Carotenoid Cleavage Dioxygenase Genes of Chimonanthus praecox, CpCCD7 and CpCCD8, Regulate Shoot Branching in Arabidopsis
Abstract
:1. Introduction
2. Results
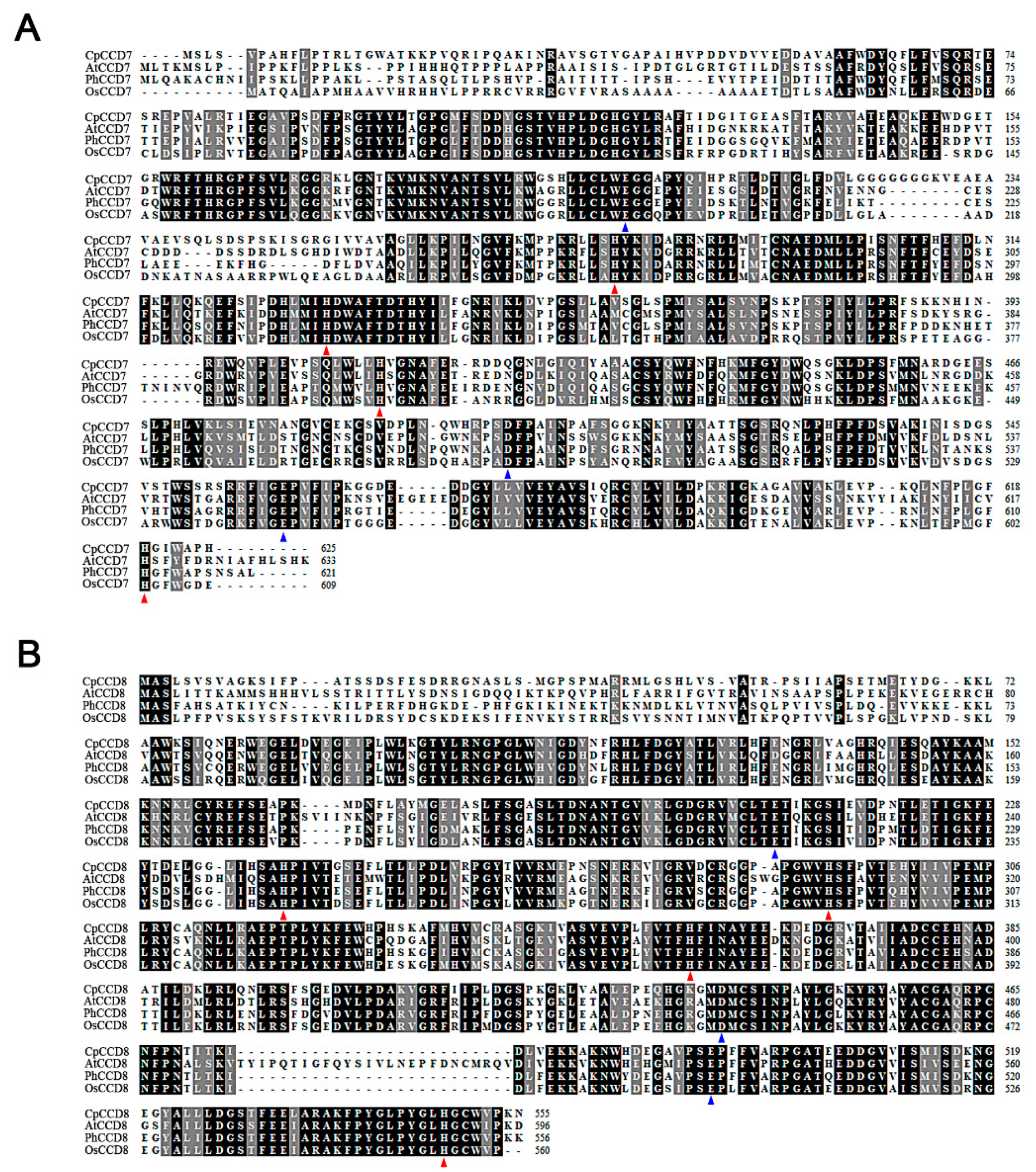
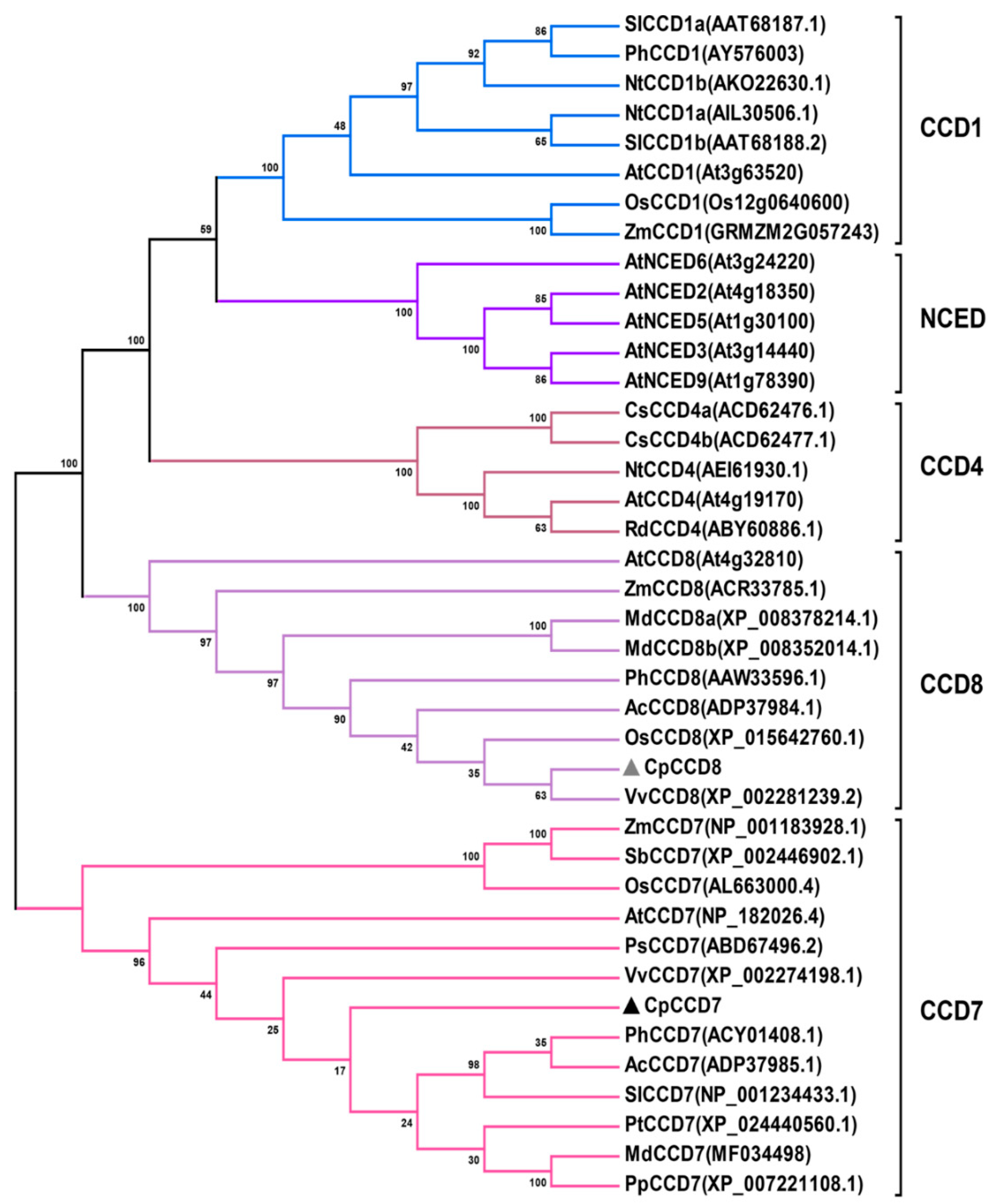
2.1. Cloning and Phylogenetic Analysis of CpCCD7 and CpCCD8
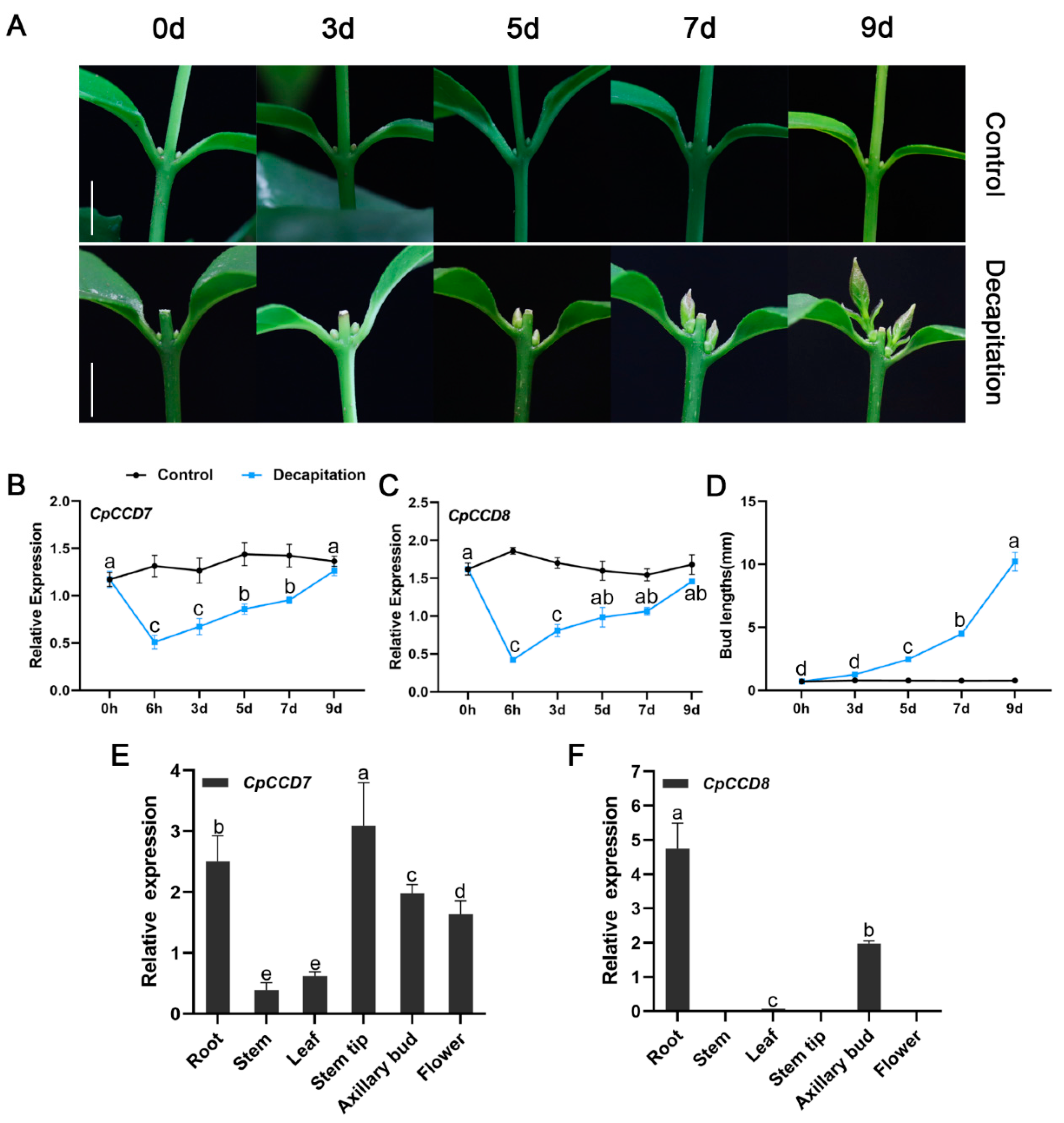
2.2. Expression Patterns of CpCCD7 and CpCCD8
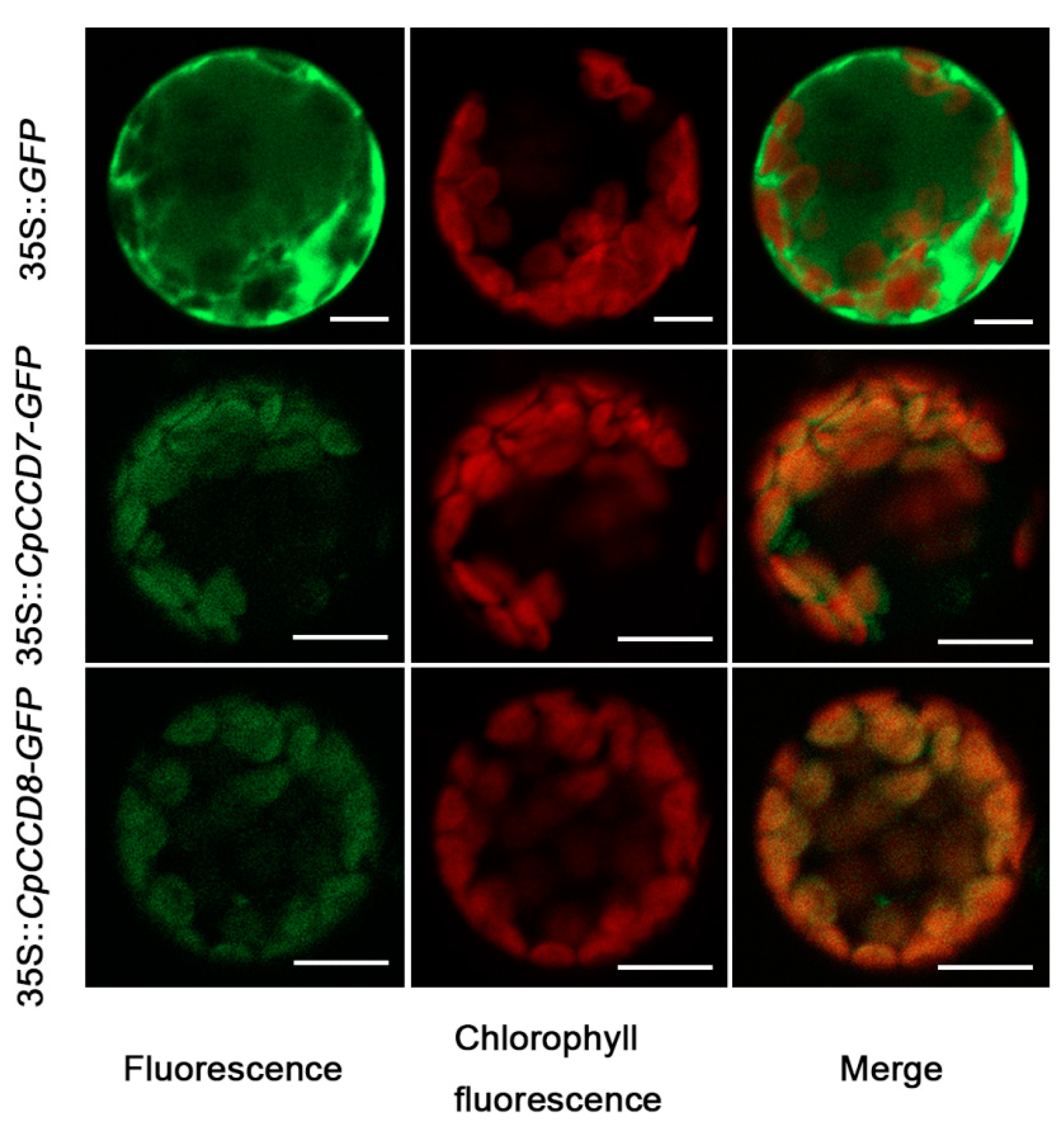
2.3. Subcellular Localization of CpCCD7 and CpCCD8
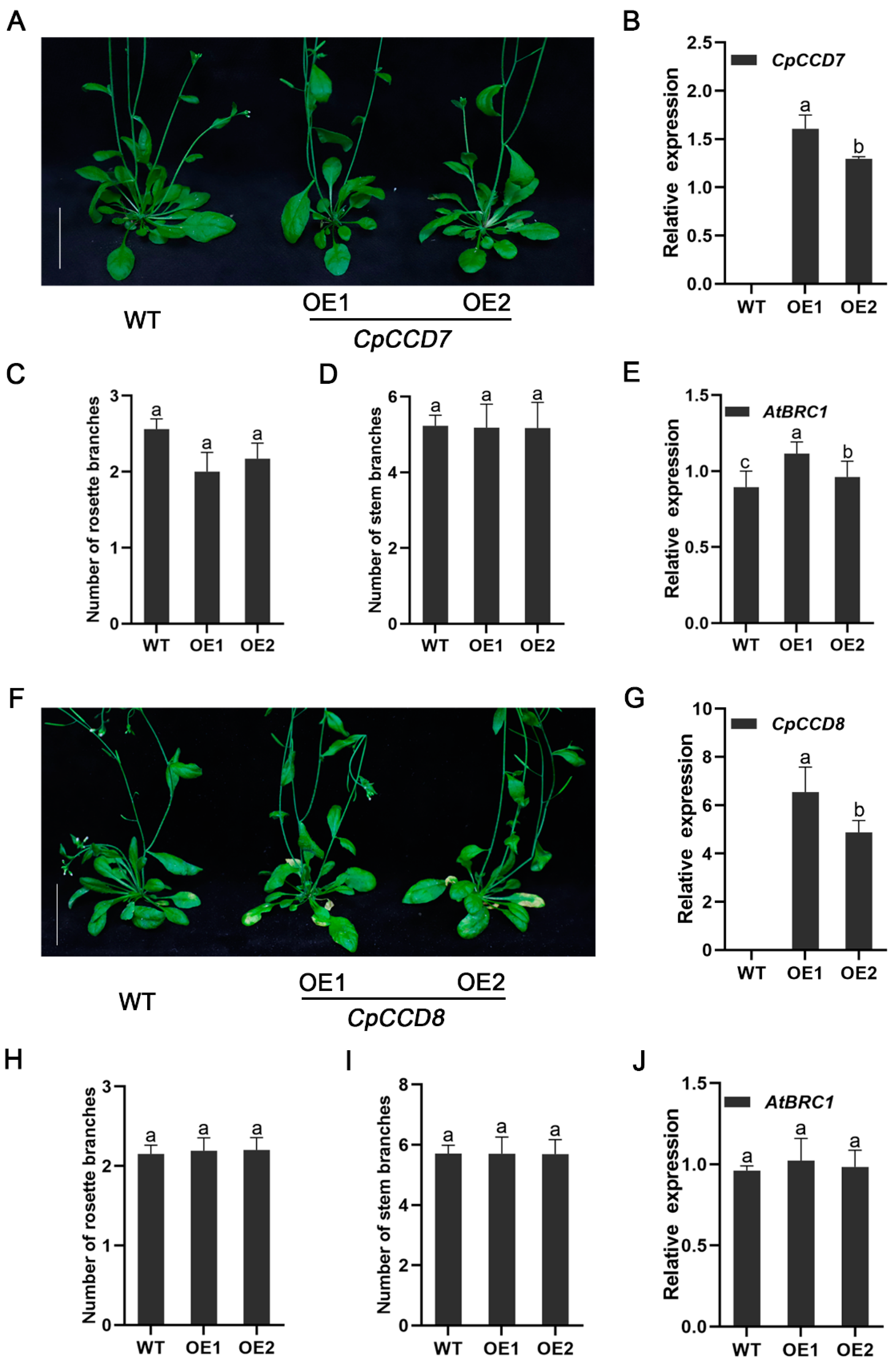
2.4. Effect of CpCCD7 and CpCCD8 Overexpression on the Branching Phenotype of Arabidopsis
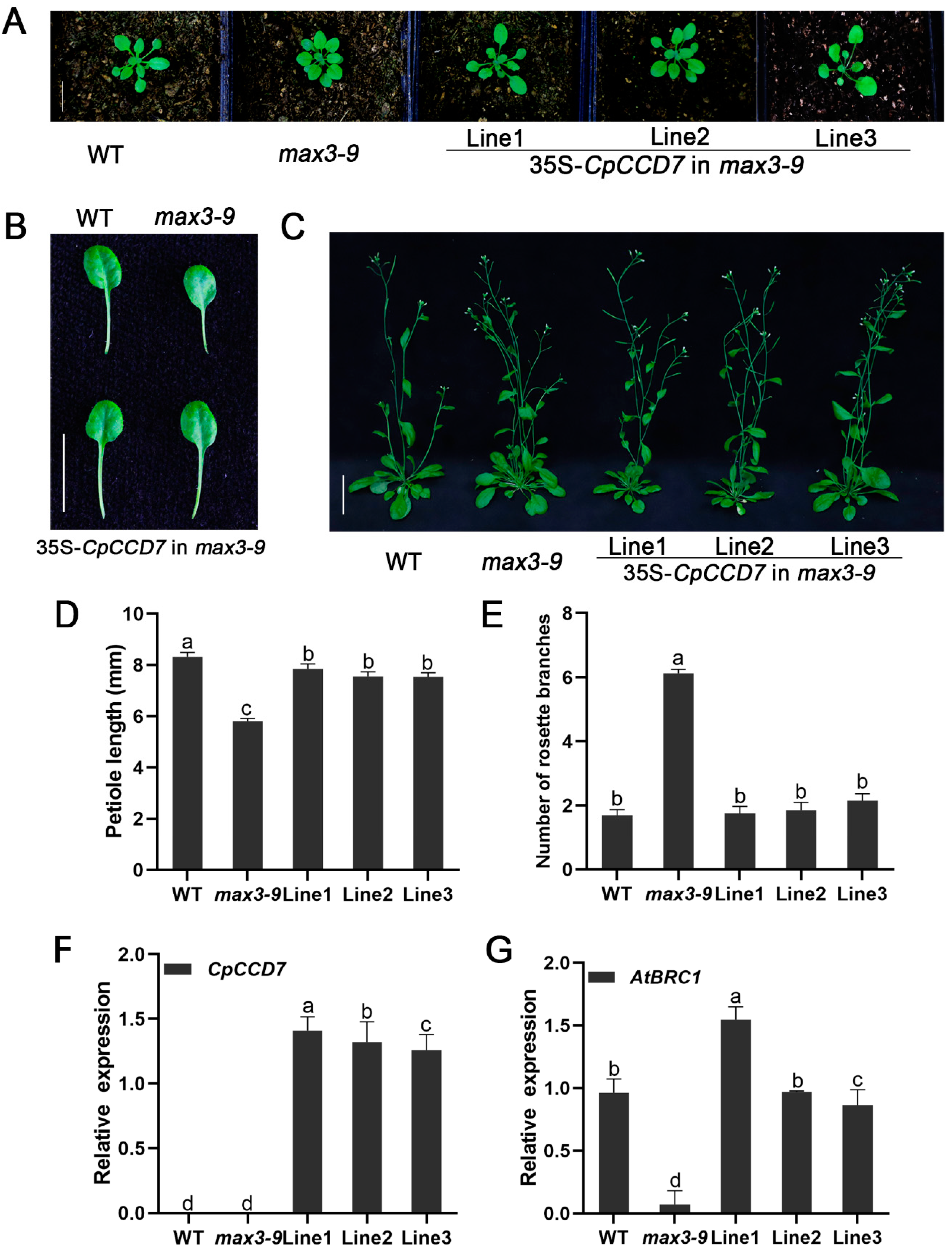
2.5. CpCCD7 and CpCCD8 Genes Restore the Branching Phenotype of Arabidopsis Max Mutants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cloning of CpCCD7 and CpCCD8 Genes
4.3. Gene Expression Analysis
4.4. Subcellular Localization Analysis of CpCCD7 and CpCCD8 Proteins
4.5. Overexpression Plasmid Construction and Arabidopsis Transformation
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, J.; Ni, B.; Zeng, Y.; He, C.; Zhang, J. Transcriptomic Analysis Reveals Hormonal Control of Shoot Branching in Salix matsudana. Forest 2020, 11, 287. [Google Scholar] [CrossRef] [Green Version]
- Ledger, S.; Janssen, B.; Karunairetnam, S.; Wang, T.; Snowden, K.C. Modified CAROTENOID CLEAVAGE DIOXYGENASE8 expression correlates with altered branching in kiwifruit (Actinidia chinensis). New Phytol. 2010, 188, 803–813. [Google Scholar] [CrossRef]
- Wang, M.; Le Moigne, M.-A.; Bertheloot, J.; Crespel, L.; Perez-Garcia, M.-D.; Ogé, L.; Demotes-Mainard, S.; Hamama, L.; Davière, J.-M.; Sakr, S. BRANCHED1: A Key Hub of Shoot Branching. Front. Plant Sci. 2019, 10, 76. [Google Scholar] [CrossRef] [Green Version]
- Umehara, M.; Hanada, A.; Magome, H.; Takeda-Kamiya, N.; Yamaguchi, S. Contribution of Strigolactones to the Inhibition of Tiller Bud Outgrowth under Phosphate Deficiency in Rice. Plant Cell Physiol. 2010, 51, 1118–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Sachs, T.; Thimann, K.V. The Role of Auxins and Cytokinins in the Release of Buds from Dominance. Am. J. Bot. 1967, 54, 136–144. [Google Scholar] [CrossRef]
