Expression Profile of Genes Encoding Proteins Involved in Regulation of Vasculature Development and Heart Muscle Morphogenesis—A Transcriptomic Approach Based on a Porcine Model
Abstract
:1. Introduction
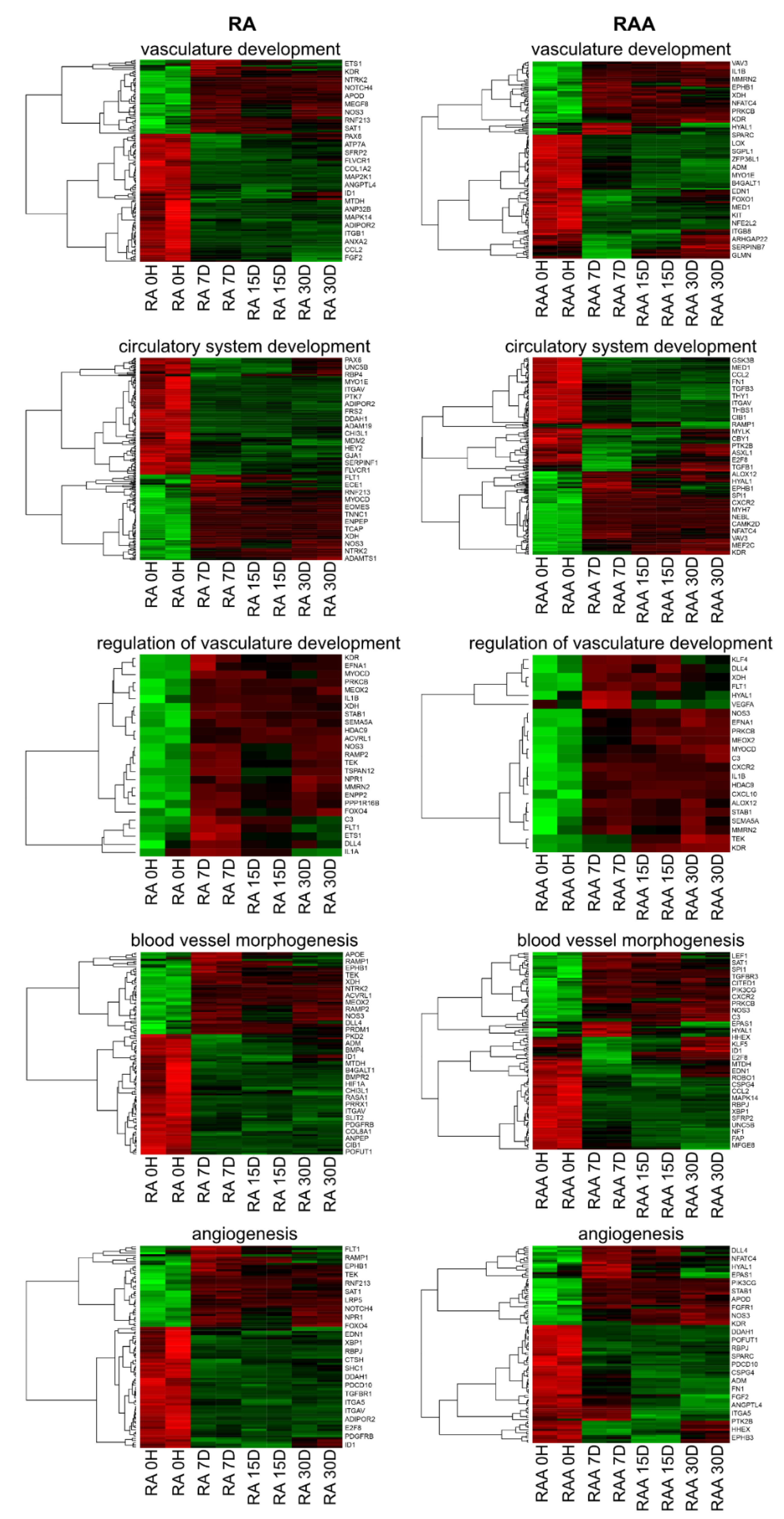
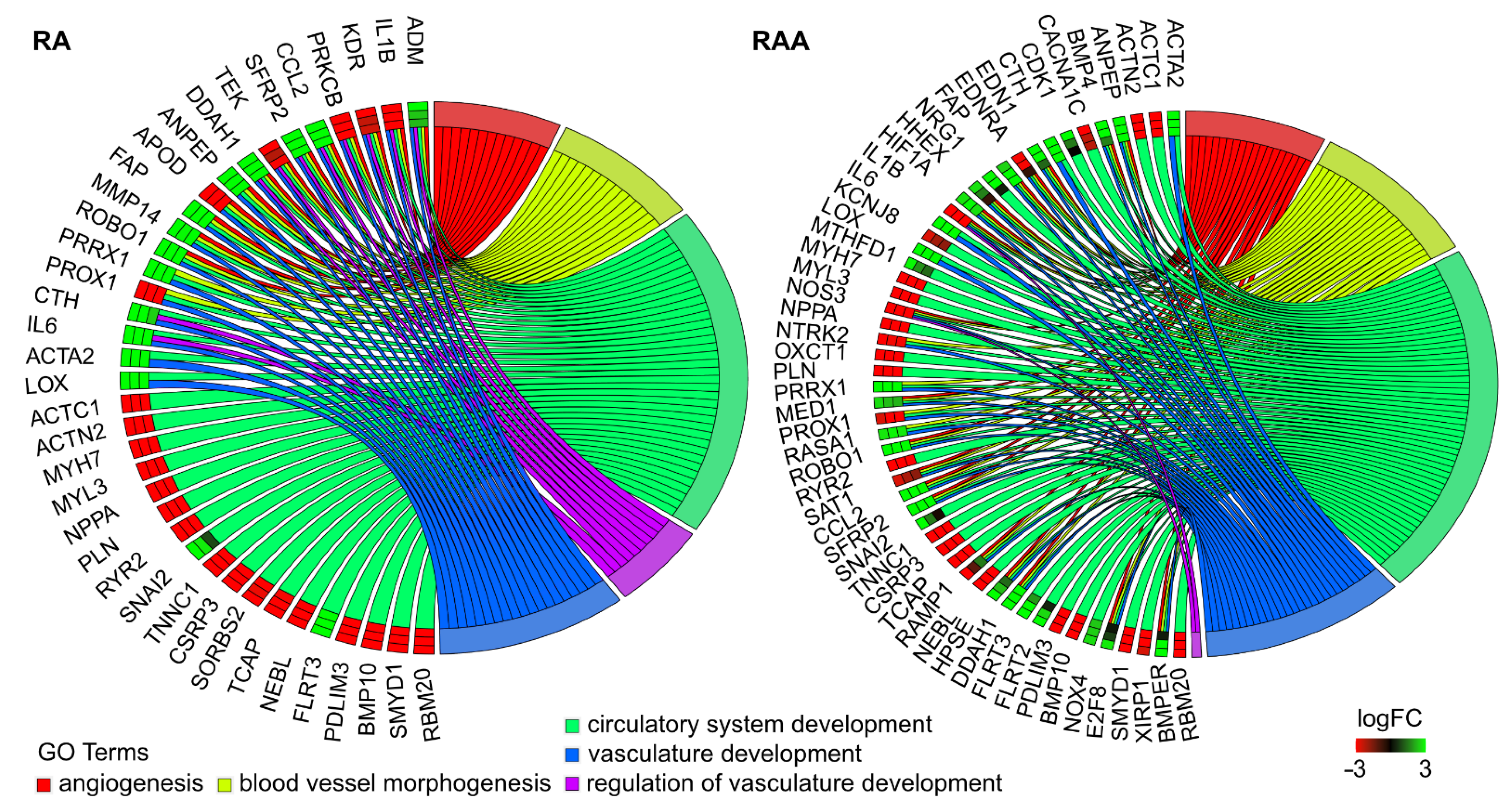
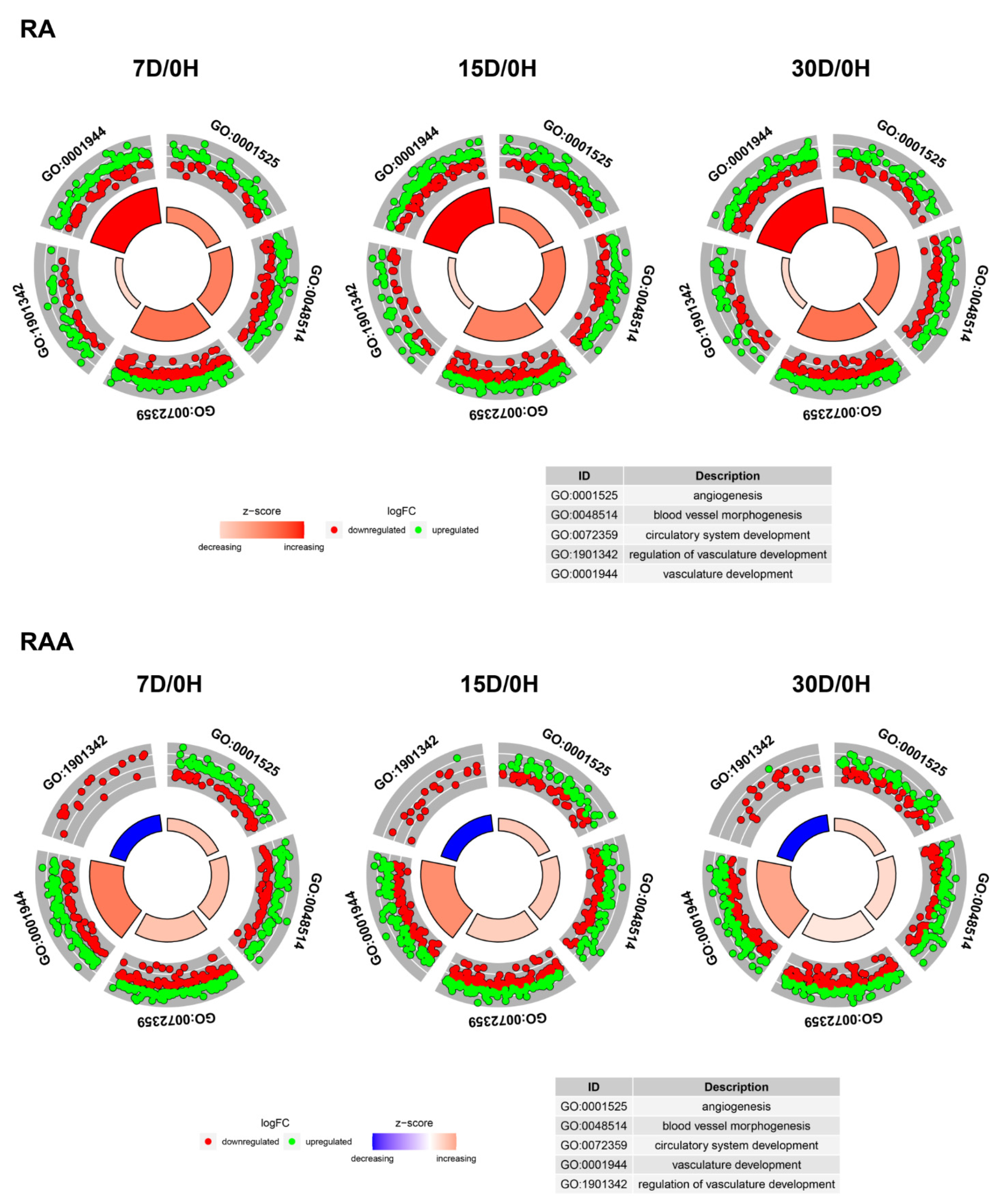
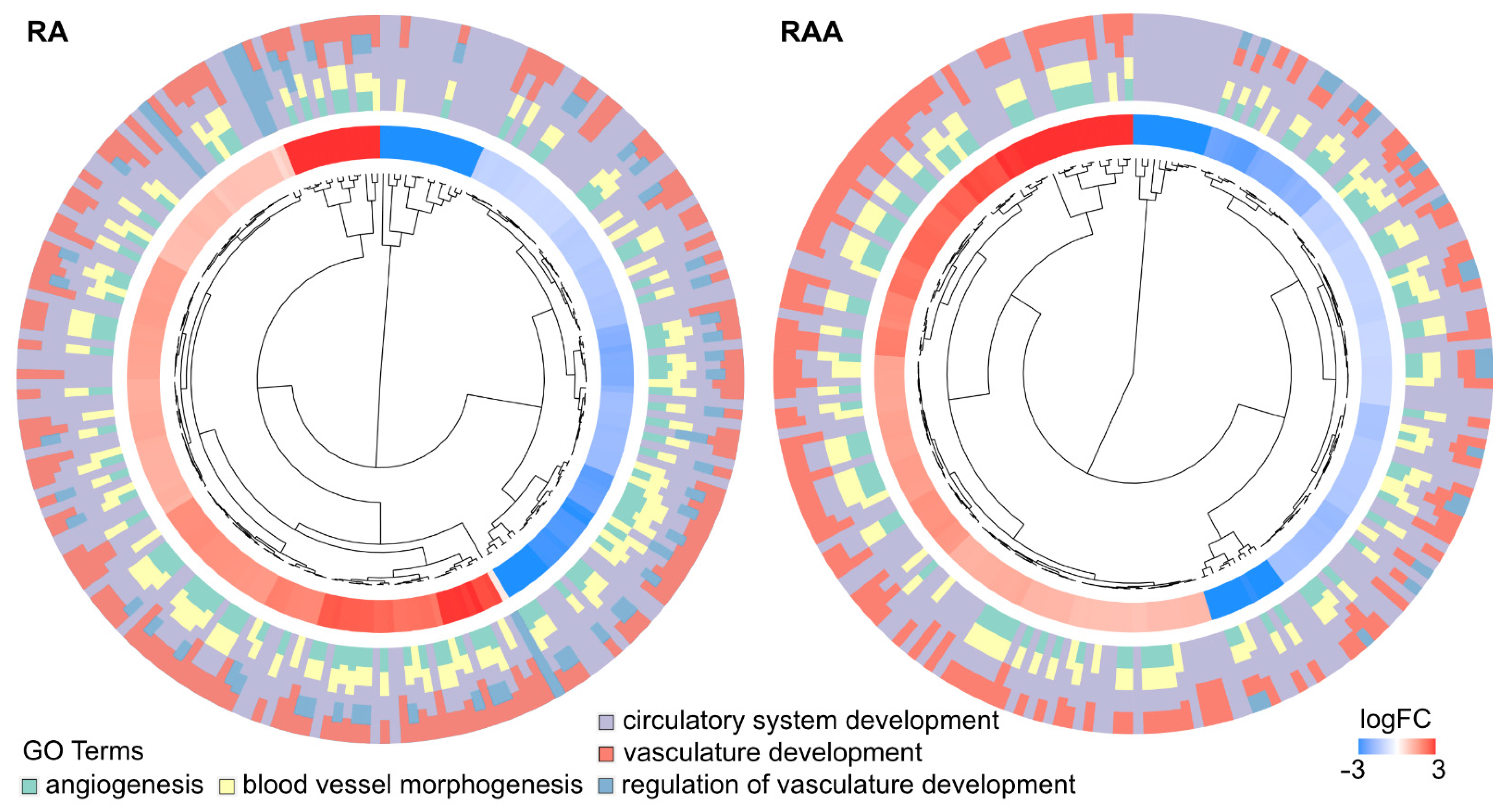
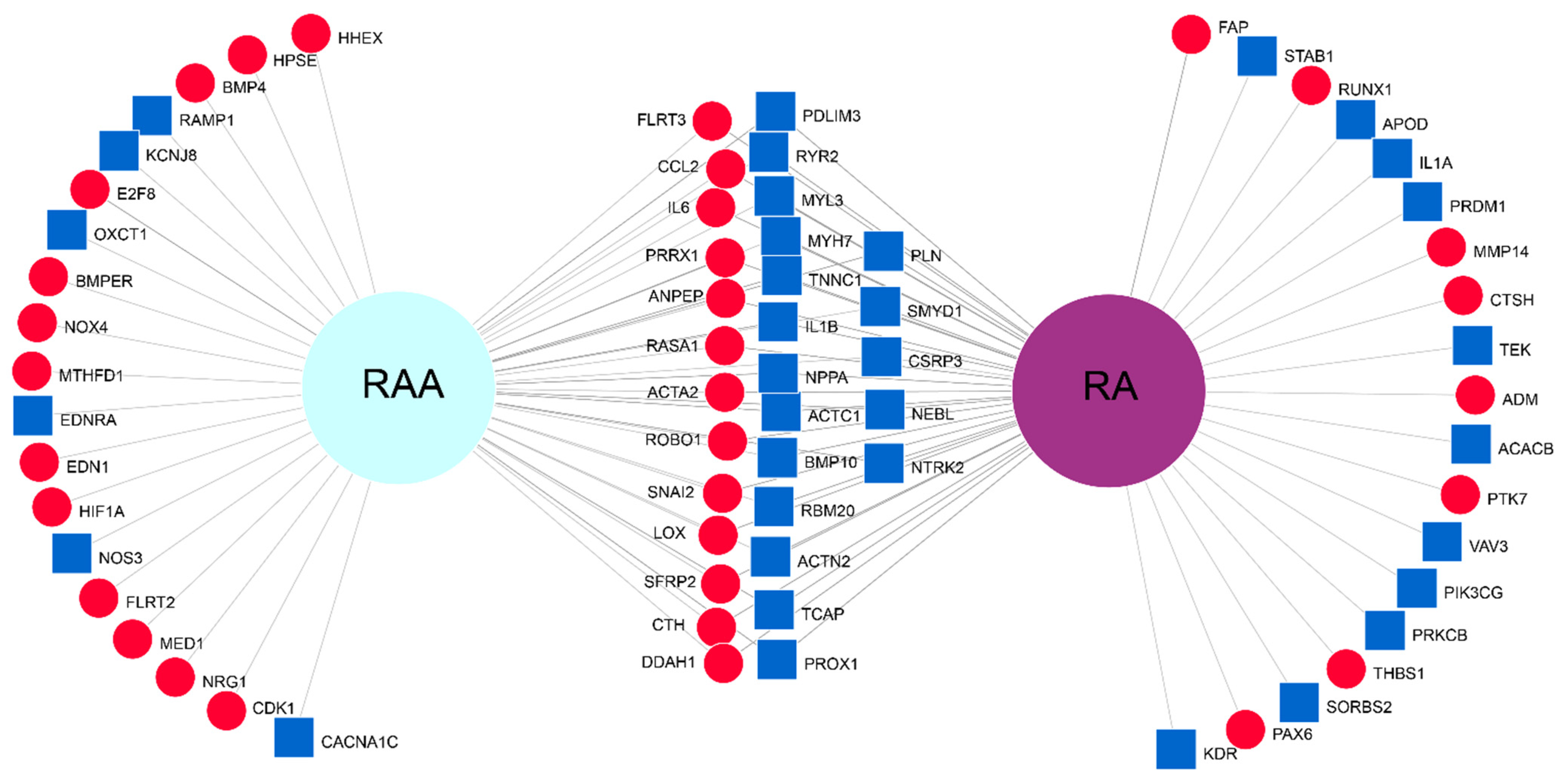
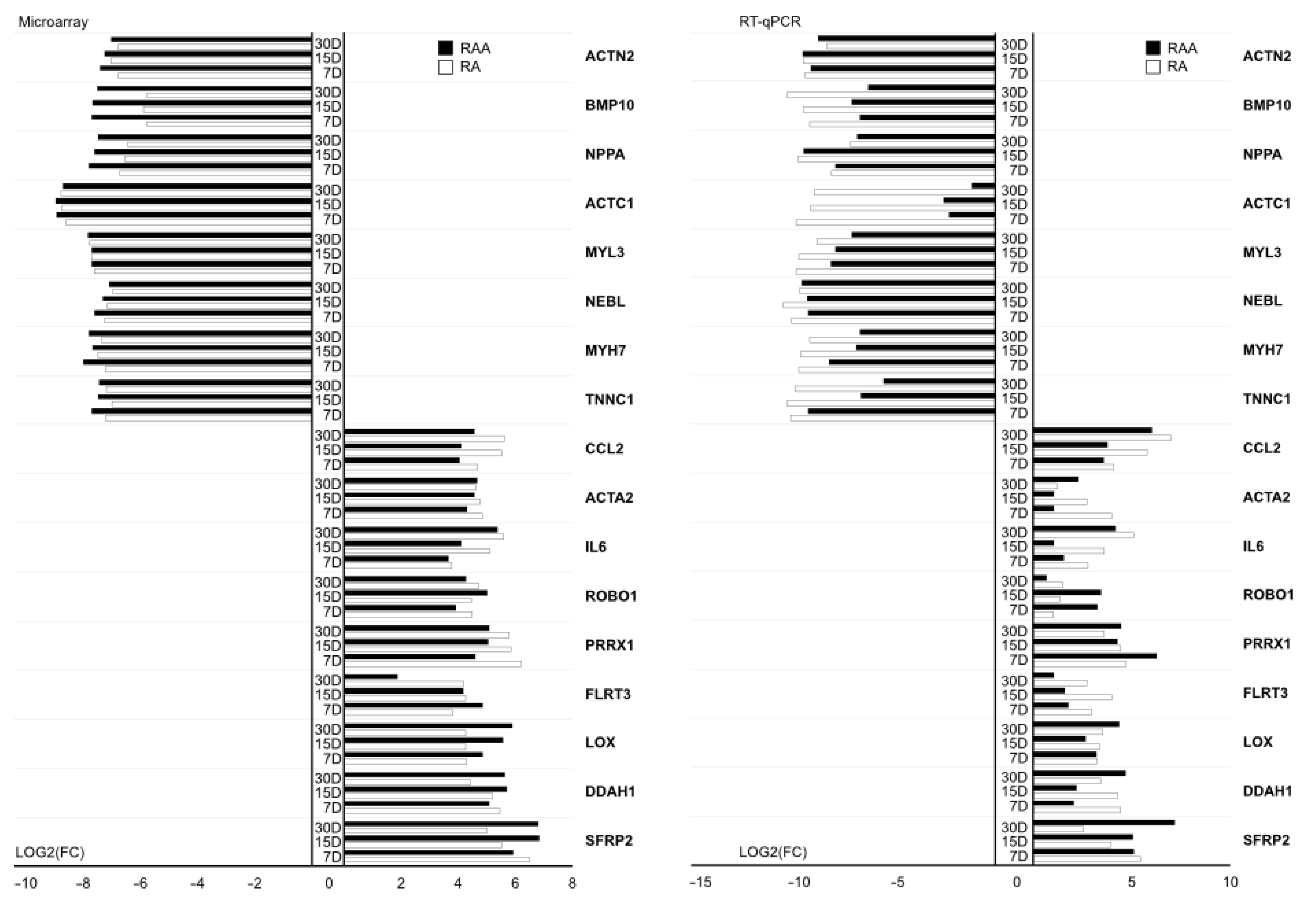
2. Results
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Tissue Collection from Porcine Hearts
4.3. Enzymatic Dissociation and Primary Cell Culture
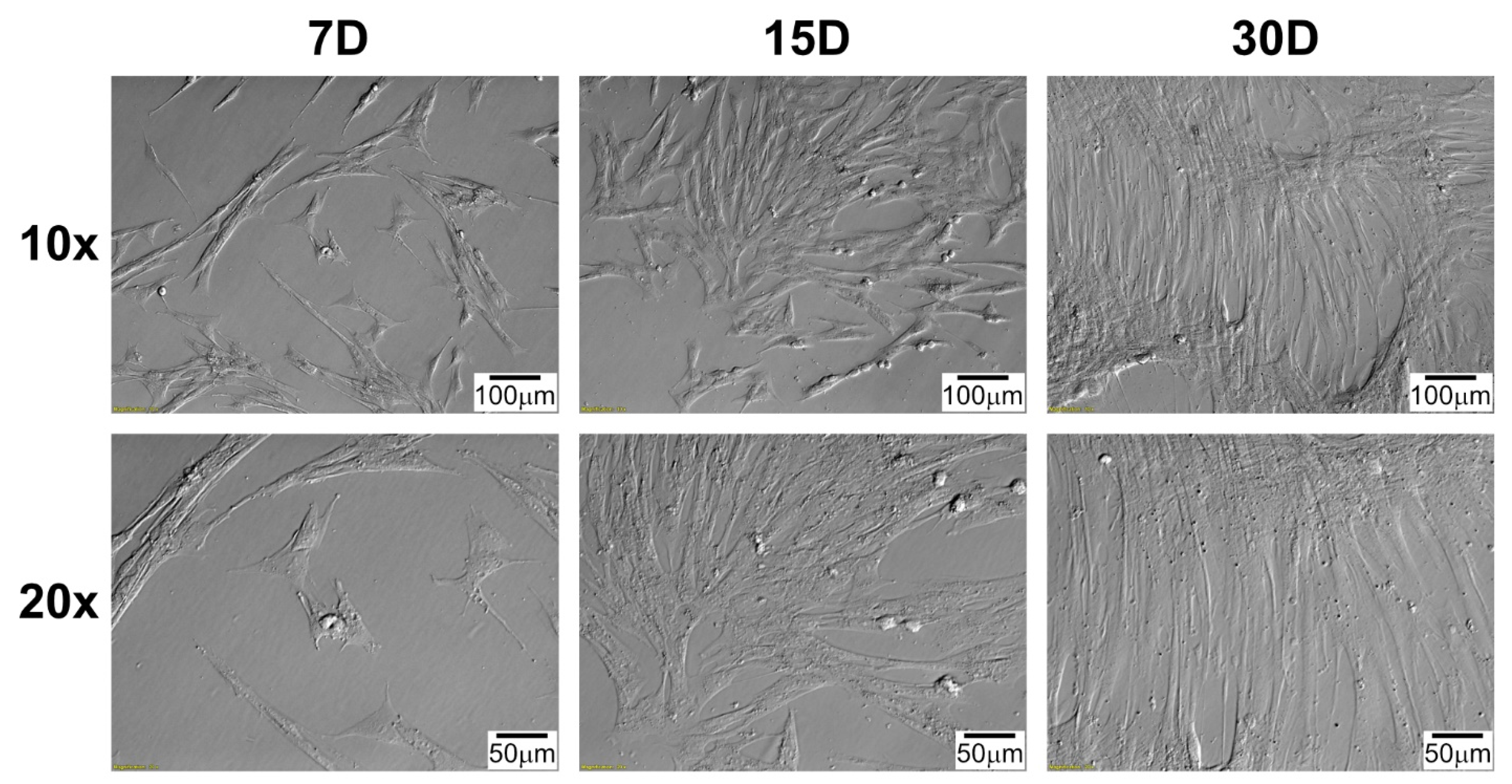
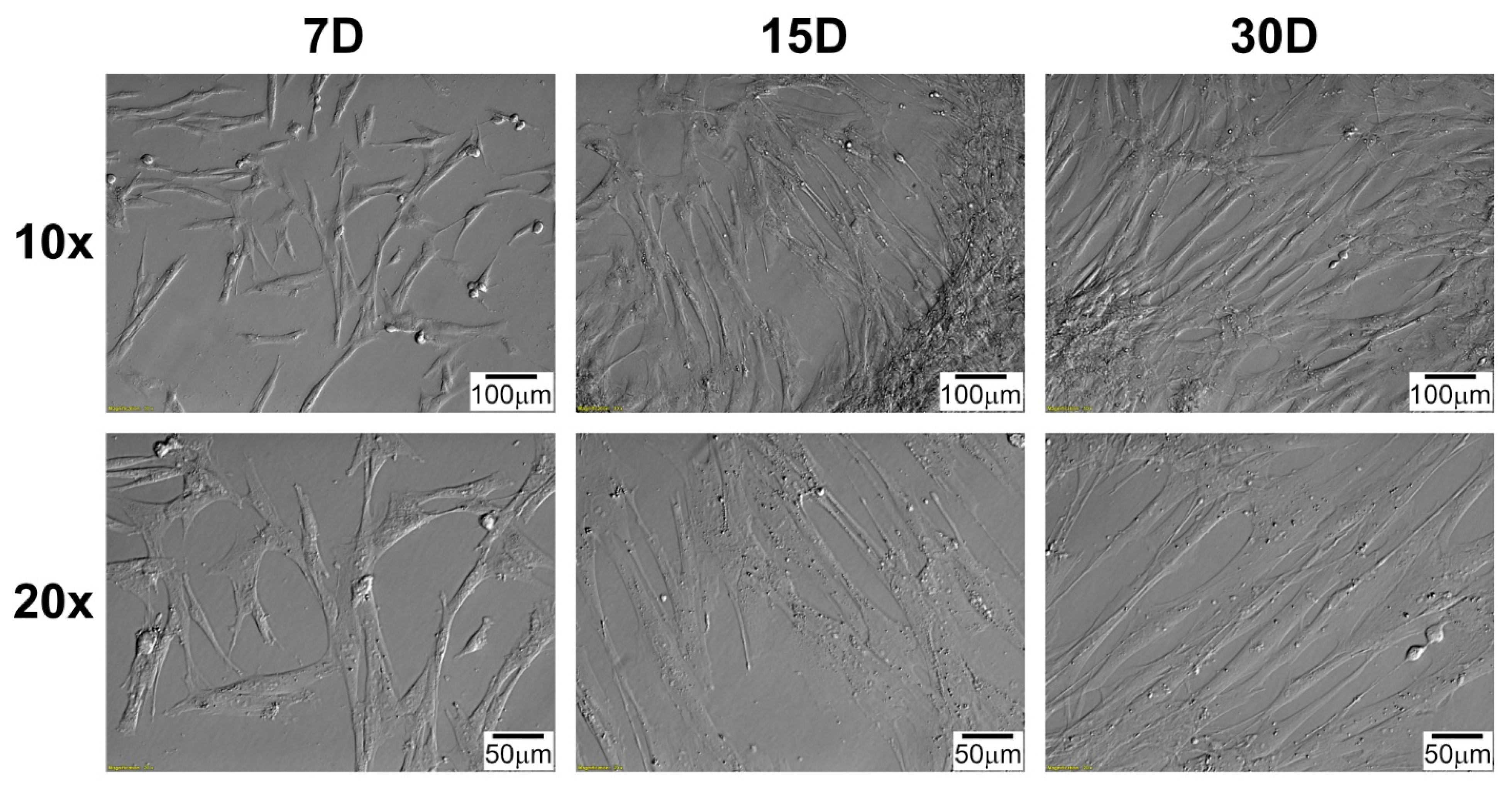
4.4. Morphological Observation of Cells during Long-Term Primary In Vitro Culture
4.5. RNA Extraction and Reverse Transcription
4.6. Microarray Expression Study and Data Analysis
4.7. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gummert, J.; Barten, M.; Rahmel, A.; Doll, S.; Krakor, R.; Garbade, J.; Autschbach, R. Therapy for Heart Failure: The Leipzig Approach. Thorac. Cardiovasc. Surg. 2017, 65, S205–S208. [Google Scholar] [CrossRef]
- He, L.; Zhou, B. The Development and Regeneration of Coronary Arteries. Curr. Cardiol. Rep. 2018, 20, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Hu, T.; Zhang, H.; He, L.; Huang, X.; Liu, Q.; Yu, W.; He, L.; Yang, Z.; Yan, Y.; et al. De novo formation of a distinct coronary vascular population in neonatal heart. Science 2014, 345, 90–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzi, A.; Benagiano, V.; Ribatti, D. Angiogenesis versus arteriogenesis. Rom. J. Morphol. Embryol. 2017, 58, 15–19. [Google Scholar] [PubMed]
- Krenning, G.; Zeisberg, E.M.; Kalluri, R. The origin of fibroblasts and mechanism of cardiac fibrosis. J. Cell. Physiol. 2010, 225, 631–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore-Morris, T.; Guimarães-Camboa, N.; Yutzey, K.E.; Pucéat, M.; Evans, S.M. Cardiac fibroblasts: From development to heart failure. J. Mol. Med. 2015, 93, 823–830. [Google Scholar] [CrossRef] [Green Version]
