteins of the CsrA family in the genus Pseudomonas. Front. Mol. Biosci. 2020, 7, 127. [Google Scholar] [CrossRef]
- Janssen, K.H.; Diaz, M.R.; Golden, M.; Graham, J.W.; Sanders, W.; Wolfgang, M.C.; Yahr, T.L. Functional analyses of the RsmY and RsmZ small noncoding regulatory RNAs in Pseudomonas aeruginosa. J. Bacteriol. 2018, 200, e00736-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, K.H.; Diaz, M.R.; Gode, C.J.; Wolfgang, M.C.; Yahr, T.L. RsmV, a small noncoding regulatory RNA in Pseudomonas aeruginosa that sequesters RsmA and RsmF from target mRNAs. J. Bacteriol. 2018, 200, e00277-18. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, H.; Callaghan, J.; Grady, E.P.; Maciá, M.D.; Borrell, N.; Gómez, C.; Casey, P.G.; Hill, C.; Adams, C.; Gahan, C.G.M.; et al. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect. Immun. 2008, 76, 632–638. [Google Scholar] [CrossRef] [Green Version]
- Yahr, T.L.; Wolfgang, M.C. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 2006, 62, 631–640. [Google Scholar] [CrossRef]
- Brutinel, E.D.; Vakulskas, C.A.; Brady, K.M.; Yahr, T.L. Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 2008, 68, 657–671. [Google Scholar] [CrossRef]
- Intile, P.J.; Diaz, M.R.; Urbanowski, M.L.; Wolfgang, M.C.; Yahr, T.L. The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 2014, 196, 357–366. [Google Scholar] [CrossRef] [Green Version]
- de Bentzmann, S.; Giraud, C.; Bernard, C.S.; Calderon, V.; Ewald, F.; Plésiat, P.; Nguyen, C.; Grunwald, D.; Attree, I.; Jeannot, K.; et al. Unique biofilm signature, drug susceptibility and decreased virulence in Drosophila through the Pseudomonas aeruginosa two-component system PprAB. PLoS Pathog. 2012, 8, e1003052. [Google Scholar] [CrossRef]
- Baynham, P.J.; Ramsey, D.M.; Gvozdyev, B.V.; Cordonnier, E.M.; Wozniak, D.J. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 2006, 188, 132–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, A.; Zhang, M.; Du, W.; Wang, D.; Ma, L.Z. A molecular mechanism for how sigma factor AlgT and transcriptional regulator AmrZ inhibit twitching motility in Pseudomonas aeruginosa. Environ. Microbiol. 2021, 23, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Allsopp, L.P.; Wood, T.E.; Howard, S.A.; Maggiorelli, F.; Nolan, L.M.; Wettstadt, S.; Filloux, A. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 7707–7712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baynham, P.J.; Wozniak, D.J. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 1996, 22, 97–108. [Google Scholar] [CrossRef]
