Abstract
Gram-negative bacteria release Outer Membrane Vesicles (OMVs) into the extracellular environment. Recent studies recognized these vesicles as vectors to horizontal gene transfer; however, the parameters that mediate OMVs transfer within bacterial communities remain unclear. The present study highlights for the first time the transfer of plasmids containing resistance genes via OMVs derived from Klebsiella pneumoniae (K. pneumoniae). This mechanism confers DNA protection, it is plasmid copy number dependent with a ratio of 3.6 times among high copy number plasmid (pGR) versus low copy number plasmid (PRM), and the transformation efficiency was 3.6 times greater. Therefore, the DNA amount in the vesicular lumen and the efficacy of horizontal gene transfer was strictly dependent on the identity of the plasmid. Moreover, the role of K. pneumoniae-OMVs in interspecies transfer was described. The transfer ability was not related to the phylogenetic characteristics between the donor and the recipient species. K. pneumoniae-OMVs transferred plasmid to Escherichia coli, Salmonella enterica, Pseudomonas aeruginosa and Burkholderia cepacia. These findings address the pivotal role of K. pneumoniae-OMVs as vectors for antimicrobial resistance genes spread, contributing to the development of antibiotic resistance in the microbial communities.
1. Introduction
Horizontal gene transfer (HGT) represents the main source of genetic material transfer among microorganisms [1]. Indeed, HGT provides a driving force for bacterial evolution, increasing bacterial survival, adjustment rate in the harshest environments, and pathogenicity [2,3,4]. Current knowledge of HGT is based on three widely described mechanisms for the exchange of genetic material between bacteria: transformation, conjugation and transduction [5,6,7]. Transformation involves the natural uptake of naked DNA from an extracellular environment; this phenomenon occurs when cells are in a physiological state of competence, regulated by 20–50 proteins [8,9]. Conjugation is a DNA transfer mechanism through the sexual pilus and requires cell-to-cell contact [10]. Conjugative systems are frequently associated with plasmid transfer [11]. Transduction entails the transfer of DNA between bacteria through the bacteriophage infections [12]. The recombinant phage particle can contain up to 100 kilobases of DNA and the infection is limited to host specificity [13]. Recently, several studies reported that HGT processes are facilitated by Outer Membrane Vesicles (OMVs) [14,15,16,17].
OMVs are spherical nanostructures, 50–250 nm in diameter, released naturally and constitutively by Gram-negative bacteria during their growth [18]. OMVs originate from the outer membrane and include in the vesicular lumen lipopolysaccharide, peptidoglycan, phospholipids, genetic material (DNA and RNA) and periplasmic and cytoplasmic protein components, during their biogenesis [19]. Although many aspects of vesicular biogenesis and regulation of their composition remain unclear, the biological functions associated with OMVs release have been extensively described [20,21]. These vesicles play a key role in the bacteria–environment, bacteria–bacteria and bacteria–host interactions [22,23]. OMVs are recognized for their role in nutrient acquisition, response to stress, biofilm formation and toxins release, adhesion and virulence factors, and in host defense system evasion [24,25]. OMVs role in HGT was reported in E. coli, Acinetobacter baumannii, Acinetobacter baylyi, Porphyromonas gingivalis, P. aeruginosa and Thermus thermophilus [25,26,27,28]. Yaron et al. demonstrated the transfer of virulence genes through E. coli-OMVs between bacteria of different species. Moreover, they proved that the genetic material was protected from digestion with DNase, confirming the packaging in the vesicular lumen [29,30]. OMVs derived from A. baumannii were also identified as vectors for antibiotic resistance gene transfer. In the study of Rumbo et al., plasmid-borne blaOXA-24 gene conferred carbapenems resistance to sensitive Acinetobacter strains [31]. These evidences highlight the potential OMVs contribution to the spread of virulence and antibiotic resistance which represents, to date, a serious risk to human health.
In this scenario, K. pneumoniae represents one of the most worrying pathogens involved in nosocomial infections [32]. The constant antibiotics treatment induces selective pressures, causing the evolution of multidrug-resistant (MDR) bacteria [33]. Our previous studies demonstrated that OMVs derived from K. pneumoniae play a crucial role in the microorganism–host interaction, modulating miRNAs genetic transcription and influencing the inflammatory response [14,34]. Currently, no study showed the role of K. pneumoniae-OMVs as a carrier for HGT, allowing the transport of genetic material and the spread of resistance genes. Therefore, this study demonstrates, for the first time, K. pneumoniae-OMVs HGT role. We investigated OMVs contribution in the genetic material cargo and in intra- and inter-species transfer. After, we collected evidence to demonstrate that plasmid copy number (PCN) might play an important role in the biogenesis, cargo and in the HGT mechanisms. Finally, we verified OMVs stability over time and whether storage conditions might influence gene transfer.