- Kanisicak, O.; Khalil, H.; Ivey, M.J.; Karch, J.; Maliken, B.D.; Correll, R.N.; Brody, M.J.; Lin, S.C.J.; Aronow, B.J.; Tallquist, M.D.; et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kaur, H.; Takefuji, M.; Ngai, C.Y.; Carvalho, J.; Bayer, J.; Wietelmann, A.; Poetsch, A.; Hoelper, S.; Conway, S.J.; Möllmann, H.; et al. Targeted Ablation of Periostin-Expressing Activated Fibroblasts Prevents Adverse Cardiac Remodeling in Mice. Circ. Res. 2016, 118, 1906–1917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeisberg, E.M.; Tarnavski, O.; Zeisberg, M.; Dorfman, A.L.; McMullen, J.R.; Gustafsson, E.; Chandraker, A.; Yuan, X.; Pu, W.T.; Roberts, A.B.; et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 2007, 13, 952–961. [Google Scholar] [CrossRef] [PubMed]
- Widyantoro, B.; Emoto, N.; Nakayama, K.; Anggrahini, D.W.; Adiarto, S.; Iwasa, N.; Yagi, K.; Miyagawa, K.; Rikitake, Y.; Suzuki, T.; et al. Endothelial cell-derived endothelin-1 promotes cardiac fibrosis in diabetic hearts through stimulation of endothelial-to-mesenchymal transition. Circulation 2010, 121, 2407–2418. [Google Scholar] [CrossRef] [Green Version]
- Ubil, E.; Duan, J.; Pillai, I.C.L.; Rosa-Garrido, M.; Wu, Y.; Bargiacchi, F.; Lu, Y.; Stanbouly, S.; Huang, J.; Rojas, M.; et al. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature 2014, 514, 585–590. [Google Scholar] [CrossRef] [Green Version]
- Moore-Morris, T.; Pucéat, M.; Evans, S.M. Targeting cardiac fibroblasts: The pressure is on. Cell Cycle 2014, 13, 2647–2648. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Huang, X.; Kanisicak, O.; Li, Y.; Wang, Y.; Li, Y.; Pu, W.; Liu, Q.; Zhang, H.; Tian, X.; et al. Preexisting endothelial cells mediate cardiac neovascularization after injury. J. Clin. Investig. 2017, 127, 2968–2981. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, G.; Kruszyna, Ł.; Nawrocki, M.J.; Strauss, E.; Bryl, R.; Spaczyńska, J.; Perek, B.; Jemielity, M.; Mozdziak, P.; Kempisty, B.; et al. Molecular mechanisms associated with ROS-dependent angiogenesis in lower extremity artery disease. Antioxidants 2021, 10, 735. [Google Scholar] [CrossRef]