- Nordström, A.; Tarkowski, P.; Tarkowska, D.; Norbaek, R.; Åstot, C.; Dolezal, K.; Sandberg, G. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 2004, 101, 8039–8044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.; Puech-Pagès, V.; Dun, E.; Pillot, J.-P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.-C.; et al. Strigolactone inhibition of shoot branching. Nat. Cell Biol. 2008, 455, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Bhoi, A.; Yadu, B.; Chandra, J.; Keshavkant, S. Contribution of strigolactone in plant physiology, hormonal interaction and abiotic stresses. Planta 2021, 254, 1–21. [Google Scholar] [CrossRef]
- Arite, T.; Iwata, H.; Ohshima, K.; Maekawa, M.; Nakajima, M.; Kojima, M.; Sakakibara, H.; Kyozuka, J. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007, 51, 1019–1029. [Google Scholar] [CrossRef]
- Beveridge, C. Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regul. 2000, 32, 193–203. [Google Scholar] [CrossRef]
- Morris, S.E.; Turnbull, C.G.; Murfet, I.C.; Beveridge, C.A. Mutational Analysis of Branching in Pea. Evidence that Rms1 and Rms5 Regulate the Same Novel Signal. Plant Physiol. 2001, 126, 1213. [Google Scholar] [CrossRef] [Green Version]
- Rameau, C.; Murfet, I.C.; Laucou, V.; Floyd, R.S.; Morris, S.; Beveridge, C.A. Pea rms6 mutants exhibit increased basal branching. Physiol. Plant 2002, 115, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Sorefan, K.; Booker, J.; Haurogné, K.; Goussot, M.; Bainbridge, K.; Foo, E.; Chatfield, S.; Ward, S.; Beveridge, C.; Rameau, C.; et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003, 17, 1469–1474. [Google Scholar] [CrossRef] [Green Version]
- Booker, J.; Auldridge, M.; Wills, S.; McCarty, D.; Klee, H.; Leyser, O. MAX3/CCD7 Is a Carotenoid Cleavage Dioxygenase Required for the Synthesis of a Novel Plant Signaling Molecule. Curr. Biol. 2004, 14, 1232–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stirnberg, P.; van De Sande, K.; Leyser, H.O. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 2002, 129, 1131–1141. [Google Scholar] [CrossRef]