- Baynham, P.J.; Brown, A.L.; Hall, L.L.; Wozniak, D.J. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 1999, 33, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Waligora, E.A.; Ramsey, D.M.; Pryor, E.E.J.; Lu, H.; Hollis, T.; Sloan, G.P.; Deora, R.; Wozniak, D.J. AmrZ beta-sheet residues are essential for DNA binding and transcriptional control of Pseudomonas aeruginosa virulence genes. J. Bacteriol. 2010, 192, 5390–5401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryor, E.E.J.; Waligora, E.A.; Xu, B.; Dellos-Nolan, S.; Wozniak, D.J.; Hollis, T. The transcription factor AmrZ utilizes multiple DNA binding modes to recognize activator and repressor sequences of Pseudomonas aeruginosa virulence genes. PLoS Pathog. 2012, 8, e1002648. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.J.; Newsom, D.; Kelly, B.; Irie, Y.; Jennings, L.K.; Xu, B.; Limoli, D.H.; Harrison, J.J.; Parsek, M.R.; White, P.; et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 2014, 10, e1003984. [Google Scholar] [CrossRef]
- Tart, A.H.; Blanks, M.J.; Wozniak, D.J. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 2006, 188, 6483–6489. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.J.; Ryder, C.R.; Mann, E.E.; Wozniak, D.J. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J. Bacteriol. 2013, 195, 1637–1644. [Google Scholar] [CrossRef] [Green Version]
- Rocchetta, H.L.; Burrows, L.L.; Lam, J.S. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 1999, 63, 523–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cross, A.R.; Goldberg, J.B. Remodeling of O antigen in mucoid Pseudomonas aeruginosa via transcriptional repression of wzz2. MBio 2019, 10, e02914-18. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Shao, X.; Xie, Y.; Wang, T.; Zhang, Y.; Wang, X.; Deng, X. An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa. Nat. Commun. 2019, 10, 2931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, A.L.; Kulasekara, B.; Rietsch, A.; Boyd, D.; Smith, R.S.; Lory, S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 2004, 7, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, S.; Kuchma, S.L.; O’Toole, G.A. Keeping their options open: Acute versus persistent infections. J. Bacteriol. 2006, 188, 1211–1217. [Google Scholar] [CrossRef] [Green Version]
- Wagner, V.E.; Iglewski, B.H. P. aeruginosa biofilms in CF infection. Clin. Rev. Allergy Immunol. 2008, 35, 124–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franklin, M.J.; Nivens, D.E.; Weadge, J.T.; Howell, P.L. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2011, 2, 167. [Google Scholar] [CrossRef] [Green Version]
- Silverman, J.M.; Brunet, Y.R.; Cascales, E.; Mougous, J.D. Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 2012, 66, 453–472. [Google Scholar] [CrossRef] [Green Version]