2. Results
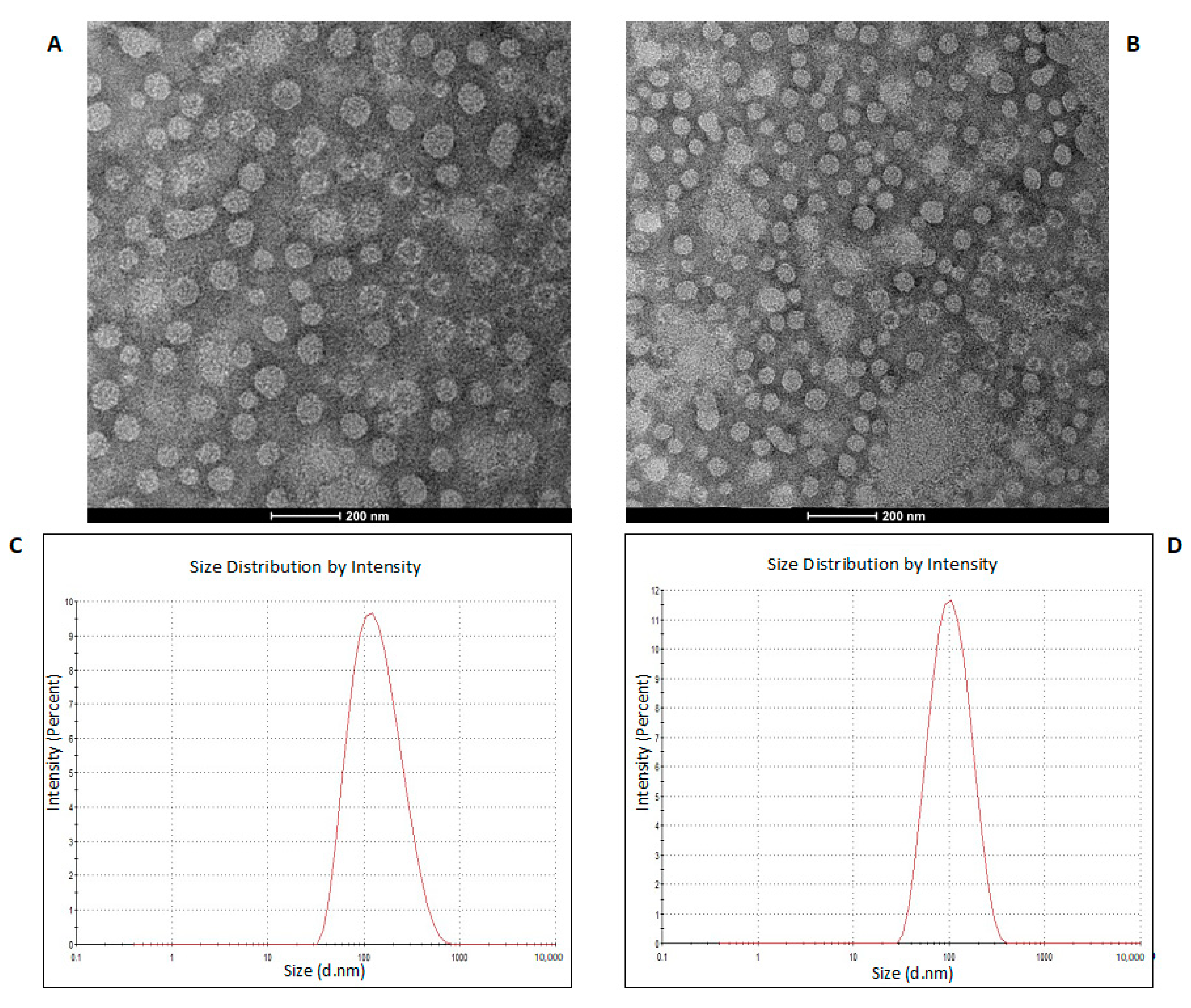
2.1. Characterization of Isolated K. pneumoniae-OMVs
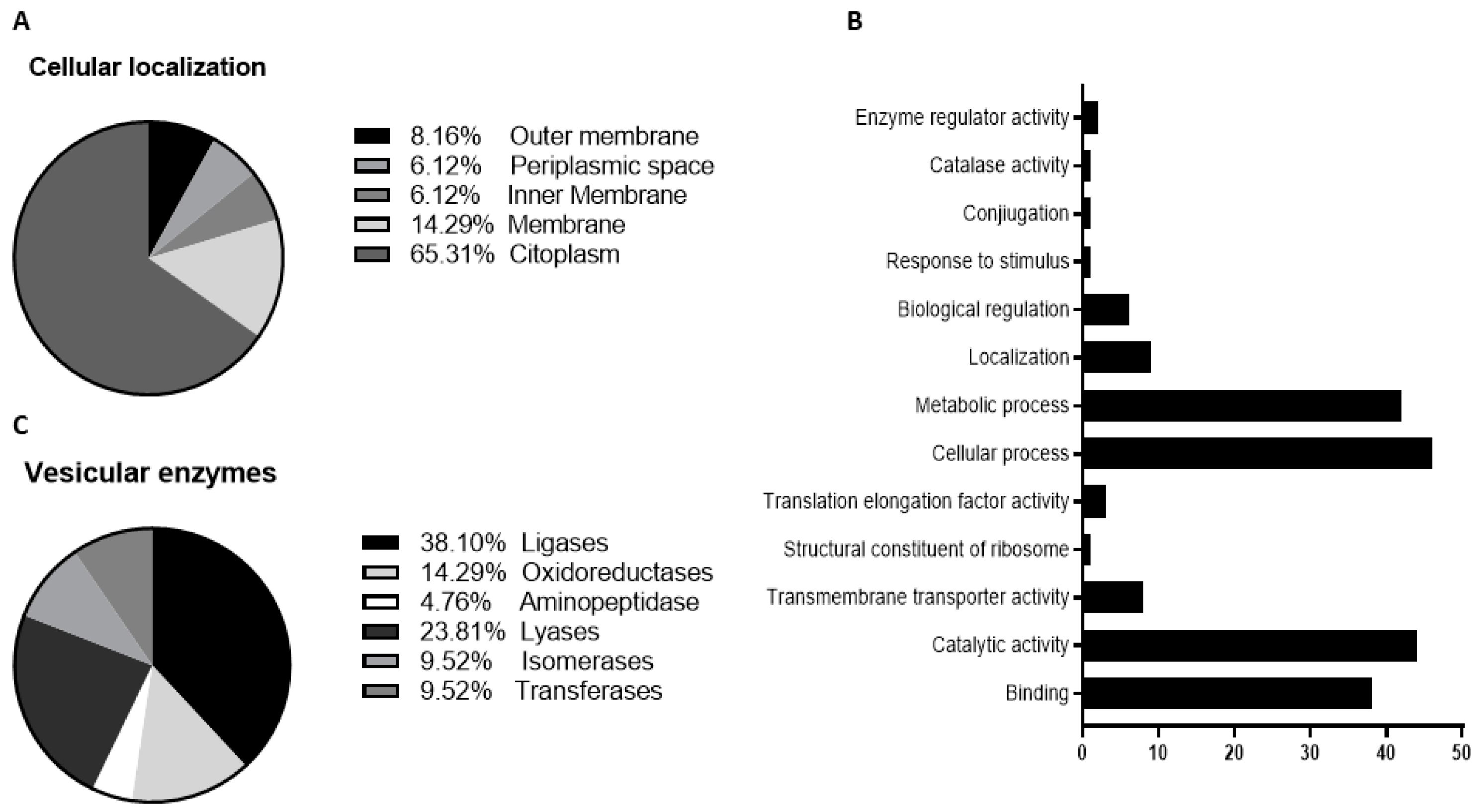
To purify the OMVs derived from K. pneumoniae-pGR and K. pneumoniae-PRM, bacteria were grown in LB supplemented with ampicillin up to the late logarithmic-phase of the bacterial growth curve (see Materials and Methods for specifications). Vesicles were collected from culture supernatants and characterized in terms of morphology, size and polydispersity index (PDI). Purified OMVs appeared at TEM as electron-dense particles, with uniform spherical morphology (Figure 1, panel A and B). No bacterial contaminant was visualized, demonstrating the total sterility of the vesicular suspensions used. Dynamic light scattering (DLS) analysis showed that OMVs derived from K. pneumoniae-pGR measured a size of 113.8 ± 53.7 nm and were characterized by a slightly heterogeneous size distribution, represented by the PDI of 0.223 (Figure 1C). OMVs purified from K. pneumoniae-PRM showed a reduction in size and higher vesicular populations homogeneity, recording a size of 94.13 ± 41.10 nm and a PDI of 0.191 (Figure 1D). Purified OMVs were also characterized based on protein profile. The total vesicular proteins were extracted from K. pneumoniae-OMVs via lysis buffer and then quantified by Bradford assay. The protein amount was 35.77 mg and 30.00 mg for K. pneumoniae-pGR and K. pneumoniae-PRM, respectively, obtained from 600 mL of culture supernatant. Five micrograms of protein were loaded on 10% SDS-PAGE and the gel was stained with Blue Coomassie (Supplementary Figure S1). The corresponding bands were excised and subjected to in-situ digestion protocols. Peptides were analyzed by high resolution nanoLC- MS/MS. Mass spectra analysis allowed identifying with high confidence proteins common to both purified OMVs samples (Supplementary Table S1). These proteins were classified according to the subcellular localization site and biological function (Figure 2A,B). The vesicles contained 14 membrane-associated proteins (28.57%), 3 periplasmic proteins (6.12%) and 32 cytosolic proteins (65.31%). In addition, 3 DNA-binding proteins (6.12%) were identified among the proteins annotated for their binding function. Twenty-one enzymes were revealed, including 3 oxidoreductases (14.29%), 2 transferases (9.52%), 1 aminopeptidase (4.76%), 5 lyases (23.81%), 2 isomerases (9.52%) and 8 ligases (38.10%) (Figure 2C). To confirm accurate reliability and reproducibility of data, three independent OMVs purifications were performed and analyzed for each strain.
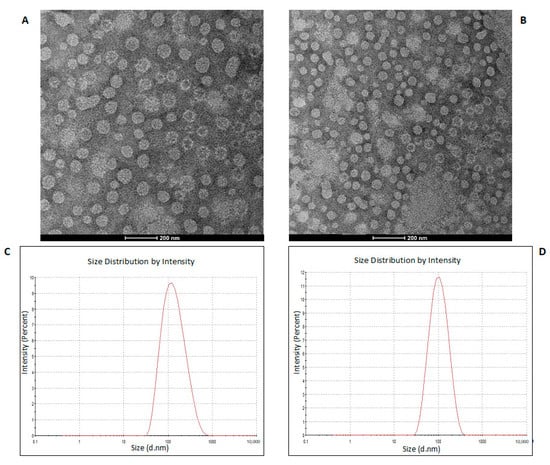
Figure 1.
TEM of OMVs purified from K. pneumoniae-pGR (A) and K. pneumoniae-PRM (B) (scale bar = 200 nm). DLS intensity-weighed distribution of OMVs derived from K. pneumoniae-pGR (C) and K. pneumoniae-PRM (D).
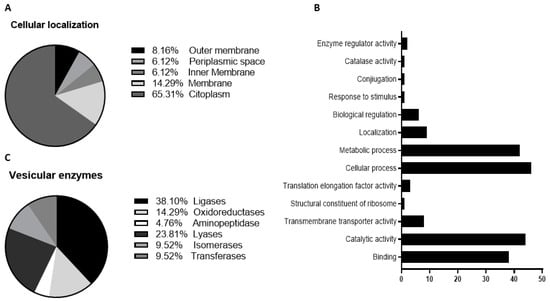
Figure 2.
Classification of cellular localization (A), functional annotation (B) and enzymatic classes (C) of protein extracted from K. pneumoniae-OMVs.
2.2. DNA Packaging in K. pneumoniae OMVs
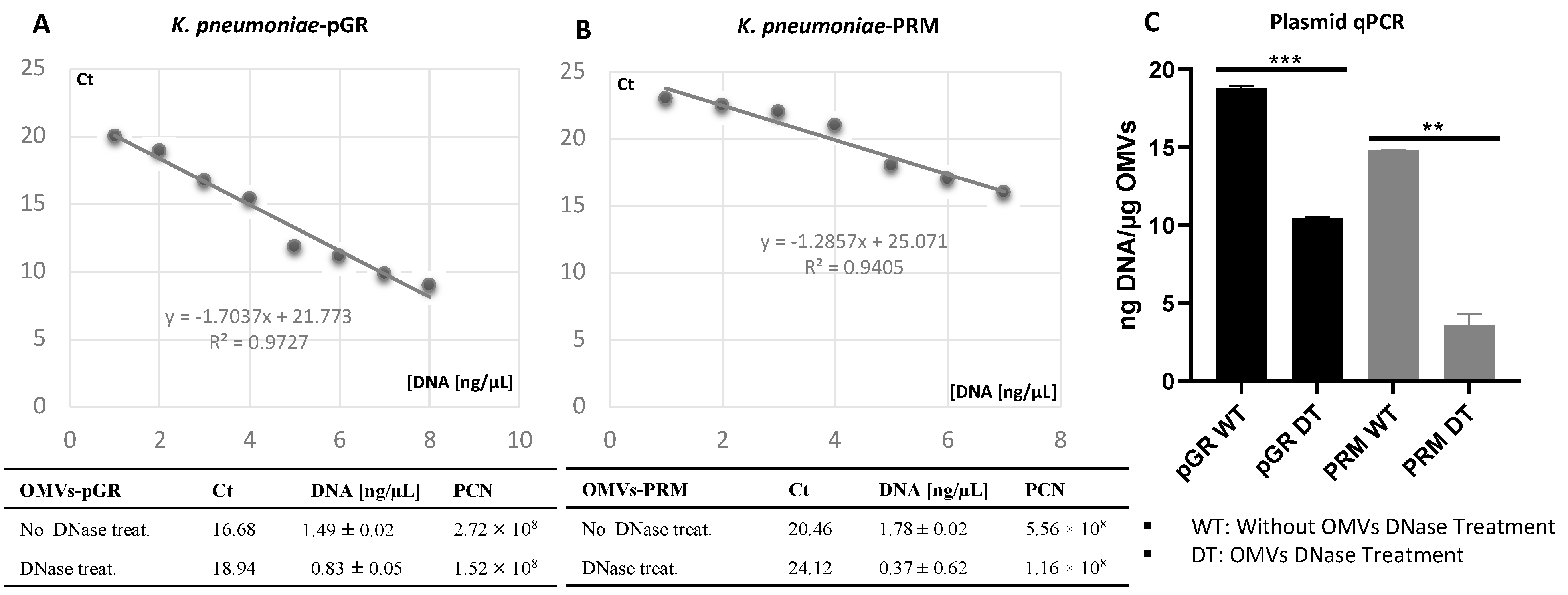
The propensity of K. pneumoniae OMVs to incorporate genetic material during the biogenesis process was evaluated by transforming bacteria with pGR and PRM plasmids. K. pneumoniae-pGR and K. pneumoniae-PRM were grown on LB supplemented with 100 µg mL−1 of ampicillin for selection of transformants. Plasmid DNA extraction and enzymatic digestion profile confirmed the plasmids presence in the bacterial strains (Supplemental Figure S2A,B). The presence of pGR and PRM plasmids in K. pneumoniae-OMVs was evaluated by absolute qPCR. To demonstrate that DNA was present in the vesicular lumen and protected from the extracellular nucleases action, qPCR was performed using OMVs samples either treated or untreated with DNase. In untreated OMVs, plasmid concentration was 18.91 ± 0.53 and 14.78 ± 0.91 ng DNA/µg OMVs, for pGR and PRM, respectively. In OMVs treated with DNase before vesicular lysis, pGR recorded a higher loading density, measuring 10.4 ± 0.05 ng DNA/µg OMVs, which corresponded approximately to 1.9 × 109 PCN/µg OMVs. Otherwise, PRM measured a plasmid concentration of 3.08 ± 0.62 ng DNA/µg OMVs, corresponding to 9.6 × 108 PCN/µg OMVs (Figure 3).