- Zou, J.; Chen, Z.; Zhang, S.; Zhang, W.; Jiang, G.; Zhao, X.; Zhai, W.; Pan, X.; Zhu, L. Characterizations and fine mapping of a mutant gene for high tillering and dwarf in rice (Oryza sativa L.). Planta 2005, 222, 604–612. [Google Scholar] [CrossRef]
- Napoli, C. Highly Branched Phenotype of the Petunia dadl-7 Mutant 1s Reversed by Grafting. Plant Phys. 1996, 111, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Napoli, C.A.; Ruehle, J. New Mutations Affecting Meristem Growth and Potential in Petunia hybrida Vilm. J. Hered. 1996, 87, 371–377. [Google Scholar] [CrossRef] [Green Version]
- Johnson, X.; Brcich, T.; Dun, E.A.; Goussot, M.; Haurogné, K.; Beveridge, C.A.; Rameau, C. Branching Genes Are Conserved across Species. Genes Controlling a Novel Signal in Pea Are Coregulated by Other Long-Distance Signals. Plant Physiol. 2006, 142, 1014–1026. [Google Scholar] [CrossRef] [Green Version]
- Waters, M.T.; Brewer, P.; Bussell, J.D.; Smith, S.; Beveridge, C.A. The Arabidopsis Ortholog of Rice DWARF27 Acts Upstream of MAX1 in the Control of Plant Development by Strigolactones. Plant Physiol. 2012, 159, 1073–1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Wang, R.; Qian, Q.; Yan, M.; Meng, X.; Fu, Z.; Yan, C.; Jiang, B.; Su, Z.; Li, J.; et al. DWARF27, an Iron-Containing Protein Required for the Biosynthesis of Strigolactones, Regulates Rice Tiller Bud Outgrowth. Plant Cell 2009, 21, 1512–1525. [Google Scholar] [CrossRef] [Green Version]
- Harrison, P.J.; Bugg, T.D. Enzymology of the carotenoid cleavage dioxygenases: Reaction mechanisms, inhibition and biochemical roles. Arch. Biochem. Biophys. 2014, 544, 105–111. [Google Scholar] [CrossRef]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [Green Version]
- Booker, J.; Sieberer, T.; Wright, W.; Williamson, L.; Willett, B.; Stirnberg, P.; Turnbull, C.; Srinivasan, M.; Goddard, P.; Leyser, O. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 2005, 8, 443–449. [Google Scholar] [CrossRef] [Green Version]
- Donald, C.M. The breeding of crop ideotypes. Euphytica 1968, 17, 385–403. [Google Scholar] [CrossRef]