- Irie, Y.; Starkey, M.; Edwards, A.N.; Wozniak, D.J.; Romeo, T.; Parsek, M.R. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 2010, 78, 158–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wozniak, D.J.; Ohman, D.E. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 1994, 176, 6007–6014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hershberger, C.D.; Ye, R.W.; Parsek, M.R.; Xie, Z.D.; Chakrabarty, A.M. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (sigma E). Proc. Natl. Acad. Sci. USA 1995, 92, 7941–7945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stacey, S.D.; Pritchett, C.L. Pseudomonas aeruginosa AlgU contributes to posttranscriptional activity by increasing rsmA expression in a mucA22 Strain. J. Bacteriol. 2016, 198, 1812–1826. [Google Scholar] [CrossRef] [Green Version]
- Lee, V.T.; Matewish, J.M.; Kessler, J.L.; Hyodo, M.; Hayakawa, Y.; Lory, S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 2007, 65, 1474–1484. [Google Scholar] [CrossRef] [Green Version]
- Coggan, K.A.; Wolfgang, M.C. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 2012, 14, 47–70. [Google Scholar]
- Moscoso, J.A.; Jaeger, T.; Valentini, M.; Hui, K.; Jenal, U.; Filloux, A. The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 4081–4088. [Google Scholar] [CrossRef] [Green Version]
- Colley, B.; Dederer, V.; Carnell, M.; Kjelleberg, S.; Rice, S.A.; Klebensberger, J. SiaA/D interconnects c-di-GMP and RsmA signaling to coordinate cellular aggregation of Pseudomonas aeruginosa in response to rnvironmental conditions. Front. Microbiol. 2016, 7, 179. [Google Scholar] [CrossRef]
- Huertas-Rosales, Ó.; Romero, M.; Heeb, S.; Espinosa-Urgel, M.; Cámara, M.; Ramos-González, M.I. The Pseudomonas putida CsrA/RsmA homologues negatively affect c-di-GMP pools and biofilm formation through the GGDEF/EAL response regulator CfcR. Environ. Microbiol. 2017, 19, 3551–3566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.; Tolker-Nielsen, T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006, 59, 1114–1128. [Google Scholar] [CrossRef]
- Kay, E.; Humair, B.; Dénervaud, V.; Riedel, K.; Spahr, S.; Eberl, L.; Valverde, C.; Haas, D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 6026–6033. [Google Scholar] [CrossRef] [Green Version]
- Burrowes, E.; Abbas, A.; O’Neill, A.; Adams, C.; O’Gara, F. Characterisation of the regulatory RNA RsmB from Pseudomonas aeruginosa PAO1. Res. Microbiol. 2005, 156, 7–16. [Google Scholar] [CrossRef]
- Valverde, C.; Heeb, S.; Keel, C.; Haas, D. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 2003, 50, 1361–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, C.L.; Romero, M.; Karna, S.L.R.; Chen, T.; Heeb, S.; Leung, K.P. RsmW, Pseudomonas aeruginosa small non-coding RsmA-binding RNA upregulated in biofilm versus planktonic growth conditions. BMC Microbiol. 2016, 16, 155. [Google Scholar] [CrossRef] [PubMed]