- Hutchings, G.; Nawrocki, M.J.; Mozdziak, P.; Kempisty, B. Cardiac Stem Cell Therapy, Resident Progenitor Cells and the role of Cellular Signalling; a Review. Med. J. Cell Biol. 2019, 7, 112–118. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, Y.; Yeung, A.C.; Ikeno, F. The representative porcine model for human cardiovascular disease. J. Biomed. Biotechnol. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Lelovas, P.P.; Kostomitsopoulos, N.G.; Xanthos, T.T. A comparative anatomic and physiologic overview of the porcine heart. J. Am. Assoc. Lab. Anim. Sci. 2014, 53, 432–438. [Google Scholar]
- van Loon, K.; Huijbers, E.J.M.; Griffioen, A.W. Secreted frizzled-related protein 2: A key player in noncanonical Wnt signaling and tumor angiogenesis. Cancer Metastasis Rev. 2021, 40, 191–203. [Google Scholar] [CrossRef]
- Hsueh, Y.C.; Hodgkinson, C.P.; Gomez, J.A. The role of Sfrp and DKK proteins in cardiomyocyte development. Physiol. Rep. 2021, 9, e14678. [Google Scholar] [CrossRef] [PubMed]
- Pate, K.T.; Stringari, C.; Sprowl-Tanio, S.; Wang, K.; TeSlaa, T.; Hoverter, N.P.; McQuade, M.M.; Garner, C.; Digman, M.A.; Teitell, M.A.; et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 2014, 33, 1454–1473. [Google Scholar] [CrossRef] [PubMed]
- Vanhollebeke, B.; Stone, O.A.; Bostaille, N.; Cho, C.; Zhou, Y.; Maquet, E.; Gauquier, A.; Cabochette, P.; Fukuhara, S.; Mochizuki, N.; et al. Tip cell-specific requirement for an atypical Gpr124- and Reck-dependent Wnt/β-catenin pathway during brain angiogenesis. eLife 2015, 4, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, X.; Zheng, H.; Zhu, H.; Mai, W.; Huang, X.; Huang, Y. Multiple roles of sFRP2 in cardiac development and cardiovascular disease. Int. J. Biol. Sci. 