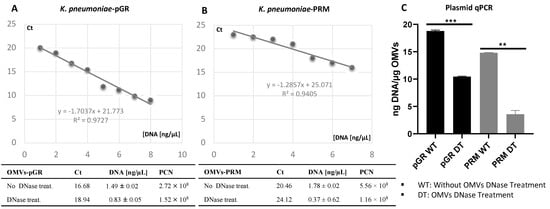
Figure 3.
Determination of PCN in OMVs, using quantitative PCR standard curves. The standard curves were generated by qPCR of the purified pGR (A) and PRM (B) plasmids. Histogram Graph of PGR and PRM cargo efficiency, before and after DNase treatment (C) ** p value < 0.005, *** p value < 0.0005.
2.3. OMVs Mediate the Plasmid Intraspecies Transfer
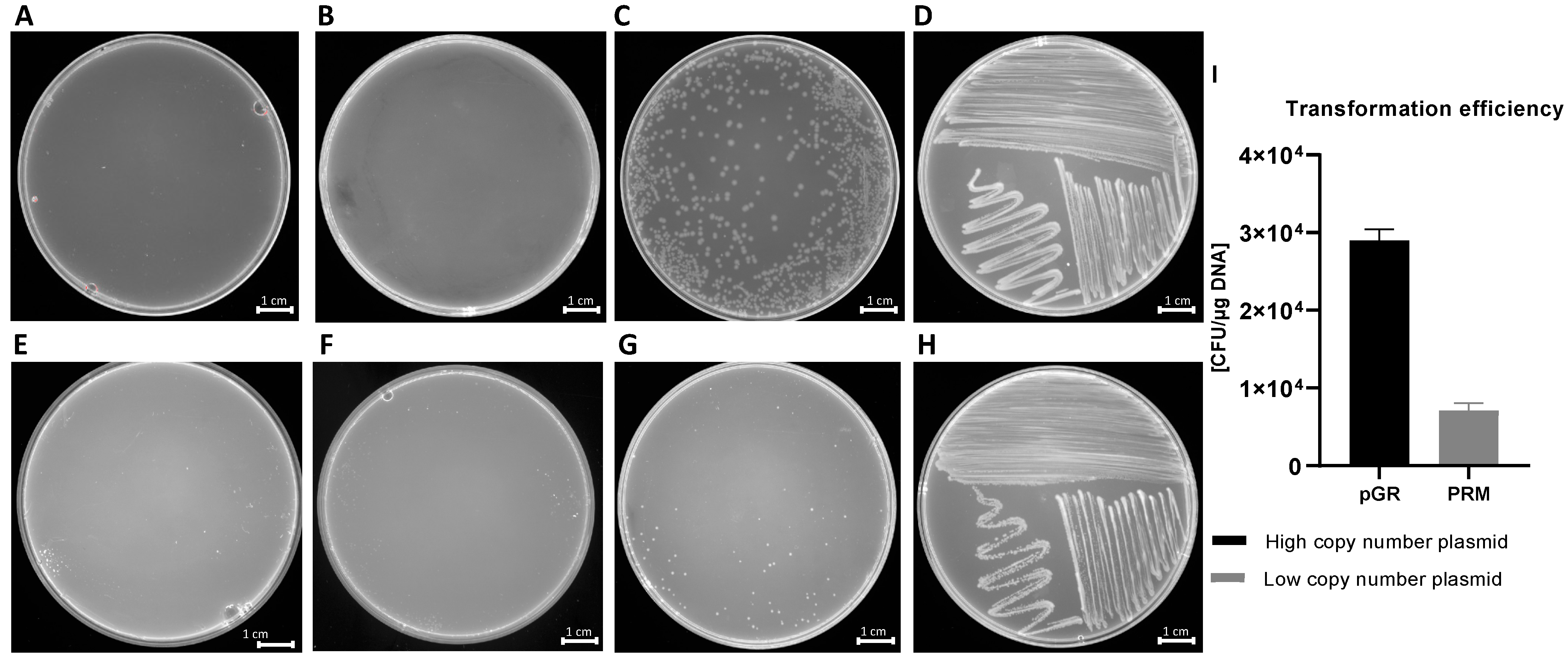
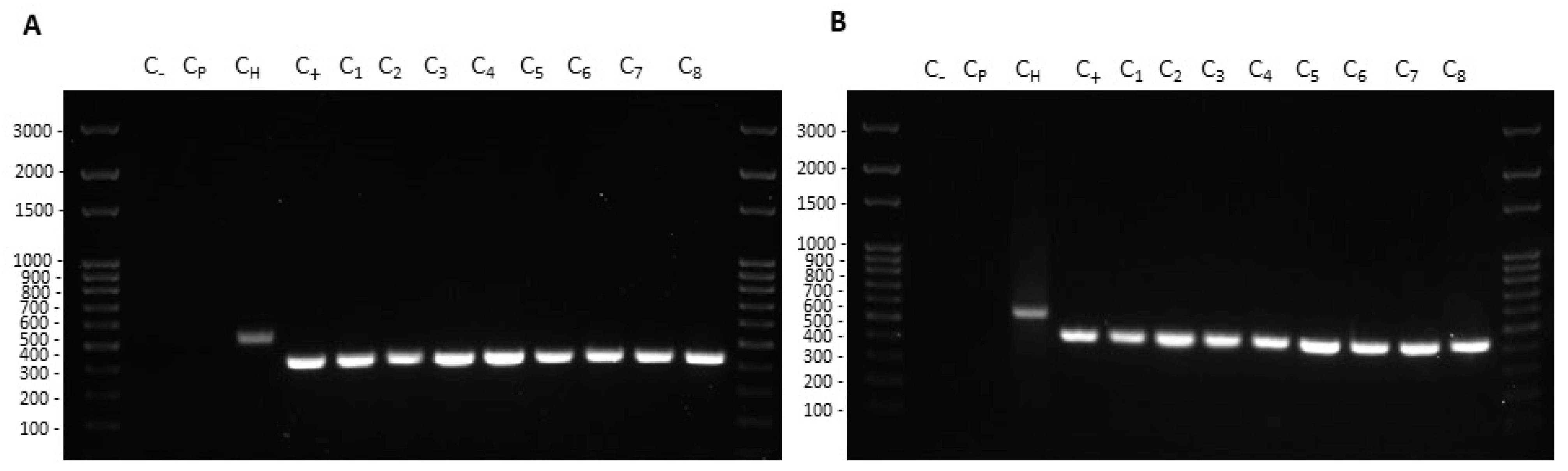
Transformation experiments were performed by isolating OMVs from K. pneumoniae-pGR and K. pneumoniae-PRM. K. pneumoniae ATCC recipient cells were incubated with 10 μg of OMVs derived from K. pneumoniae-pGR and K. pneumoniae-PRM. After 24 h, treated cells were plated on LB-ampicillin agar to detect the plasmid resistance marker in the recipient bacteria. OMVs purified from K. pneumoniae-pGR induced a transformation efficiency of 2.8 ± 0.1 × 104 CFU/μg. HGT mediated by K. pneumoniae-PRM OMVs occurred with a transformation efficiency of 7.8 ± 0.9 × 103 CFU/μg. In both conditions, no plasmid acquisition occurred when recipient cells were incubated with free plasmid (Figure 4A–H). Therefore, HGT via OMVs derived from K. pneumoniae-pGR was 3.6 times more efficient than the K. pneumoniae-PRM OMVs transfer, with the same vesicular concentration (Figure 4I). Colony-PCR was used to confirm that resistant acquisition. PGR and PRM were determined by amplifying a region of the β-lactamase gene and the amplicon was visualized by agarose gel electrophoresis (Figure 5A,B). PCR analysis showed that all grown and selected colonies on LB-ampicillin plates contained pGR and PRM plasmids. Pre-transformation colonies of K. pneumoniae did not show amplification, demonstrating the absence of resistance. Amplification of the 16S ribosomal gene region was used as a housekeeping control.
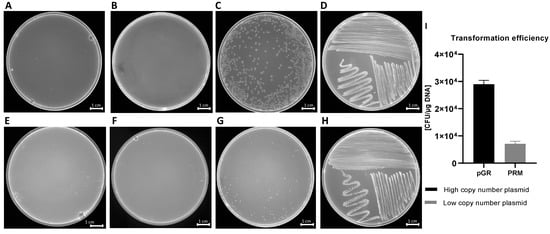
Figure 4.