- Pan, X.; Zheng, H.; Zhao, J.; Xu, Y.; Li, X. ZmCCD7/ZpCCD7 encodes a carotenoid cleavage dioxygenase mediating shoot branching. Planta 2016, 243, 1407–1418. [Google Scholar] [CrossRef]
- Zhao, B.; Wu, T.T.; Ma, S.S.; Jiang, D.J.; Bie, X.M.; Sui, N.; Zhang, X.S.; Wang, F. TaD27-B gene controls the tiller number in hexaploid wheat. Plant Biotechnol. J. 2019, 18, 513–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, R.; Rowland, L.J.; Tanino, K. Induction and Release of Bud Dormancy in Woody Perennials: A Science Comes of Age. HortScience 2003, 38, 911–921. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Li, Z.; Wang, B.; Sui, S.; Li, M. Cloning of an Expansin Gene from Chimonanthus praecox Flowers and Its Expression in Flowers Treated with Ethephon or 1-Methylcyclopropene. HortScience 2012, 47, 1472–1477. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Huang, R.; Ma, J.; Sui, S.; Guo, Y.; Liu, D.; Li, Z.; Lin, Y.; Li, M. Two C3H Type Zinc Finger Protein Genes, CpCZF1 and CpCZF2, from Chimonanthus praecox Affect Stamen Development in Arabidopsis. Genes 2017, 8, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, K.G.; Zhou, M.Q.; Chen, L.Q.; Zhang, D.; Robert, G.W. Genetic Diversity and Discrimination of Chimonanthus praecox (L.) Link Germplasm Using ISSR and RAPD Markers. Hortscience 2007, 42, 1144–1148. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Liu, D.; Ma, J.; Li, M.; Sui, S. CpWRKY71, a WRKY Transcription Factor Gene of Wintersweet (Chimonanthus praecox), Promotes Flowering and Leaf Senescence in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 5325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Jiang, Y.; Liu, D.; Ma, J.; Li, J.; Li, M.; Sui, S. Floral Scent Emission from Nectaries in the Adaxial Side of the Innermost and Middle Petals in Chimonanthus praecox. Int. J. Mol. Sci. 2018, 19, 3278. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Ma, Z.; Zhao, K.; Zhang, J.; Xiang, L.; Chen, L. Transcriptomic and proteomic approaches to explore the differences in monoterpene and benzenoid biosynthesis between scented and unscented genotypes of wintersweet. Physiol. Plant 2018, 166, 478–493. [Google Scholar] [CrossRef]