2020, 16, 730–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galli, L.M.; Barnes, T.; Cheng, T.; Acosta, L.; Anglade, A.; Willert, K.; Nusse, R.; Burrus, L.W. Differential inhibition of Wnt-3a by Strp-1, Sfrp-2, and Sfrp-3. Dev. Dyn. 2006, 235, 681–690. [Google Scholar] [CrossRef] [Green Version]
- Alfaro, M.P.; Vincent, A.; Saraswati, S.; Thorne, C.A.; Hong, C.C.; Lee, E.; Young, P.P. sFRP2 suppression of Bone Morphogenic Protein (BMP) and Wnt signaling mediates Mesenchymal Stem Cell (MSC) self-renewal promoting engraftment and myocardial repair. J. Biol. Chem. 2010, 285, 35645–35653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, Y.; Yang, Y.; Li, Q.; He, X.; Zhu, W.; Wang, J.; Gan, X. Oligomerization of Frizzled and LRP5/6 protein initiates intracellular signaling for the canonical WNT/β-catenin pathway. J. Biol. Chem. 2018, 293, 19710–19724. [Google Scholar] [CrossRef] [Green Version]
- Skah, S.; Nadjar, J.; Sirakov, M.; Plateroti, M. The secreted Frizzled-Related Protein 2 modulates cell fate and the Wnt pathway in the murine intestinal epithelium. Exp. Cell Res. 2015, 330, 56–65. [Google Scholar] [CrossRef]
- Xavier, C.P.; Melikova, M.; Chuman, Y.; Üren, A.; Baljinnyam, B.; Rubin, J.S. Secreted Frizzled-related protein potentiation versus inhibition of Wnt3a/β-catenin signaling. Cell. Signal. 2014, 26, 94–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufourcq, P.; Leroux, L.; Ezan, J.; Descamps, B.; Lamazière, J.M.D.; Costet, P.; Basoni, C.; Moreau, C.; Deutsch, U.; Couffinhal, T.; et al. Regulation of endothelial cell cytoskeletal reorganization by a secreted frizzled-related protein-1 and frizzled 4-and frizzled 7-dependent pathway: Role in neovessel formation. Am. J. Pathol. 2008, 172, 37–49. [Google Scholar] [CrossRef] [Green Version]
- Courtwright, A.; Siamakpour-Reihani, S.; Arbiser, J.L.; Banet, N.; Hilliard, E.; Fried, L.; Livasy, C.; Ketelsen, D.; Nepal, D.B.; Perou, C.M.; et al. Secreted frizzle-related protein 2 stimulates angiogenesis via a calcineurin/NFAT signaling pathway. Cancer Res. 2009, 69, 4621–4628. [Google Scholar] [CrossRef] [Green Version]