HGT via OMVs derived from K. pneumoniae-pGR. Untreated cells (A) and cells treated with free plasmid (B) did not record transformants. K. pneumoniae ATCC treated with 10 µg of OMVs (C) and bacteria control on LB-plates (D). HGT-OMVs from K. pneumoniae-PRM. Transformation in untreated (E) and treated with free plasmid (F) bacteria did not occur. K. pneumoniae ATCC incubated with 10 µg of OMVs (G) and bacteria control on LB-plates (H). Comparison of intra-species HGT efficiency via purified OMVs from K. pneumoniae-pGR and K. pneumoniae-PRM (I).
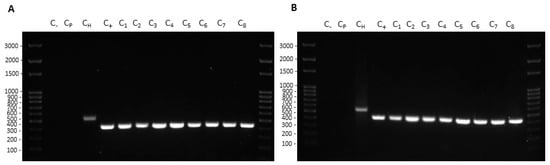
Figure 5.
Colony-PCR from recipient cells treated with K. pneumoniae-pGR (A) and K. pneumoniae-PRM (B) OMVs. DNA gel showed PCR products with expected lengths: ß-lactamase product~424 bp (C1–8), ribosomal 16S product~550 bp (CH). Control water (C−) and untreated bacteria (Cp) did not show amplification.
2.4. OMVs Induce the Generalized Resistance Spread
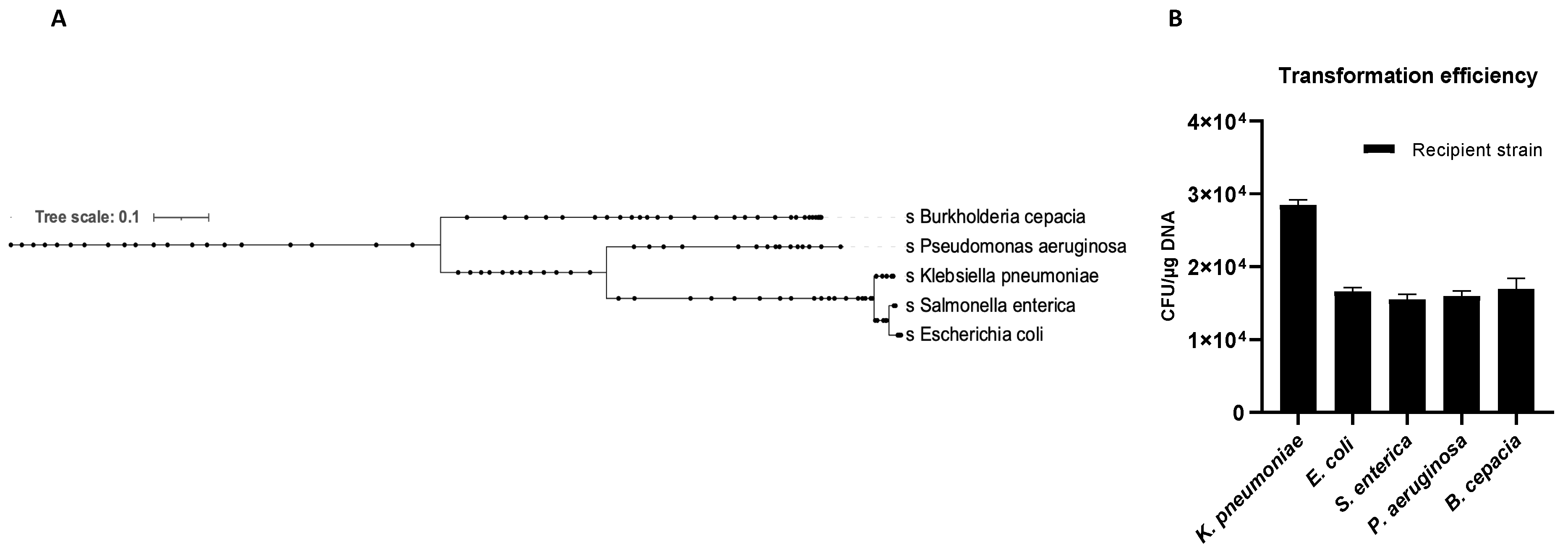
The OMVs potential to transfer genetic material between different microbial species was evaluated. Five recipient bacterial species were selected based on taxonomic differences (Figure 6A). Cultures of K. pneumoniae, E. coli, S. enterica, P. aeruginosa and B. cepacia were treated with K. pneumoniae-pGR OMVs. After 24 h of incubation, recipient cells were plated on LB-ampicillin agar plates and counted to define the transformation efficiency. OMVs derived from K. pneumoniae-pGR transferred plasmid DNA with a transformation efficiency of 2.8 ± 0.1 × 104, 1.7 ± 0.2 × 104, 1.5 ± 0.9 × 104, 1.6 ± 0.1 × 104, and 1.8 ± 0.8 × 104 CFU/μg for K. pneumoniae, E. coli, S. enterica, P. aeruginosa and B. cepacia, respectively (Figure 6B). Colonies of each recipient bacterial species were selected and subjected to PCR analysis to confirm the presence of the β-lactamase gene in the recipient species (Supplementary Figure S3A–D). Recipient cells incubated with free plasmid pGR and untreated cells did not acquire antibiotic resistance in any condition.
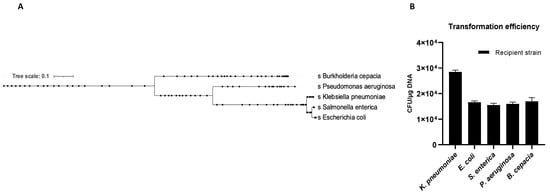
Figure 6.
Phylogenetic relationship of the recipient species (A). K. pneumoniae-pGR OMVs inter-species transformation efficiency (B).
2.5. OMVs Stability over Time
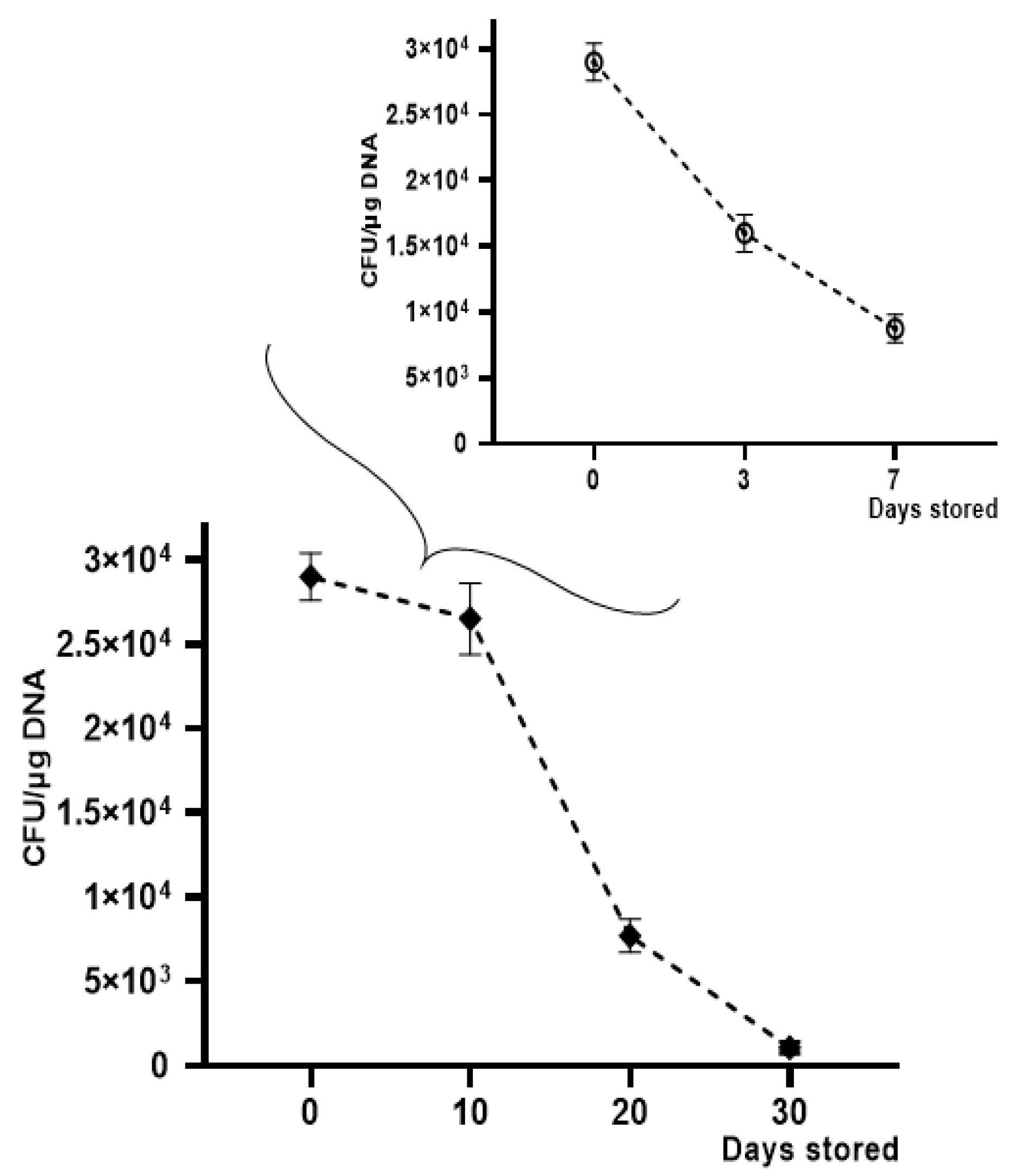
OMV-HGT experiments continued to evaluate transformation efficiency over time, by storing OMVs derived from K. pneumoniae-pGR at −20 °C for 30 days and at +4 °C for 7 days. A gradual reduction in transformation efficiency was observed using OMVs treated with DNase and stored at −20 °C and +4 °C for increasing periods of time. The HGT experiment was conducted using K. pneumoniae ATCC as a recipient cell. The maximum number of transformants was obtained with OMVs used after 10 days of storage, showing an efficiency of 2.5 ± 0.1 × 104 CFU/μg. After 20 days, a reduction in efficiency was verified, recording 7.7 ± 0.9 × 103 CFU/μg. On the 30th day of storage, a drastic decrease of transformants had occurred, registering 9.1 ± 0.12 × 102 CFU/μg. The OMVs stability at +4 °C showed a slight reduction over time. The recorded transformation efficiency was 1.7 ± 0.25 × 104 and 8.0 ± 0.43 × 103 CFU/μg, after 3 and 7 days of storage, respectively (Figure 7).