- Aslam, M.Z.; Lin, X.; Li, X.; Yang, N.; Chen, L. Molecular Cloning and Functional Characterization of CpMYC2 and CpBHLH13 Transcription Factors from Wintersweet (Chimonanthus praecox L.). Plants 2020, 9, 785. [Google Scholar] [CrossRef]
- Zhao, R.; Song, X.; Yang, N.; Chen, L.; Xiang, L.; Liu, X.-Q.; Zhao, K. Expression of the subgroup IIIf bHLH transcription factor CpbHLH1 from Chimonanthus praecox (L.) in transgenic model plants inhibits anthocyanin accumulation. Plant Cell Rep. 2020, 39, 891–907. [Google Scholar] [CrossRef]
- Liu, D.; Sui, S.; Ma, J.; Li, Z.; Guo, Y.; Luo, D.; Yang, J.; Li, M. Transcriptomic Analysis of Flower Development in Wintersweet (Chimonanthus praecox). PLoS ONE 2014, 9, e86976. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1Acts as an Integrator of Branching Signals within Axillary Buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, C.A.; Kyozuka, J. New genes in the strigolactone-related shoot branching pathway. Curr. Opin. Plant Biol. 2010, 13, 34–39. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, J.; Tan, B.-C.; McCarty, D.R.; Klee, H.J. The Carotenoid Cleavage Dioxygenase 1 Enzyme Has Broad Substrate Specificity, Cleaving Multiple Carotenoids at Two Different Bond Positions. J. Biol. Chem. 2008, 283, 11364–11373. [Google Scholar] [CrossRef] [Green Version]
- Ilg, A.; Bruno, M.; Beyer, P.; Al-Babili, S. Tomato carotenoid cleavage dioxygenases 1A and 1B: Relaxed double bond specificity leads to a plenitude of dialdehydes, mono-apocarotenoids and isoprenoid volatiles. FEBS Open Bio 2014, 4, 584–593. [Google Scholar] [CrossRef] [Green Version]
- Ohmiya, A.; Kishimoto, S.; Aida, R.; Yoshioka, S.; Sumitomo, K. Carotenoid Cleavage Dioxygenase (CmCCD4a) Contributes to White Color Formation in Chrysanthemum Petals. Plant Physiol. 2006, 142, 1193–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Liu, C.; Wang, Y.; Yao, X.; Wang, F.; Wu, J.; King, G.; Liu, K. Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 2015, 206, 1513–1526. [Google Scholar] [CrossRef] [PubMed]