- Heeb, S.; Heurlier, K.; Valverde, C.; Cámara, M.; Haas, D.; Williams, P. Post-transcriptional regulation in Pseudomonas spp. via the Gac/Rsm regulatory network. In Virulence and Gene Regulation; Ramos, J.-L., Ed.; Springer: Boston, MA, USA, 2004; pp. 239–255. ISBN 978-1-4419-9084-6. [Google Scholar]
- Mulcahy, H.; O’Callaghan, J.; O’Grady, E.P.; Adams, C.; O’Gara, F. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect. Immun. 2006, 74, 3012–3015. [Google Scholar] [CrossRef] [Green Version]
- Brencic, A.; McFarland, K.A.; McManus, H.R.; Castang, S.; Mogno, I.; Dove, S.L.; Lory, S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 2009, 73, 434–445. [Google Scholar] [CrossRef] [Green Version]
- Bordi, C.; Lamy, M.-C.; Ventre, I.; Termine, E.; Hachani, A.; Fillet, S.; Roche, B.; Bleves, S.; Méjean, V.; Lazdunski, A.; et al. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 2010, 76, 1427–1443. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Yang, G.; Debru, A.B.; Li, P.; Zong, L.; Li, P.; Xu, T.; Wu, W.; Jin, S.; Bao, Q. SuhB regulates the motile-sessile switch in Pseudomonas aeruginosa through the Gac/Rsm pathway and c-di-GMP signaling. Front. Microbiol. 2017, 8, 1045. [Google Scholar] [CrossRef] [PubMed]
- Petrova, O.E.; Sauer, K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 2010, 192, 5275–5288. [Google Scholar] [CrossRef] [Green Version]
- Ventre, I.; Goodman, A.L.; Vallet-Gely, I.; Vasseur, P.; Soscia, C.; Molin, S.; Bleves, S.; Lazdunski, A.; Lory, S.; Filloux, A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 171–176. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, M.A.; Osborn, E.; Kazmierczak, B.I. A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol. Microbiol. 2004, 54, 1090–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laskowski, M.A.; Kazmierczak, B.I. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect. Immun. 2006, 74, 4462–4473. [Google Scholar] [CrossRef] [Green Version]
- Goodman, A.L.; Merighi, M.; Hyodo, M.; Ventre, I.; Filloux, A.; Lory, S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009, 23, 249–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambonnier, G.; Roux, L.; Redelberger, D.; Fadel, F.; Filloux, A.; Sivaneson, M.; de Bentzmann, S.; Bordi, C. The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PLoS Genet. 2016, 12, e1006032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhagirath, A.Y.; Pydi, S.P.; Li, Y.; Lin, C.; Kong, W.; Chelikani, P.; Duan, K. Characterization of the direct interaction between hybrid sensor kinases PA1611 and RetS that controls biofilm formation and the type III secretion system in Pseudomonas aeruginosa. ACS Infect. Dis. 2017, 3, 162–175. [Google Scholar] [CrossRef]