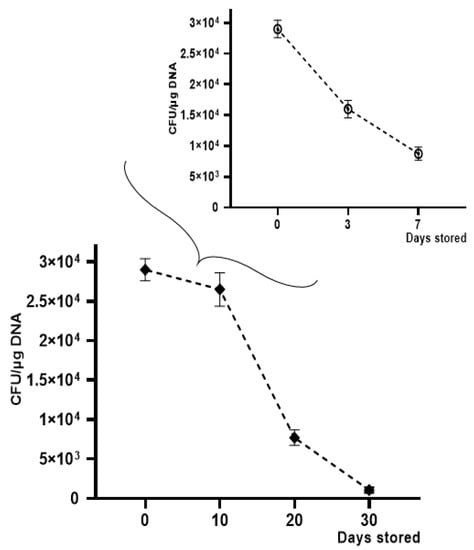
Figure 7.
Transformation frequency of K. pneumoniae-pGR OMVs storage at −20 °C and +4 °C for 30 and 7 days, respectively.
2.6. Diagnostic Transformed Strains Characterization
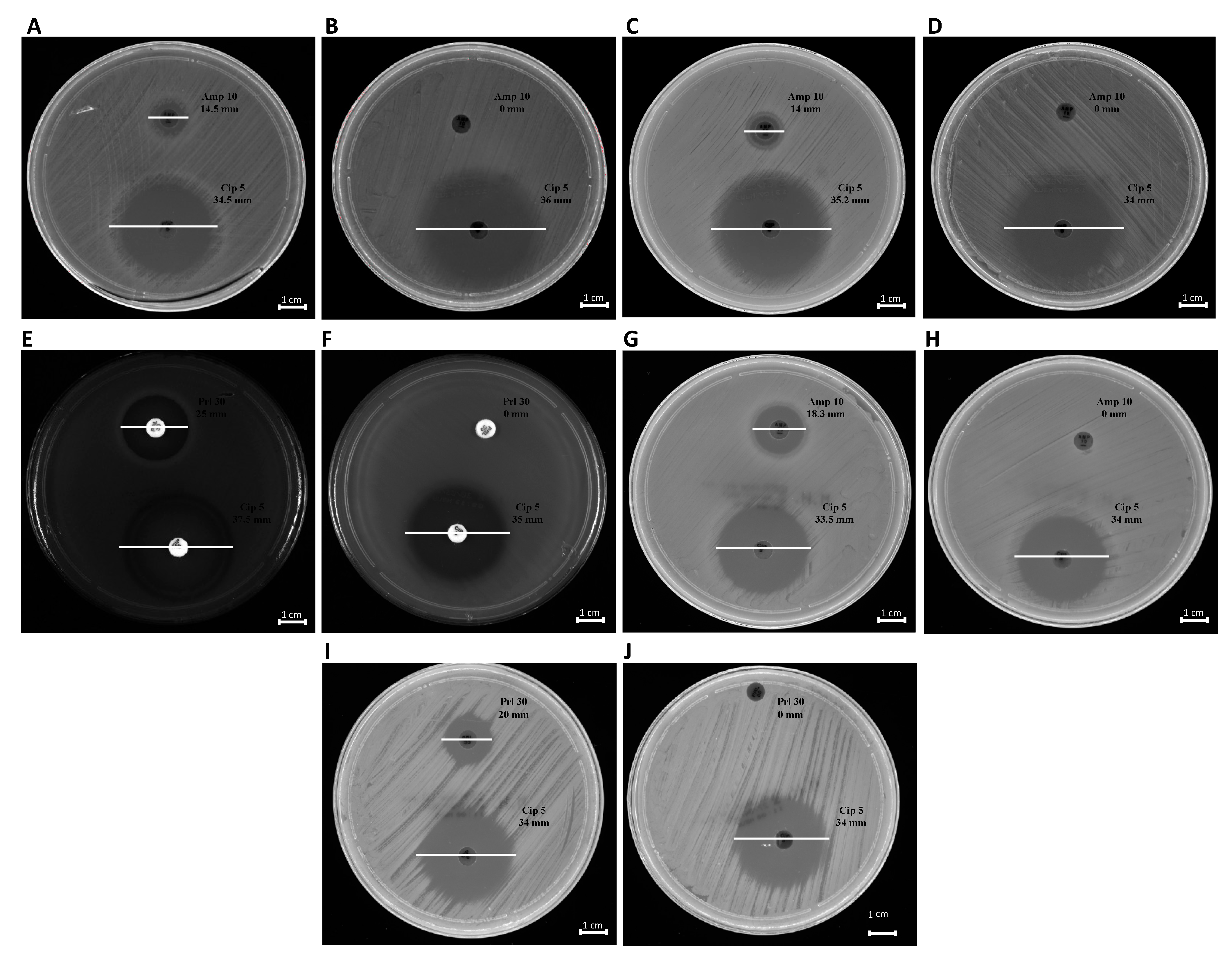
The phenotypic effect correlated to the genotypic resistance detected by PCR analysis was evaluated through antibiotic susceptibility testing. Each bacterial strain, before and after treatment with OMVs derived from K. pneumoniae-pGR, was examined. Concerning the susceptibility to β-lactams, the inhibition diameters measured before OMVs treatment were 14.5 ± 0.3, 14 ± 0.01, 18.3 ± 0.07, 25 ± 0.5, and 20 ± 0.9 mm for K. pneumoniae, E. coli, S. enterica, P. aeruginosa and B. cepacia, respectively. Inhibition zones recorded were associated with susceptible strains, in accordance with EUCAST guidelines. After the OMV-HGT, no inhibition area was identified for β-lactam antibiotics, demonstrating the acquisition of resistance. The inhibition area measured for ciprofloxacin was ≥30 mm, before and after OMVs treatment, in each bacterial species (Figure 8). The ciprofloxacin control was used to demonstrate that the acquired resistance was associated with the plasmid containing β-lactamase gene.
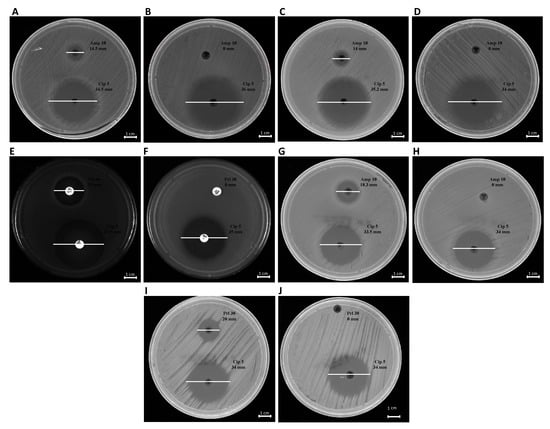
Figure 8.
Antibiotic susceptibility of bacteria to ß-lactams. K. pneumoniae pre- and post-OMVs treatment (A,B); E. coli pre and post-OMVs treatment (C,D); P. aeruginosa pre- and post-OMVs treatment (E,F); S. enterica pre- and post-OMVs treatment (G,H); B. cepacia pre- and post-OMVs treatment (I,J).
3. Discussion
Horizontal gene transfer plays an important role in promoting bacterial evolution, adaptation to environmental changes and acquisition of new metabolic capabilities [3]. Genetic pool modifications as a consequence of genetic transfer were observed in bacterial communities with high frequency rates, demonstrating the importance of this phenomenon for bacterial survival [35]. Currently, transformation, transduction and conjugation are considered the three canonical HGT mechanisms, contributing significantly to genetic diversity [36]. However, novel genetic material exchange events are under consideration and may be added to those currently known. Nowadays, the HGT mechanism should include the OMVs secretion by Gram-negative bacteria [37]. Previous studies reported that OMVs incorporated DNA into the lumen and transported it to recipient cells [38]. Currently, no studies have assessed the ability of K. pneumoniae to exploit HGT via OMVs to spread antimicrobial resistance. However, multidrug-resistant K. pneumoniae is increasingly implicated in hospital-acquired infections causing mortality rates above 50% [39,40]. This bacterial species causes a wide variety of often untreatable diseases as a consequence of a relatively frequent acquisition of antibiotic resistance genes [41]. Improved understanding of K. pneumoniae mechanisms to resistance genes spread is needed to limit such a serious threat to global health. Therefore, the focus of the present research was the preliminary characterization of HGT mechanisms mediated by OMVs derived from K. pneumoniae. In detail, we demonstrated, for the first time, that OMVs derived from K. pneumoniae were able to incorporate genetic determinants of resistance during the biogenesis process. While protected within the vesicular lumen, antibiotic resistance genes were transferred to different bacterial species, through fusion of the vesucles with the outer membrane of the recipient species, therefore conferring a novel resistance to recipient cells.