- Campbell, R.; Ducreux, L.J.; Morris, W.L.; Morris, J.A.; Suttle, J.C.; Ramsay, G.; Bryan, G.; Hedley, P.; Taylor, M.A. The Metabolic and Developmental Roles of Carotenoid Cleavage Dioxygenase4 from Potato. Plant Physiol. 2010, 154, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Rodrigo, M.J.; Alquezar, B.; Alos, E.; Medina, V.; Carmona, L.; Bruno, M.; Al-Babili, S.; Zacarias, L. A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J. Exp. Bot. 2013, 64, 4461–4478. [Google Scholar] [CrossRef] [PubMed]
- Daruwalla, A.; Kiser, P.D. Structural and mechanistic aspects of carotenoid cleavage dioxygenases (CCDs). Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2019, 1865, 158590. [Google Scholar] [CrossRef] [PubMed]
- Batra, R.; Agarwal, P.; Tyagi, S.; Saini, D.K.; Kumar, V.; Kumar, A.; Kumar, S.; Balyan, H.S.; Pandey, R.; Gupta, P.K. A study of CCD8 genes/proteins in seven monocots and eight dicots. PLoS ONE 2019, 14, e0213531. [Google Scholar] [CrossRef]
- Foster, T.M.; Ledger, S.E.; Janssen, B.J.; Luo, Z.; Drummond, R.S.M.; Tomes, S.; Karunairetnam, S.; Waite, C.N.; Funnell, K.A.; Van Hooijdonk, B.M.; et al. Expression of MdCCD7 in the scion determines the extent of sylleptic branching and the primary shoot growth rate of apple trees. J. Exp. Bot. 2017, 69, 2379–2390. [Google Scholar] [CrossRef] [Green Version]
- Drummond, R.; Martínez-Sánchez, N.M.; Janssen, B.; Templeton, K.; Simons, J.; Quinn, B.D.; Karunairetnam, S.; Snowden, K.C. Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 Is Involved in the Production of Negative and Positive Branching Signals in Petunia. Plant Physiol. 2009, 151, 1867–1877. [Google Scholar] [CrossRef] [Green Version]
- Vogel, J.; Walter, M.H.; Giavalisco, P.; Lytovchenko, A.; Kohlen, W.; Charnikhova, T.; Simkin, A.J.; Goulet, C.; Strack, D.; Bouwmeester, H.J.; et al. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J. 2009, 61, 300–311. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, L.; Zhu, M.; Han, L.; Lv, Y.; Liu, Y.; Li, P.; Jing, H.; Cai, H. Non-dormant Axillary Bud 1 regulates axillary bud outgrowth in sorghum. J. Integr. Plant Biol. 2018, 60, 938–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, J.; Zhang, S.; Zhang, W.; Li, G.; Chen, Z.; Zhai, W.; Zhao, X.; Pan, X.; Xie, Q.; Zhu, L. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 2006, 48, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Czarnecki, O.; Yang, J.; Wang, X.; Wang, S.; Muchero, W.; Tuskan, G.A.; Chen, J.-G. Characterization of MORE AXILLARY GROWTH Genes in Populus. PLoS ONE 2014, 9, e102757. [Google Scholar] [CrossRef] [Green Version]
- Snowden, K.C.; Simkin, A.J.; Janssen, B.; Templeton, K.; Loucas, H.M.; Simons, J.; Karunairetnam, S.; Gleave, A.; Clark, D.G.; Klee, H.J. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 Gene Affects Branch Production and Plays a Role in Leaf Senescence, Root Growth, and Flower Development. Plant Cell 2005, 17, 746–759. [Google Scholar] [CrossRef] [Green Version]
- Kohlen, W.; Charnikhova, T.; Lammers, M.; Pollina, T.; Tóth, P.; Haider, I.; Pozo, M.J.; De Maagd, R.A.; Ruyter-Spira, C.; Bouwmeester, H.J.; et al. The tomato CAROTENOID CLEAVAGE DIOXYGENASE 8 (S l CCD 8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 2012, 196, 535–547. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, T.; Xu, B.; Jia, L.; Xiao, B.; Liu, H.; Liu, L.; Yan, H.; Xia, Q. CRISPR/Cas9-Mediated Mutagenesis of Carotenoid Cleavage Dioxygenase 8 (CCD8) in Tobacco Affects Shoot and Root Architecture. Int. J. Mol. Sci. 2018, 19, 1062. [Google Scholar] [CrossRef] [Green Version]
- Pasare, S.A.; Ducreux, L.J.; Morris, W.L.; Campbell, R.; Sharma, S.K.; Roumeliotis, E.; Kohlen, W.; van der Krol, S.; Bramley, P.M.; Roberts, A.G.; et al. The role of the potato (Solanum tuberosum) CCD8 gene in stolon and tuber development. New Phytol. 2013, 198, 1108–1120. [Google Scholar] [CrossRef]
- Guan, J.C.; Koch, K.E.; Suzuki, M.; Wu, S.; Latshaw, S.; Petruff, T.; Goulet, C.; Klee, H.J.; McCarty, D.R. Diverse Roles of Strigolactone Signaling in Maize Architecture and the Uncoupling of a Branching-Specific Subnetwork. Plant Physiol. 2012, 160, 1303–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bainbridge, K.; Sorefan, K.; Ward, S.; Leyser, O. Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J. 2005, 44, 569–580. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Qin, X.; Loewen, M.C. The Biochemical Characterization of Two Carotenoid Cleavage Enzymes from Arabidopsis Indicates That a Carotenoid-derived Compound Inhibits Lateral Branching. J. Biol. Chem. 2004, 279, 46940–46945. [Google Scholar] [CrossRef] [Green Version]