- Jean-Pierre, F.; Tremblay, J.; Déziel, E. Broth versus surface-grown cells: Differential regulation of RsmY/Z small RNAs in Pseudomonas aeruginosa by the Gac/HptB system. Front. Microbiol. 2017, 7, 2168. [Google Scholar] [CrossRef]
- Bouillet, S.; Ba, M.; Houot, L.; Iobbi-Nivol, C.; Bordi, C. Connected partner-switches control the life style of Pseudomonas aeruginosa through RpoS regulation. Sci. Rep. 2019, 9, 6496. [Google Scholar] [CrossRef]
- Li, K.; Xu, C.; Jin, Y.; Sun, Z.; Liu, C.; Shi, J.; Chen, G.; Chen, R.; Jin, S.; Wu, W. SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. MBio 2013, 4, e00419-13. [Google Scholar] [CrossRef] [Green Version]
- O’Callaghan, J.; Reen, F.J.; Adams, C.; O’Gara, F. Low oxygen induces the type III secretion system in Pseudomonas aeruginosa via modulation of the small RNAs rsmZ and rsmY. Microbiology 2011, 157, 3417–3428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravarty, S.; Melton, C.N.; Bailin, A.; Yahr, T.L.; Anderson, G.G. Pseudomonas aeruginosa magnesium transporter MgtE inhibits type III secretion system gene expression by stimulating rsmYZ transcription. J. Bacteriol. 2017, 199, e00268-17. [Google Scholar] [CrossRef] [Green Version]
- Janssen, K.H.; Corley, J.M.; Djapgne, L.; Cribbs, J.T.; Voelker, D.; Slusher, Z.; Nordell, R.; Regulski, E.E.; Kazmierczak, B.I.; McMackin, E.W.; et al. Hfq and sRNA 179 inhibit expression of the Pseudomonas aeruginosa cAMP-Vfr and type III secretion regulons. MBio 2020, 11, e00363-20. [Google Scholar] [CrossRef]
- Liu, W.; Li, M.; Jiao, L.; Wang, P.; Yan, Y. PmrA/PmrB two-component system regulation of lipA expression in Pseudomonas aeruginosa PAO1. Front. Microbiol. 2018, 8, 2690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castang, S.; McManus, H.R.; Turner, K.H.; Dove, S.L. H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. USA 2008, 105, 18947–18952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Ye, F.; Kumar, V.; Gao, Y.-G.; Zhang, L.-H. BswR controls bacterial motility and biofilm formation in Pseudomonas aeruginosa through modulation of the small RNA rsmZ. Nucleic Acids Res. 2014, 42, 4563–4576. [Google Scholar] [CrossRef] [PubMed]
- Reimmann, C.; Valverde, C.; Kay, E.; Haas, D. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 2005, 187, 276–285. [Google Scholar] [CrossRef] [Green Version]
- Sorger-Domenigg, T.; Sonnleitner, E.; Kaberdin, V.R.; Bläsi, U. Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem. Biophys. Res. Commun. 2007, 352, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, J.A.; Chopra, A.K. The exoribonuclease polynucleotide phosphorylase influences the virulence and stress responses of yersiniae and many other pathogens. Front. Cell. Infect. Microbiol. 2013, 3, 81. [Google Scholar] [CrossRef]