Firstly, OMVs were isolated from K. pneumoniae-pGR and K. pneumoniae-PRM, respectively. TEM and DLS analysis revealed that the vesicles featured a spherical morphology, in accordance with our previously published data, but with a reduced diameter compared with the OMVs collected from K. pneumoniae ATCC [14]. The different vesicular size could be attributed to the antibiotic presence during bacterial growth. Indeed, Fulsundar et al. showed that antibiotic and environmental stresses determined a significant effect on the OMVs production, size and DNA content [17]. These evidences confirm that OMVs release is a physiologically controlled process, dependent on environmental factors. The proteomic characterization of OMVs derived from K. pneumoniae-pGR and K. pneumoniae-PRM identified more than 55 proteins, mainly from the outer membrane and the periplasmic space. Inner membrane and cytosolic proteins were also detected, demonstrating that, although the mechanism of inclusion is unclear, cytoplasmic components and portions of membrane were incorporated into OMVs during the biogenesis process. Finally, the presence of proteins capable of interacting with DNA could confirm the ability of OMVs to also incorporate genetic material. These results may suggest, in addition to the OMVs originating from outer membrane budding, the possible existence of another vesiculation pattern. Indeed, Cruez et al. showed in S. vesiculosa M7T, N. gonorrhoeae, P. aeruginosa PAO1 e, and A. baumannii AB41 the presence of vesicles containing a bilayer of membrane and highly electrodense cytoplasmic material. These vesicles were classified as outer-inner membrane vesicles (O-IMVs) [42]. The possible secretion of two vesicle types, OMVs and O-IMVs, could explain how DNA is incorporated in OMVs, since it is not properly clarified. Currently, three models have been proposed: (i) the DNA present in the extracellular environment was internalized according with a mechanism similar to bacterial transformation; (ii) DNA was transported through the inner membrane and the cell wall up to the periplasmic space, where it was included in the OMVs; (iii) the DNA inclusion in the vesicles occurred through the secretion of O-IMV, which incorporate cytoplasmic components and DNA. The third model is the most accredited and supported by experimental evidence [20]. Although the DNA inclusion mechanism is not known with absolute certainty, our findings demonstrated that OMVs secreted by K. pneumoniae were involved in HGT, allowing the spread of resistance genes in microbial communities.
Contextually, our manuscript demonstrated that K. pneumoniae transferred genetic material, incorporating DNA within the OMVs and protecting it from the extracellular exonucleases action. The DNA in the vesicular lumen was transferred to the recipient cell by determining the acquisition of resistance genes present in the plasmid. The recipient cell K. pneumoniae, after contact with OMVs, acquired and expressed resistance to ampicillin, proving the OMVs ability to promote intraspecies HGT. Plasmid transfer did not occur when cells were incubated with free plasmid, suggesting that vesicles could represent a physiological mechanism that exceeds environmental limits (exonuclease degradation, dilution of gene material, long-distance transfer, etc.) and associated with the donor/recipient cell (state of competence, high vesicle-OM affinity, correlation phylogenetics, etc.). Moreover, the transfer efficiency over time of the stored OMVs was evaluated. The transfer rates remained unchanged for up to 10 days. Thereafter, the number of transformants gradually decreased for up to 30 days. Similar trends were shown in the study conducted by Chatterjee et al. on OMVs derived from A. baumannii, confirming the long-lasting stability without cryopreservatives [27]. Subsequently, it was investigated whether plasmid identity affected incorporation and transfer rate. The transfer of two different plasmids via K. pneumoniae-OMVs was examined, showing that the plasmid type induced changes in packaging and transformation rate. The high copy number plasmid (pGR) was loaded and transferred with greater efficiency compared to the low copy number plasmid (PRM). Our results were in line with a study conducted by Tran and Boedicker, in which the low copy number plasmid (pZS2501) had a low loading capacity (0.49 × 103 copies per pg of OMVs), while the high copy number plasmids (pLC291 and pUC19) showed a high loading potential (2.58 × 103 and 482.7 × 103 copies per pg of OMVs) [16]. Therefore, the plasmid cargo in the OMVs was strictly dependent on the copy number; the higher the PCN, the greater the plasmid amount in the OMVs and consequently the transformation efficiency. OMVs-mediated transfer exceeds the limits observed in other HGT mechanisms [43]. Chatterjee et al. have already reported the ability of A. baumannii-OMVs to allow interspecies gene transfer [27]. For this reason, interspecies gene exchange was observed via K. pneumoniae-OMVs, using four different recipient species: E. coli, S. enterica, P. aeruginosa and B. cepacia. The generalized transfer to the different bacterial genera highlighted the HGT-OMVs efficiency, which verified independently of the phylogenetic correlation between the donor and recipient cell. Our experimental evidence showed that OMVs contributed to genetic exchange in microbial communities even among distantly related bacteria, without specific exchange mechanisms. Future studies will examine the possibility of OMVs to exchange DNA between different Gram-positive species.
In summary, the present study demonstrates, for the first time, the resistance gene to β-lactams spreads through OMVs secreted by K. pneumoniae. This innovative HGT mechanism allows for intra-species or inter-species diffusion, persistent over time and apparently not associated with specific limitations. Our study reveals a novel gene transfer mechanism of resistance genes that may contribute to the spread of antibiotic resistance between different bacterial species. Understanding the distribution of resistance determinants in several bacterial populations and clarifying the factors that promote their spread could be useful for identifying new pharmacological targets to limit the spread of antibiotic resistance.
4. Materials and Methods
4.1. Bacterial Strains, Plasmids and Growing Conditions
The strains used in this study were obtained from the American Type Culture Collection (ATCC) (Manassan, VA, USA). K. pneumoniae ATCC 10031 was used for the OMVs purification. K. pneumoniae was transformed using the calcium chloride method with pGR (K. pneumoniae-pGR) (Addgene, Watertown, MA, USA) and PRM-GFP (K. pneumoniae-PRM) (Addgene, Watertown, MA, USA), respectively [44,45]. The first one was a high copy number plasmid (500~600 copies) containing genes for green fluorescent protein (GFP) and β-lactamase which conferred resistance to ampicillin. PRM was a plasmid containing the same genes and differed in copy number (10~12 copies). After transformation, K. pneumoniae-pGR and K. pneumoniae-PRM were cultured on Luria-Bertani agar (LB) (Sigma-Aldrich, St. Louis, MO, USA) containing 100 μg mL−1 of ampicillin (Sigma-Aldrich, St. Louis, MO, USA). E. coli ATCC 25922, S. enterica ATCC 14028, P. aeruginosa ATCC 13388 and Burkholderia cepacia ATCC 25416 were used as recipient strains for the HGT mediated by OMVs. All bacterial strains were cultured in LB (Sigma-Aldrich, St. Louis, MO, USA) medium at 37 °C under orbital shaking at 180 rpm.
4.2. OMVs Purification
OMVs were isolated from liquid cultures of K. pneumoniae-pGR and K. pneumoniae-PRM-GFP as previously described with modifications [46]. First, 10 mL of overnight (O/N) bacterial culture was inoculated in 600 mL of LB containing 100 µg mL−1 ampicillin. The bacterial inoculum was cultured at 37 °C under orbital shaking (180 rpm) for 8–12 h, up to the OD600 nm value of 1. The cultures were centrifuged at 4000× g at 4 °C for 20 min, to remove bacterial cells. Supernatants were decanted and filtered using vacuum Stericup™ 0.45 μm and 0.22 μm pore size polyethersulfone (PES) top filter (Millipore, Burlington, MA, USA), to deflect remaining bacteria and cell debris. Vesicles were collected from cell-free supernatant culture by ultracentrifugation at 100,000× g (centrifuge Optima XPN-100 Beckman Coulter and rotor SW28) at 4 °C for 1.5 h. Pellets were washed in sterile phosphate buffered saline 1X (PBS) by ultracentrifugation (100,000× g at 4 °C for 1.5 h). Vesicular pellets were suspended in 250 μL of PBS 1X and OMVs sterility was checked by inoculating 10 μL of vesicles on LB-agar plates. OMV samples were treated with DNase (Applied Biological Materials (abm), Richmond, BC, Canada) according to the manufacturer’s protocol and stored at −20 °C until use.
4.3. Transmission Electron Microscopy (TEM)
Purified OMVs were visualized by TEM, using negative staining. Five microliters of sample were adsorbed on carbon-coated copper/palladium grids for 30 min. A drop of sterile deionized water was used to wash the grids and a negative staining was realized by addition of 5 µL of 1% (w/v) uranyl acetate. TEM images were acquired using an EM 208 S transmission electron microscope (Philips, Amsterdam, The Netherlands).
4.4. OMVs Size Characterization by Dynamic Light Scattering (DLS)
Vesicle diameter size (Z-ave) and PDI analysis were performed using Zetasizer Nano-ZS (Malvern Instruments, Worcestershire, UK). For DLS, 40 µL of OMVs aliquot were mixed gently and transferred to sterile cuvettes. All measurements were conducted at 25 °C and three independent experiments for each purification were performed. DLS data were processed using Zetasizer software (V 7.11) provided by Malvern Panalytical (Malvern, UK).