- Crowley, R.K.; O’Reilly, M.W.; Bujalska, I.J.; Hassan-Smith, Z.K.; Hazlehurst, J.M.; Foucault, D.R.; Stewart, P.M.; Tomlinson, J.W. SFRP2 is associated with increased adiposity and VEGF expression. PLoS ONE 2016, 11, e0163777. [Google Scholar] [CrossRef]
- Kim, H.; Yoo, S.; Zhou, R.; Xu, A.; Bernitz, J.M.; Yuan, Y.; Gomes, A.M.; Daniel, M.G.; Su, J.; Demicco, E.G.; et al. Oncogenic role of SFRP2 in p53-mutant osteosarcoma development via autocrine and paracrine mechanism. Proc. Natl. Acad. Sci. USA 2018, 115, E11128–E11137. [Google Scholar] [CrossRef] [Green Version]
- Garcia, D.; Nasarre, P.; Bonilla, I.V.; Hilliard, E.; Peterson, Y.K.; Spruill, L.; Broome, A.M.; Hill, E.G.; Yustein, J.T.; Mehrotra, S.; et al. Development of a Novel Humanized Monoclonal Antibody to Secreted Frizzled-Related Protein-2 That Inhibits Triple-Negative Breast Cancer and Angiosarcoma Growth In Vivo. Ann. Surg. Oncol. 2019, 26, 4782–4790. [Google Scholar] [CrossRef]
- Vatner, D.E.; Oydanich, M.; Zhang, J.; Babici, D.; Vatner, S.F. Secreted frizzled-related protein 2, a novel mechanism to induce myocardial ischemic protection through angiogenesis. Basic Res. Cardiol. 2020, 115, 1–12. [Google Scholar] [CrossRef]
- Higuchi, M.; Yoshida, S.; Ueharu, H.; Chen, M.; Kato, T.; Kato, Y. PRRX1 and PRRX2 distinctively participate in pituitary organogenesis and a cell-supply system. Cell Tissue Res. 2014, 357, 323–335. [Google Scholar] [CrossRef]
- Ocaña, O.H.; Córcoles, R.; Fabra, Á.; Moreno-Bueno, G.; Acloque, H.; Vega, S.; Barrallo-Gimeno, A.; Cano, A.; Nieto, M.A. Metastatic Colonization Requires the Repression of the Epithelial-Mesenchymal Transition Inducer Prrx1. Cancer Cell 2012, 22, 709–724. [Google Scholar] [CrossRef] [Green Version]