- Chen, R.; Weng, Y.; Zhu, F.; Jin, Y.; Liu, C.; Pan, X.; Xia, B.; Cheng, Z.; Jin, S.; Wu, W. Polynucleotide phosphorylase regulates multiple virulence factors and the stabilities of small RNAs RsmY/Z in Pseudomonas aeruginosa. Front. Microbiol. 2016, 7, 247. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Förstner, K.U.; Holmqvist, E.; Otto, A.; Günster, R.; Becher, D.; Reinhardt, R.; Vogel, J. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. USA 2016, 113, 11591–11596. [Google Scholar] [CrossRef] [Green Version]
- Smirnov, A.; Schneider, C.; Hör, J.; Vogel, J. Discovery of new RNA classes and global RNA-binding proteins. Curr. Opin. Microbiol. 2017, 39, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Gerovac, M.; El Mouali, Y.; Kuper, J.; Kisker, C.; Barquist, L.; Vogel, J. Global discovery of bacterial RNA-binding proteins by RNase-sensitive gradient profiles reports a new FinO domain protein. RNA 2020, 26, 1448–1463. [Google Scholar] [CrossRef] [PubMed]
- Hör, J.; Garriss, G.; Di Giorgio, S.; Hack, L.-M.; Vanselow, J.T.; Förstner, K.U.; Schlosser, A.; Henriques-Normark, B.; Vogel, J. Grad-seq in a Gram-positive bacterium reveals exonucleolytic sRNA activation in competence control. EMBO J. 2020, 39, e103852. [Google Scholar] [CrossRef] [PubMed]
- Hör, J.; Vogel, J. Analysis of the RNA and protein complexome by Grad-seq. Methods Mol. Biol. 2021, 2300, 183–201. [Google Scholar] [CrossRef]
- Gerovac, M.; Wicke, L.; Chihara, K.; Schneider, C.; Lavigne, R.; Vogel, J. A Grad-seq view of RNA and protein complexes in Pseudomonas aeruginosa under standard and bacteriophage predation conditions. MBio 2021, 12, e03454-20. [Google Scholar] [CrossRef] [PubMed]
- Gerovac, M.; Vogel, J.; Smirnov, A. The world of stable ribonucleoproteins and its mapping with Grad-seq and related approaches. Front. Mol. Biosci. 2021, 8, 661448. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet. Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Woolhouse, M.; Waugh, C.; Perry, M.R.; Nair, H. Global disease burden due to antibiotic resistance state of the evidence. J. Glob. Health 2016, 6, 10306. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Dong, B.; Liu, W.; Liu, Z. Recent advances in peptide nucleic acids as antibacterial agents. Curr. Med. Chem. 2021, 28, 1104–1125. [Google Scholar] [CrossRef]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef] [Green Version]
- Pifer, R.; Greenberg, D.E. Antisense antibacterial compounds. Transl. Res. 2020, 223, 89–106. [Google Scholar] [CrossRef]
- Vogel, J. An RNA biology perspective on species-specific programmable RNA antibiotics. Mol. Microbiol. 2020, 113, 550–559. [Google Scholar] [CrossRef] [Green Version]