4.5. OMVs Protein Profile by Tandem Mass Spectrometry (MS/MS)
For protein profile, OMVs were incubated with 1% Triton X-100 for 1 h at 4 °C. Lysed vesicles were centrifuged at 14,000× g at 4 °C for 30 min and the supernatant was examined for protein amount by Bradford assay (HIMEDIA, Maharashtra, India). The protein extract was subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The gel was stained with Coomassie Brilliant blue G250 (Sigma-Aldrich, St. Louis, MO, USA) and different bands were cut to perform MS and MS/MS analysis, as previously described [14]. Briefly, protein bands were extracted from the gel and digested with trypsin. NanoUPLC-hr MS/MS analysis of the resulting peptide mixtures were performed on a Q-Exactive orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA), coupled with a nanoUltimate300 UHPLC system (Thermo Fisher Scientific, USA). For protein identification, mass spectra were subjected to analysis by Mascot software (v2.5, Matrix Science, Boston, MA, USA), using the non-database redundant UniprotKB/Swiss-Prot (version 2020_03). The identified proteins were analyzed by subcellular localization, biological processes and molecular functions using Uniprot software (https://www.uniprot.org/ (accessed on 14 March 2021)).
4.6. Intra-Vesicular DNA Analysis
Plasmid concentration in OMVs was determined by Real-time PCR (qPCR) using BrightGreen qPCR MasterMix Kits (abm, San Francisco, CA, USA), according to the manufacturer’s instructions. For DNA extraction, vesicles were lysed by boiling at 100 °C for 10 min. Two microliters of OMVs were added to 0.2 μM of primer, 1X mastermix in a final reaction volume of 20 μL. Primers used for qPCR were: β-lactamase Fw 5′-AACTTTATCCGCCTCCATCC-3′, and β-lactamase Rev 3′-GCTATGTGGCGCGGTATTAT-5′. The amplification was performed in CFX96 Touch Real-Time PCR Detection System (Bio-Rad, California, USA), using the following amplification program: denaturation at 95 °C for 15 s, annealing at 60 °C for 20 s and extension at 72 °C for 15 s (40 cycles). The standard curves were constructed using purified plasmids from K. pneumoniae-pGR and K. pneumoniae-PRM, respectively. Plasmid concentration in OMVs was converted in plasmid copy number (PCN), according to the formula:
http://cels.uri.edu/gsc/cndna.html (accessed on 29 January 2004) Subsequently, plasmid loading was estimated based on OMVs protein concentrations.
4.7. OMVs-Mediated Gene Transfer
For gene transfer experiments through OMVs, the recipient strains K. pneumoniae ATCC, E. coli ATCC, P. aeruginosa ATCC, B. cepacia ATCC and S. enterica ATCC were inoculated in LB-broth up to OD600 nm value of 0.4. Cells were diluted in cold LB at final concentration of 107 CFU/mL. Bacterial suspensions (60 μL) were incubated with 10 µg of OMVs statically for 4 h at 37 °C and, subsequently, for 4 h under orbital shaking (180 rpm) at 37 °C. Fresh LB-medium was added to each bacterial suspension and then incubated O/N under orbital shaking (180 rpm) at 37 °C. To further confirm that the plasmid transfer was mediated by OMVs, two separate experiments were performed with: (i) free plasmid and (ii) untreated cells. The following day, a 100 μL aliquot of bacteria was plated on LB-agar supplemented with 100 μg mL−1 ampicillin and incubated O/N at 37 °C. The bacterial colonies (C1–8) were counted to define the transformation efficiency, according to the formula:
The same transformation experiments were performed using OMVs stored at −20 °C for 10, 20 and 30 days and OMVs stored at +4 °C for 3 and 7 days.
4.8. Polymerase Chain Reaction (PCR) Screening
After OMVs gene transfer, bacterial colonies grown on LB-agar supplemented with 100 μg mL−1 ampicillin were selected and subjected to molecular investigation for the presence of the plasmid by colony-PCR. Each bacterial colony was lysed by heat-shock and then centrifuged at 16,000× g at 4 °C for 10 min. The supernatant was transferred to a new Eppendorf and the DNA concentration was examined by NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). PCR was performed in a total volume of 50 μL containing 1 μM each primer, 1X Taq Master Mix (abm, San Francisco, CA, USA) and 100 ng of DNA. The primers used to amplify the 424 bp region of β-lactamase gene were: Fw 5′-AACTTTATCCGCCTCCATCC-3′, and Rev 3′-GCTATGTGGCGCGGTATTAT-5′. The amplification was conducted in Thermal Cycler UNO96 (VWR International, Pennsylvania, USA) according to the following program: initial denaturation at 94 °C for 3 min, 35 cycles of amplification in which each cycle was denatured at 94 °C for 30 s, annealed at 57.3 °C for 30 s and extended at 72 °C 1 min; the final extension at 72 °C for 5 min. As a housekeeping gene control, 16S rRNA gene was amplified, using the primers: Fw 5′-GGTAGAGTTTGATCCTGGCTCAG-3′, and Rev 3′-ATTACCGCGGCTGCTGG-5′. The used program was: initial denaturation at 94 °C for 1 min, 30 cycles of amplification in which each cycle was denatured at 94 °C for 1 min, annealed at 58 °C for 1 min and extended at 72 °C for 1.5 min; the final extension at 72 °C for 10 min. To visualize the amplification product, 1% agarose gel electrophoresis was performed.
4.9. Antibiotic Susceptibility Test
The disk diffusion assay was performed according to the National Committee on Clinical Laboratory Standards (NCCLS) [47,48]. Fresh colonies, before and after OMVs treatment, were inoculated in physiological solutions to 0.5 McFarland turbidity. With a cotton swab dipped in the bacterial inoculum, the solution was homogeneously plated into Mueller-Hinton (MH) agar plates. Disks of ampicillin (10 μg) (Thermo Fisher Scientific, Waltham, MA, USA), piperacillin (30 μg) (Thermo Fisher Scientific, Waltham, MA, USA) and ciprofloxacin (5 μg) (Thermo Fisher Scientific, Waltham, MA, USA) were placed on the plates and were incubated at 37 °C O/N. The antibiotic susceptibility was examined by measuring the zone of inhibition diameter, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22168732/s1.
Author Contributions
Conceptualization, F.D. and V.F.; methodology, N.D. and C.D.; software, G.D. and V.I.; validation, A.D.F. and L.A.; formal analysis, V.F. and M.G. (Marilena Galdiero); investigation, C.D. and F.D.; resources, G.B. and G.D.; data curation, G.F.; writing—original draft preparation, M.G. (Marilena Galdiero) and G.F.; writing—review and editing, G.F. and M.G. (Massimiliano Galdiero); visualization, A.D.F. and F.D.P.; supervision, M.G. (Massimiliano Galdiero) and V.I.; project administration, L.A.; funding acquisition, G.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bedhomme, S.; Amorós-Moya, D.; Valero, L.M.; Bonifaci, N.; Pujana, M.-À.; Bravo, I.G. Evolutionary Changes after Translational Challenges Imposed by Horizontal Gene Transfer. Genome Biol. Evol. 2019, 11, 814–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bello-López, J.M.; Cabrero-Martínez, O.A.; Ibáñez-Cervantes, G.; Hernández-Cortez, C.; Pelcastre-Rodríguez, L.I.; Gonzalez-Avila, L.U.; Castro-Escarpulli, G. Horizontal Gene Transfer and Its Association with Antibiotic Resistance in the Genus Aeromonas spp. Microorganisms 2019, 7, 363. [Google Scholar] [CrossRef] [Green Version]
- Emamalipour, M.; Seidi, K.; Zununi Vahed, S.; Jahanban-Esfahlan, A.; Jaymand, M.; Majdi, H.; Amoozgar, Z.; Chitkushev, L.T.; Javaheri, T.; Jahanban-Esfahlan, R.; et al. Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression. Front. Cell Dev. Biol. 2020, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, R.J.; Whelan, F.J.; McInerney, J.O.; Ou, Y.; Domingo-Sananes, M.R. Horizontal Gene Transfer as a Source of Conflict and Cooperation in Prokaryotes. Front. Microbiol. 2020, 11, 1569. [Google Scholar] [CrossRef]
- Ely, B. Recombination and Gene Loss Occur Simultaneously during Bacterial Horizontal Gene Transfer. PLoS ONE 2020, 15, e0227987. [Google Scholar] [CrossRef]
- Redondo-Salvo, S.; Fernández-López, R.; Ruiz, R.; Vielva, L.; de Toro, M.; Rocha, E.P.C.; Garcillán-Barcia, M.P.; de la Cruz, F. Pathways for Horizontal Gene Transfer in Bacteria Revealed by a Global Map of Their Plasmids. Nat. Commun. 2020, 11, 3602. [Google Scholar] [CrossRef]
- Riva, F.; Riva, V.; Eckert, E.M.; Colinas, N.; Di Cesare, A.; Borin, S.; Mapelli, F.; Crotti, E. An Environmental Escherichia coli Strain Is Naturally Competent to Acquire Exogenous DNA. Front. Microbiol. 2020, 11, 574301. [Google Scholar] [CrossRef]
- Salvadori, G.; Junges, R.; Morrison, D.A.; Petersen, F.C. Competence in Streptococcus pneumoniae and Close Commensal Relatives: Mechanisms and Implications. Front. Cell. Infect. Microbiol. 2019, 9, 94. [Google Scholar] [CrossRef] [Green Version]
- Headd, B.; Bradford, S.A. The Conjugation Window in an Escherichia coli K-12 Strain with an IncFII Plasmid. Appl. Environ. Microbiol. 2020, 86, e00948-20. [Google Scholar] [CrossRef]