- Ren, C.; Guo, Y.; Kong, J.; Lecourieux, F.; Dai, Z.; Li, S.; Liang, Z. Knockout of VvCCD8 gene in grapevine affects shoot branching. BMC Plant Biol. 2020, 20, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dun, E.; Germain, A.D.S.; Rameau, C.; Beveridge, C.A. Antagonistic Action of Strigolactone and Cytokinin in Bud Outgrowth Control. Plant Physiol. 2011, 158, 487–498. [Google Scholar] [CrossRef] [Green Version]
- Drummond, R.S.; Janssen, B.J.; Luo, Z.; Oplaat, C.; Ledger, S.E.; Wohlers, M.; Snowden, K.C. Environmental Control of Branching in Petunia. Plant Physiol. 2015, 168, 735–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Ma, J.; Liu, H.; Wang, X.; Li, J.; Li, Z.; Li, M.; Sui, S.; Liu, D. Overexpression of CpSIZ1, a SIZ/PIAS-Type SUMO E3 Ligase from Wintersweet (Chimonanthus praecox), Delays Flowering, Accelerates Leaf Senescence and Enhances Cold Tolerance in Arabidopsis. Plant Mol. Biol. Rep. 2020, 39, 301–316. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Liu, D.; Lin, J.; Zhu, T.; Liu, N.; Yang, X.; Ma, J.; Sui, S. Carotenoid Cleavage Dioxygenase Genes of Chimonanthus praecox, CpCCD7 and CpCCD8, Regulate Shoot Branching in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 8750. https://doi.org/10.3390/ijms22168750
Wang X, Liu D, Lin J, Zhu T, Liu N, Yang X, Ma J, Sui S. Carotenoid Cleavage Dioxygenase Genes of Chimonanthus praecox, CpCCD7 and CpCCD8, Regulate Shoot Branching in Arabidopsis. International Journal of Molecular Sciences. 2021; 22(16):8750. https://doi.org/10.3390/ijms22168750
Chicago/Turabian StyleWang, Xia, Daofeng Liu, Jie Lin, Ting Zhu, Ning Liu, Ximeng Yang, Jing Ma, and Shunzhao Sui. 2021. "Carotenoid Cleavage Dioxygenase Genes of Chimonanthus praecox, CpCCD7 and CpCCD8, Regulate Shoot Branching in Arabidopsis" International Journal of Molecular Sciences 22, no. 16: 8750. https://doi.org/10.3390/ijms22168750