- Good, L.; Nielsen, P.E. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 1998, 16, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Forster, A.C.; Altman, S. External guide sequences for an RNA enzyme. Science 1990, 249, 783–786. [Google Scholar] [CrossRef]
- Wojciechowska, M.; Równicki, M.; Mieczkowski, A.; Miszkiewicz, J.; Trylska, J. Antibacterial peptide nucleic acids-facts and perspectives. Molecules 2020, 25, 559. [Google Scholar] [CrossRef] [Green Version]
- Castillo, J.I.; Równicki, M.; Wojciechowska, M.; Trylska, J. Antimicrobial synergy between mRNA targeted peptide nucleic acid and antibiotics in E. coli. Bioorg. Med. Chem. Lett. 2018, 28, 3094–3098. [Google Scholar] [CrossRef]
- Martínez-Guitián, M.; Vázquez-Ucha, J.C.; Álvarez-Fraga, L.; Conde-Pérez, K.; Bou, G.; Poza, M.; Beceiro, A. Antisense inhibition of lpxB gene expression in Acinetobacter baumannii by peptide-PNA conjugates and synergy with colistin. J. Antimicrob. Chemother. 2020, 75, 51–59. [Google Scholar] [CrossRef]
- Ayhan, D.H.; Tamer, Y.T.; Akbar, M.; Bailey, S.M.; Wong, M.; Daly, S.M.; Greenberg, D.E.; Toprak, E. Sequence-specific targeting of bacterial resistance genes increases antibiotic efficacy. PLoS Biol. 2016, 14, e1002552. [Google Scholar] [CrossRef]
- Howard, J.J.; Sturge, C.R.; Moustafa, D.A.; Daly, S.M.; Marshall-Batty, K.R.; Felder, C.F.; Zamora, D.; Yabe-Gill, M.; Labandeira-Rey, M.; Bailey, S.M.; et al. Inhibition of Pseudomonas aeruginosa by peptide-conjugated phosphorodiamidate morpholino oligomers. Antimicrob. Agents Chemother. 2017, 61, e01938-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosal, A.; Nielsen, P.E. Potent antibacterial antisense peptide-peptide nucleic acid conjugates against Pseudomonas aeruginosa. Nucleic Acid Ther. 2012, 22, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Aunins, T.R.; Erickson, K.E.; Chatterjee, A. Transcriptome-based design of antisense inhibitors potentiates carbapenem efficacy in CRE Escherichia coli. Proc. Natl. Acad. Sci. USA 2020, 117, 30699–30709. [Google Scholar] [CrossRef]
- Moustafa, D.A.; Wu, A.W.; Zamora, D.; Daly, S.M.; Sturge, C.R.; Pybus, C.; Geller, B.L.; Goldberg, J.B.; Greenberg, D.E. Peptide-conjugated phosphorodiamidate morpholino oligomers retain activity against multidrug-resistant Pseudomonas aeruginosa in vitro and in vivo. MBio 2021, 12, e02411-20. [Google Scholar] [CrossRef] [PubMed]
- Mondhe, M.; Chessher, A.; Goh, S.; Good, L.; Stach, J.E.M. Species-selective killing of bacteria by antimicrobial peptide-PNAs. PLoS ONE 2014, 9, e89082. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, T.; Brauer, A.L.; Kirkham, C.; Sully, E.K.; Pettigrew, M.M.; Kong, Y.; Geller, B.L.; Murphy, T.F. Antimicrobial activity of antisense peptide-peptide nucleic acid conjugates against non-typeable Haemophilus influenzae in planktonic and biofilm forms. J. Antimicrob. Chemother. 2017, 72, 137–144. [Google Scholar] [CrossRef] [Green Version]