- Grohmann, E.; Muth, G.; Espinosa, M. Conjugative Plasmid Transfer in Gram-Positive Bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 277–301. [Google Scholar] [CrossRef] [Green Version]
- Fillol-Salom, A.; Alsaadi, A.; Sousa, J.A.M.D.; Zhong, L.; Foster, K.R.; Rocha, E.P.C.; Penadés, J.R.; Ingmer, H.; Haaber, J. Bacteriophages Benefit from Generalized Transduction. PLoS Pathog. 2019, 15, e1007888. [Google Scholar] [CrossRef]
- Gómez-Gómez, C.; Blanco-Picazo, P.; Brown-Jaque, M.; Quirós, P.; Rodríguez-Rubio, L.; Cerdà-Cuellar, M.; Muniesa, M. Infectious Phage Particles Packaging Antibiotic Resistance Genes Found in Meat Products and Chicken Feces. Sci. Rep. 2019, 9, 13281. [Google Scholar] [CrossRef]
- Dell’Annunziata, F.; Ilisso, C.P.; Dell’Aversana, C.; Greco, G.; Coppola, A.; Martora, F.; Dal Piaz, F.; Donadio, G.; Falanga, A.; Galdiero, M.; et al. Outer Membrane Vesicles Derived from Klebsiella pneumoniae Influence the MiRNA Expression Profile in Human Bronchial Epithelial BEAS-2B Cells. Microorganisms 2020, 8, 1985. [Google Scholar] [CrossRef]
- Domingues, S.; Nielsen, K.M. Membrane Vesicles and Horizontal Gene Transfer in Prokaryotes. Curr. Opin. Microbiol. 2017, 38, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Tran, F.; Boedicker, J.Q. Genetic Cargo and Bacterial Species Set the Rate of Vesicle-Mediated Horizontal Gene Transfer. Sci. Rep. 2017, 7, 8813. [Google Scholar] [CrossRef] [Green Version]
- Fulsundar, S.; Harms, K.; Flaten, G.E.; Johnsen, P.J.; Chopade, B.A.; Nielsen, K.M. Gene Transfer Potential of Outer Membrane Vesicles of Acinetobacter Baylyi and Effects of Stress on Vesiculation. Appl. Environ. Microbiol. 2014, 80, 3469–3483. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Gao, J.; Wang, Z. Outer Membrane Vesicles for Vaccination and Targeted Drug Delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019, 11, e1523. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H. Bacterial Outer Membrane Vesicles as a Delivery System for Virulence Regulation. J. Microbiol. Biotechnol. 2016, 26, 1343–1347. [Google Scholar] [CrossRef] [Green Version]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef] [Green Version]
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Cárcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive Cell Lysis as a Mechanism for the Biogenesis of Bacterial Membrane Vesicles and Biofilms. Nat. Commun. 2016, 7, 11220. [Google Scholar] [CrossRef] [Green Version]
- Cecil, J.D.; Sirisaengtaksin, N.; O’Brien-Simpson, N.M.; Krachler, A.M. Outer Membrane Vesicle-Host Cell Interactions. Microbiol. Spectr. 2019, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Caruana, J.C.; Walper, S.A. Bacterial Membrane Vesicles as Mediators of Microbe—Microbe and Microbe—Host Community Interactions. Front. Microbiol. 2020, 11, 432. [Google Scholar] [CrossRef] [Green Version]
- Dell’Annunziata, F.; Folliero, V.; Giugliano, R.; De Filippis, A.; Santarcangelo, C.; Izzo, V.; Daglia, M.; Galdiero, M.; Arciola, C.R.; Franci, G. Gene Transfer Potential of Outer Membrane Vesicles of Gram-Negative Bacteria. Int. J. Mol. Sci. 2021, 22, 5985. [Google Scholar] [CrossRef]
- Verhoeven, A.J.; Estrela, J.M.; Meijer, A.J. Alpha-Adrenergic Stimulation of Glutamine Metabolism in Isolated Rat Hepatocytes. Biochem. J. 1985, 230, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Kolling, G.L.; Matthews, K.R. Export of Virulence Genes and Shiga Toxin by Membrane Vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 1999, 65, 1843–1848. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, S.; Mondal, A.; Mitra, S.; Basu, S. Acinetobacter Baumannii Transfers the BlaNDM-1 Gene via Outer Membrane Vesicles. J. Antimicrob. Chemother. 2017, 72, 2201–2207. [Google Scholar] [CrossRef] [Green Version]
- Blesa, A.; Berenguer, J. Contribution of Vesicle-Protected Extracellular DNA to Horizontal Gene Transfer in Thermus spp. Int. Microbiol. 2015, 18, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.-H.; Chen, C.-H.; Goodwin, J.S.; Wang, B.-Y.; Xie, H. Functional Advantages of Porphyromonas Gingivalis Vesicles. PLoS ONE 2015, 10, e0123448. [Google Scholar] [CrossRef]
- Yaron, S.; Kolling, G.L.; Simon, L.; Matthews, K.R. Vesicle-Mediated Transfer of Virulence Genes from Escherichia coli O157:H7 to Other Enteric Bacteria. Appl. Environ. Microbiol. 2000, 66, 4414–4420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumbo, C.; Fernández-Moreira, E.; Merino, M.; Poza, M.; Mendez, J.A.; Soares, N.C.; Mosquera, A.; Chaves, F.; Bou, G. Horizontal Transfer of the OXA-24 Carbapenemase Gene via Outer Membrane Vesicles: A New Mechanism of Dissemination of Carbapenem Resistance Genes in Acinetobacter Baumannii. Antimicrob. Agents Chemother. 2011, 55, 3084–3090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A Major Worldwide Source and Shuttle for Antibiotic Resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Lopes, E.; Saavedra, M.J.; Costa, E.; de Lencastre, H.; Poirel, L.; Aires-de-Sousa, M. Epidemiology of Carbapenemase-Producing Klebsiella pneumoniae in Northern Portugal: Predominance of KPC-2 and OXA-48. J. Glob. Antimicrob. Resist. 2020, 22, 349–353. [Google Scholar] [CrossRef]
- Martora, F.; Pinto, F.; Folliero, V.; Cammarota, M.; Dell’Annunziata, F.; Squillaci, G.; Galdiero, M.; Morana, A.; Schiraldi, C.; Giovane, A.; et al. Isolation, Characterization and Analysis of pro-Inflammatory Potential of Klebsiella pneumoniae Outer Membrane Vesicles. Microb. Pathog. 2019, 136, 103719. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T.; Aminov, R. Potential Effects of Horizontal Gene Exchange in the Human Gut. Front. Immunol. 2017, 8, 1630. [Google Scholar] [CrossRef] [Green Version]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal Transfer of Antibiotic Resistance Genes in Clinical Environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Da Silva, G.J.; Domingues, S. Insights on the Horizontal Gene Transfer of Carbapenemase Determinants in the Opportunistic Pathogen Acinetobacter Baumannii. Microorganisms 2016, 4, 29. [Google Scholar] [CrossRef] [Green Version]
- Ha, J.Y.; Choi, S.-Y.; Lee, J.H.; Hong, S.-H.; Lee, H.-J. Delivery of Periodontopathogenic Extracellular Vesicles to Brain Monocytes and Microglial IL-6 Promotion by RNA Cargo. Front. Mol. Biosci. 2020, 7, 596366. [Google Scholar] [CrossRef]
- Caneiras, C.; Lito, L.; Melo-Cristino, J.; Duarte, A. Community- and Hospital-Acquired Klebsiella pneumoniae Urinary Tract Infections in Portugal: Virulence and Antibiotic Resistance. Microorganisms 2019, 7, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassetti, M.; Righi, E.; Carnelutti, A.; Graziano, E.; Russo, A. Multidrug-Resistant Klebsiella pneumoniae: Challenges for Treatment, Prevention and Infection Control. Expert Rev. Anti-Infect. Ther. 2018, 16, 749–761. [Google Scholar] [CrossRef]
- Effah, C.Y.; Sun, T.; Liu, S.; Wu, Y. Klebsiella pneumoniae: An Increasing Threat to Public Health. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1. [Google Scholar] [CrossRef]
- Pérez-Cruz, C.; Carrión, O.; Delgado, L.; Martinez, G.; López-Iglesias, C.; Mercade, E. New Type of Outer Membrane Vesicle Produced by the Gram-Negative Bacterium Shewanella Vesiculosa M7T: Implications for DNA Content. Appl. Environ. Microbiol. 2013, 79, 1874–1881. [Google Scholar] [CrossRef] [Green Version]
- Aminov, R.I. Horizontal Gene Exchange in Environmental Microbiota. Front. Microbiol. 2011, 2, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, W.-T.; Verma, C.S.; Lane, D.P.; Gan, S.K.-E. A Comparison and Optimization of Methods and Factors Affecting the Transformation of Escherichia coli. Biosci. Rep. 2013, 33, e00086. [Google Scholar] [CrossRef]
- Higuchi-Takeuchi, M.; Morisaki, K.; Numata, K. Method for the Facile Transformation of Marine Purple Photosynthetic Bacteria Using Chemically Competent Cells. MicrobiologyOpen 2020, 9, e00953. [Google Scholar] [CrossRef] [PubMed]
- De Lise, F.; Mensitieri, F.; Rusciano, G.; Dal Piaz, F.; Forte, G.; Di Lorenzo, F.; Molinaro, A.; Zarrelli, A.; Romanucci, V.; Cafaro, V.; et al. Novosphingobium Sp. PP1Y as a Novel Source of Outer Membrane Vesicles. J. Microbiol. 2019, 57, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, F.; Pignataro, D.; Lavano, M.A.; Santella, B.; Folliero, V.; Zannella, C.; Astarita, C.; Gagliano, C.; Franci, G.; Avitabile, T.; et al. Current Evidence on the Ocular Surface Microbiota and Related Diseases. Microorganisms 2020, 8, 1033. [Google Scholar] [CrossRef]
- Pironti, C.; Dell’Annunziata, F.; Giugliano, R.; Folliero, V.; Galdiero, M.; Ricciardi, M.; Motta, O.; Proto, A.; Franci, G. Comparative Analysis of Peracetic Acid (PAA) and Permaleic Acid (PMA) in Disinfection Processes. Sci. Total Environ. 2021, 797, 149206. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).