- Norris, R.A.; Scott, K.K.; Moore, C.S.; Stetten, G.; Brown, C.R.; Jabs, E.W.; Wulfsberg, E.A.; Yu, J.; Kern, M.J. Human PRRX1 and PRRX2 genes: Cloning, expression, genomic localization, and exclusion as disease genes for Nager syndrome. Mamm. Genome 2000, 11, 1000–1005. [Google Scholar] [CrossRef] [PubMed]
- Ihida-Stansbury, K.; McKean, D.M.; Gebb, S.A.; Martin, J.F.; Stevens, T.; Nemenoff, R.; Akeson, A.; Vaughn, J.; Jones, P.L. Paired-related homeobox gene Prx1 is required for pulmonary vascular development. Circ. Res. 2004, 94, 1507–1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, R.; Liu, Y.; Wang, Y.; Wang, X.; Ci, H.; Song, C.; Wu, S. Low PRRX1 expression and high ZEB1 expression are significantly correlated with epithelial-mesenchymal transition and tumor angiogenesis in non-small cell lung cancer. Medicine (Baltim.) 2021, 100, e24472. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, M.; Kato, T.; Yoshida, S.; Ueharu, H.; Nishimura, N.; Kato, Y. PRRX1- and PRRX2-positive mesenchymal stem/progenitor cells are involved in vasculogenesis during rat embryonic pituitary development. Cell Tissue Res. 2015, 361, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, R.; Wang, Q.; Wang, Y.; Ci, H.; Wu, S. Aberrant expression of vasculogenic mimicry, PRRX1, and CIP2A in clear cell renal cell carcinoma and its clinicopathological significance. Medicine 2019, 98, 36. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, T. The chemokine MCP-1 (CCL2) in the host interaction with cancer: A foe or ally? Cell Mol. Immunol. 2018, 15, 335–345. [Google Scholar] [CrossRef] [Green Version]
- Ridiandries, A.; Tan, J.T.M.; Bursill, C.A. The role of chemokines in wound healing. Int. J. Mol. Sci. 2018, 19, 3217. [Google Scholar] [CrossRef] [Green Version]