- Melamed, S.; Adams, P.P.; Zhang, A.; Zhang, H.; Storz, G. RNA-RNA Interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol. Cell 2020, 77, 411–425. [Google Scholar] [CrossRef]
- Immer, C.; Hacker, C.; Wöhnert, J. Solution structure and RNA-binding of a minimal ProQ-homolog from Legionella pneumophila (Lpp1663). RNA 2020, 26, 2031–2043. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, E.; Berggren, S.; Rizvanovic, A. RNA-binding activity and regulatory functions of the emerging sRNA-binding protein ProQ. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194596. [Google Scholar] [CrossRef]
- Stein, E.M.; Kwiatkowska, J.; Basczok, M.M.; Gravel, C.M.; Berry, K.E.; Olejniczak, M. Determinants of RNA recognition by the FinO domain of the Escherichia coli ProQ protein. Nucleic Acids Res. 2020, 48, 7502–7519. [Google Scholar] [CrossRef]
- Bauriedl, S.; Gerovac, M.; Heidrich, N.; Bischler, T.; Barquist, L.; Vogel, J.; Schoen, C. The minimal meningococcal ProQ protein has an intrinsic capacity for structure-based global RNA recognition. Nat. Commun. 2020, 11, 2823. [Google Scholar] [CrossRef]
- Pandey, S.; Gravel, C.M.; Stockert, O.M.; Wang, C.D.; Hegner, C.L.; LeBlanc, H.; Berry, K.E. Genetic identification of the functional surface for RNA binding by Escherichia coli ProQ. Nucleic Acids Res. 2020, 48, 4507–4520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, P.; Gimpel, M.; Wildenhain, T.; Brantl, S. A new role for CsrA: Promotion of complex formation between an sRNA and its mRNA target in Bacillus subtilis. RNA Biol. 2019, 16, 972–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ul Haq, I.; Müller, P.; Brantl, S. Intermolecular communication in Bacillus subtilis: RNA-RNA, RNA-protein andsmall protein-protein interactions. Front. Mol. Biosci. 2020, 7, 178. [Google Scholar] [CrossRef] [PubMed]
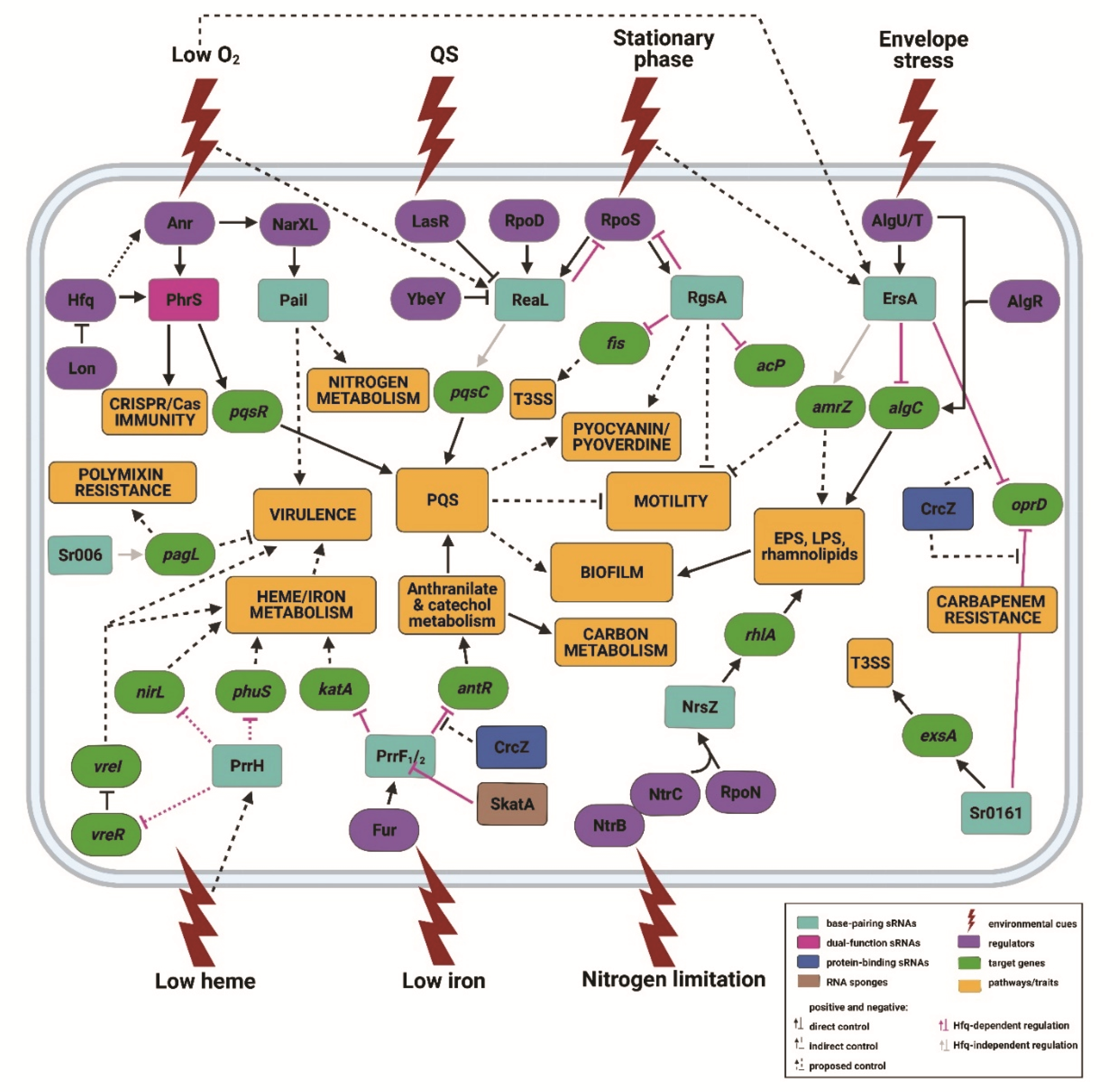
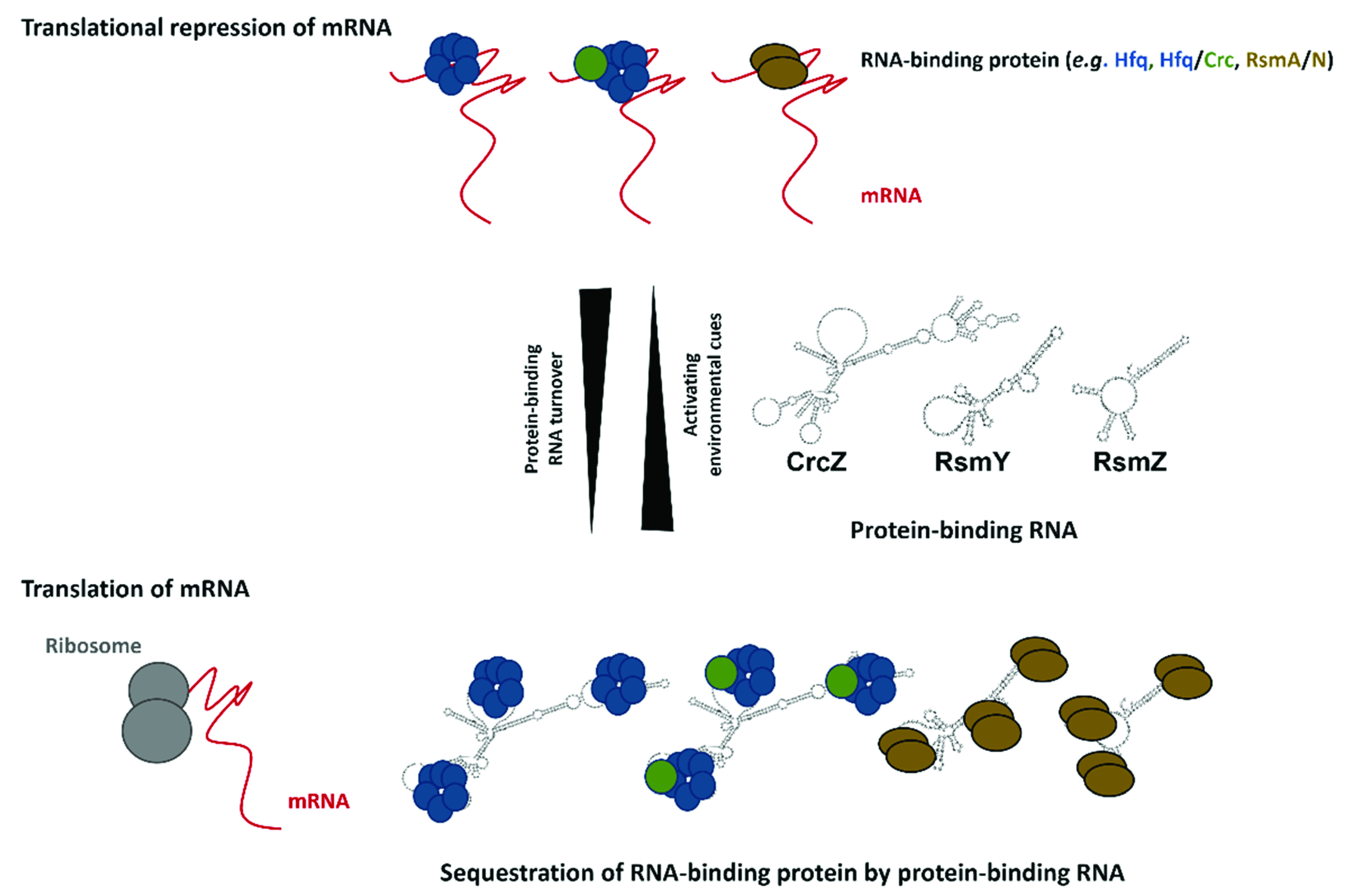
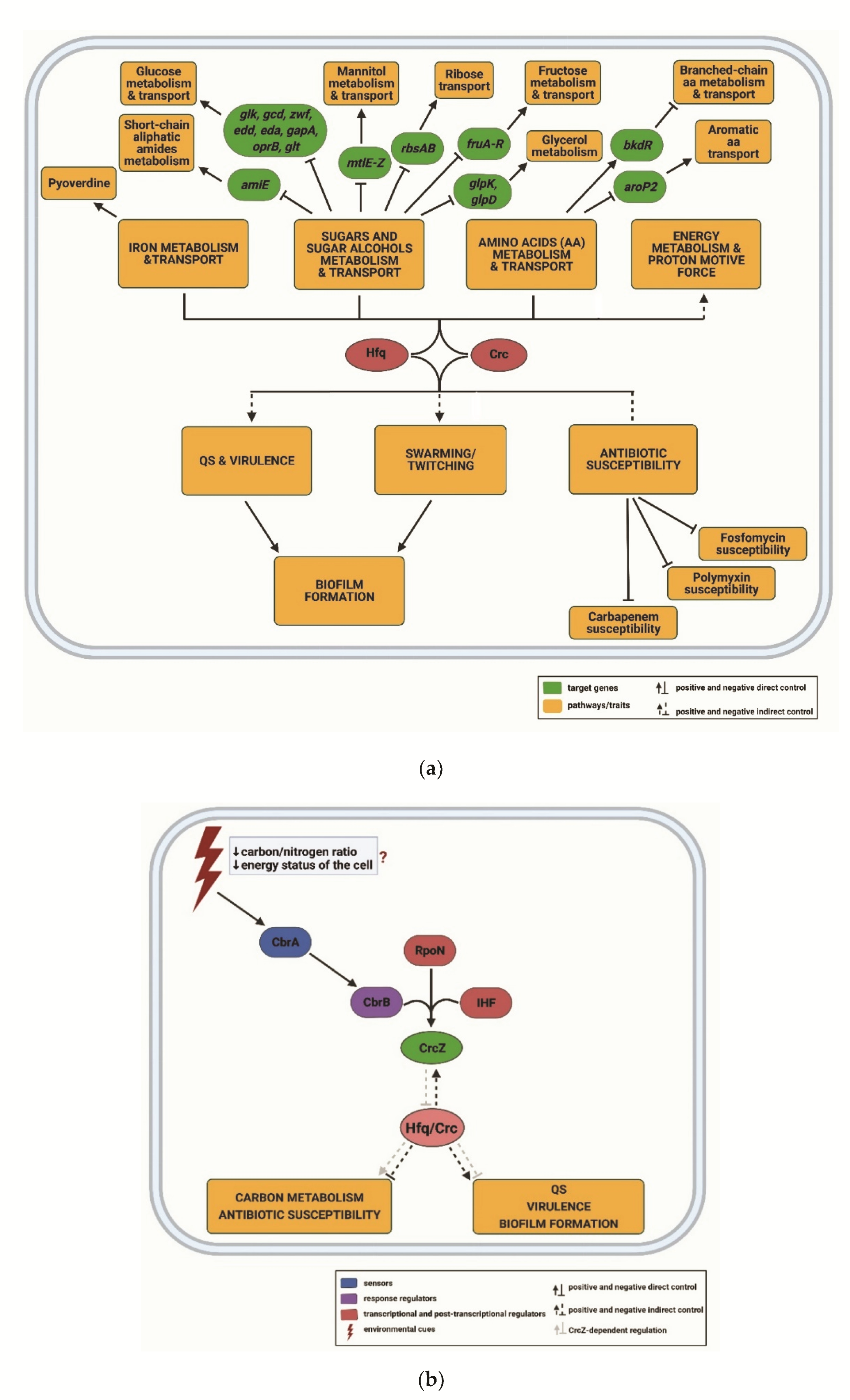

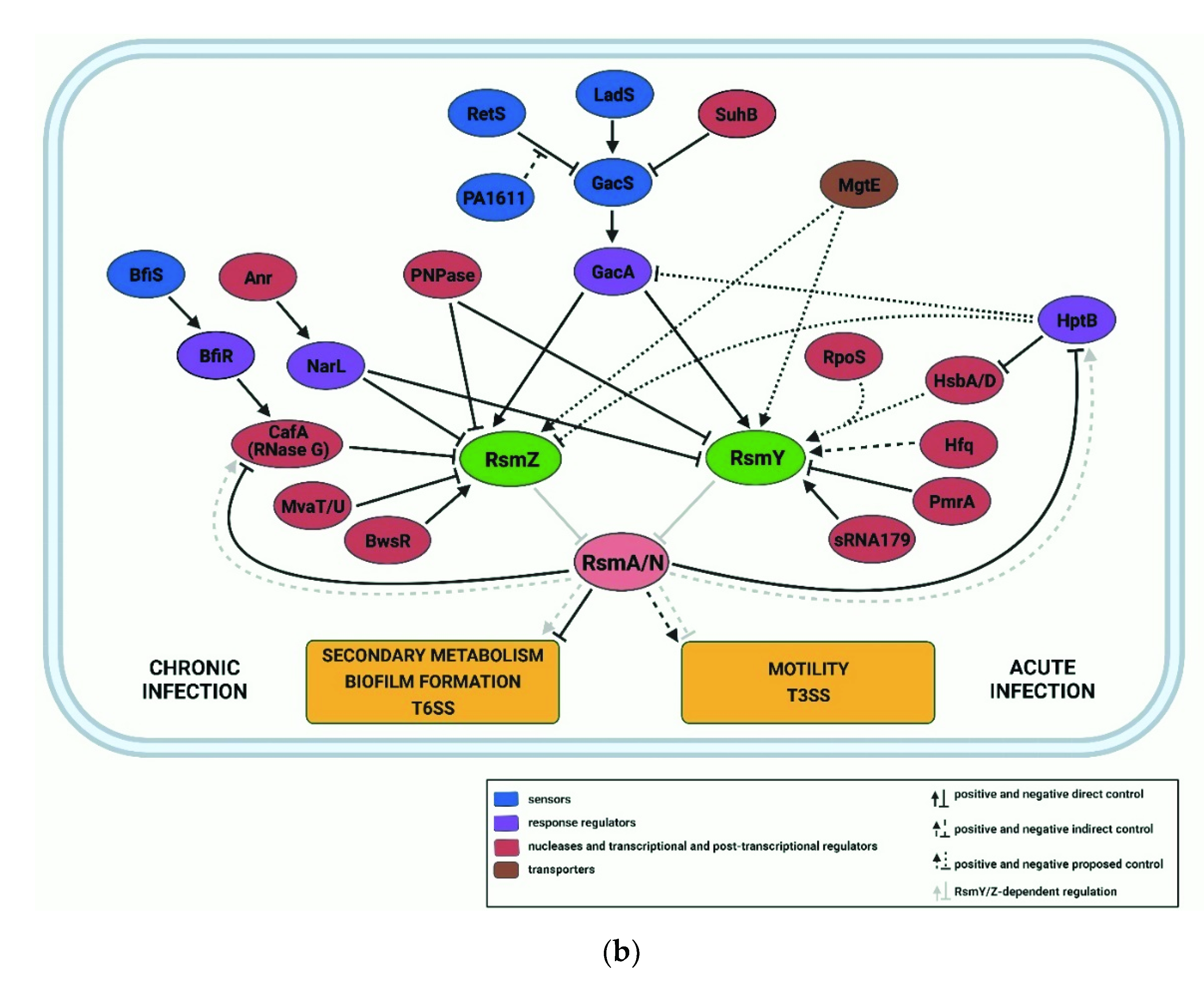
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pusic, P.; Sonnleitner, E.; Bläsi, U. Specific and Global RNA Regulators in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 8632. https://doi.org/10.3390/ijms22168632
Pusic P, Sonnleitner E, Bläsi U. Specific and Global RNA Regulators in Pseudomonas aeruginosa. International Journal of Molecular Sciences. 2021; 22(16):8632. https://doi.org/10.3390/ijms22168632
Chicago/Turabian StylePusic, Petra, Elisabeth Sonnleitner, and Udo Bläsi. 2021. "Specific and Global RNA Regulators in Pseudomonas aeruginosa" International Journal of Molecular Sciences 22, no. 16: 8632. https://doi.org/10.3390/ijms22168632