- Bonapace, L.; Coissieux, M.M.; Wyckoff, J.; Mertz, K.D.; Varga, Z.; Junt, T.; Bentires-Alj, M. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature 2014, 515, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Fisher, D.T.; Appenheimer, M.M.; Evans, S.S. The two faces of IL-6 in the tumor microenvironment. Semin. Immunol. 2014, 26, 38–47. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.H.; Kuo, M.L.; Chen, C.A.; Chou, C.H.; Lai, K.B.; Lee, C.N.; Hsieh, C.Y. Interleukin-6 promotes cervical tumor growth by VEGF-dependent angiogenesis via a STAT3 pathway. Oncogene 2003, 22, 1517–1527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kujawski, M.; Kortylewski, M.; Lee, H.; Herrmann, A.; Kay, H.; Yu, H. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J. Clin. Investig. 2008, 118, 3367–3377. [Google Scholar] [CrossRef]
- Pan, L.; Xiao, H.; Liao, R.; Chen, Q.; Peng, C.; Zhang, Y.; Mu, T.; Wu, Z. Fatty acid binding protein 5 promotes tumor angiogenesis and activates the IL6/STAT3/VEGFA pathway in hepatocellular carcinoma. Biomed. Pharmacother. 2018, 106, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Ypsilanti, A.R.; Zagar, Y.; Chédotal, A. Moving away from the midline: New developments for Slit and Robo. Development 2010, 137, 1939–1952. [Google Scholar] [CrossRef] [Green Version]
- Rama, N.; Dubrac, A.; Mathivet, T.; Ní Chárthaigh, R.A.; Genet, G.; Cristofaro, B.; Pibouin-Fragner, L.; Ma, L.; Eichmann, A.; Chédotal, A. Slit2 signaling through Robo1 and Robo2 is required for retinal neovascularization. Nat. Med. 2015, 21, 483–491. [Google Scholar] [CrossRef] [Green Version]
- Ao, J.Y.; Chai, Z.T.; Zhang, Y.Y.; Zhu, X.D.; Kong, L.Q.; Zhang, N.; Ye, B.G.; Cai, H.; Gao, D.M.; Sun, H.C. Robo1 promotes angiogenesis in hepatocellular carcinoma through the Rho family of guanosine triphosphatases’ signaling pathway. Tumor Biol. 2015, 36, 8413–8424. [Google Scholar] [CrossRef]
- Lacy, S.E.; Bönnemann, C.G.; Buzney, E.A.; Kunkel, L.M. Identification of FLRT1, FLRT2, and FLRT3: A novel family of transmembrane leucine-rich repeat proteins. Genomics 1999, 62, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Jauhiainen, S.; Laakkonen, J.P.; Ketola, K.; Toivanen, P.I.; Nieminen, T.; Ninchoji, T.; Levonen, A.L.; Kaikkonen, M.U.; Ylä-Herttuala, S. Axon guidance-related factor FLRT3 regulates VEGF-signaling and signaling and endothelial cell function. Front. Physiol. 2019, 10, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, P.S.; Schulz, R.; Maretto, S.; Costello, I.; Srinivas, S.; Bikoff, E.; Robertson, E. The fibronectin leucine-rich repeat transmembrane protein Flrt2 is required in the epicardium to promote heart morphogenesis. Development 2011, 138, 1297–1308. [Google Scholar] [CrossRef] [Green Version]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]

| Gene | Primer Sequence (5′–3′) | Product Size (bp) | |
|---|---|---|---|
| SFRP2 | F | GGCCTCAGGAATGGATAGCT | 167 |
| R | CCCCAAACATCACACCCAAG | ||
| DDAH1 | F | AGCGCGAAGGTATACGAGAA | 238 |
| R | GAAGCGATTAGACTTGGCGG | ||
| LOX | F | GTACAACCTGAGATGCGCTG | 208 |
| R | GCTGAATTCGTCCATGCTGT | ||
| FLRT3 | F | TCGCAACAATCCCTGGTACT | 216 |
| R | ACTGTGTTGGGGATCGAAGT | ||
| PRRX1 | F | GGACACACTACCCAGATGCT | 155 |
| R | TTTGAGGAGGGAAGCGTTCT | ||
| ROBO1 | F | GATGTGATTGCAGACCGACC | 222 |
| R | AGTGTCACCCAGCTTAGCAT | ||
| IL6 | F | ACCGGTCTTGTGGAGTTTCA | 170 |
| R | GCATTTGTGGTGGGGTTAGG | ||
| ACTA2 | F | CCGAGATCTCACCGACTACC | 178 |
| R | CTCGTAGCTCTTCTCCAGGG | ||
| CCL2 | F | CCACACCGAAGCTTGAATCC | 206 |
| R | CTTGCTGCTGGTGACTCTTC | ||
| TNNC1 | F | GAGCTGGGCAAAGTGATGAG | 193 |
| R | ACATGCGGAAGAGGTCAGAA | ||
| MYH7 | F | CCAACACCAACCTGTCCAAG | 173 |
| R | CAGGATGGGGCAGATCAAGA | ||
| NEBL | F | GCACGATCCAGTTTCAGGTC | 163 |
| R | GGCGTTGTCTTTATGGTGCA | ||
| MYL3 | F | TCTTCGACAAGGAGGGCAAT | 191 |
| R | TTTCCTGGGGTGAGAGGTTC | ||
| ACTC1 | F | GTCATGGTGGGTATGGGTCA | 151 |
| R | CGTTGTAGAAGGTGTGGTGC | ||
| NPPA | F | CAGCAGCCTCTATCCTCTCC | 153 |
| R | CCTGTATCCCTGGCAGTTCT | ||
| BMP10 | F | CCTGGGTCTGGGTGGTTATT | 181 |
| R | TGGGGCAATGATCCAAGAGT | ||
| ACTN2 | F | TCGGGGCTGAAGAGATTGTT | 189 |
| R | AGCTGGTGTGGAAGTTCTGA | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawrocki, M.J.; Jopek, K.; Zdun, M.; Mozdziak, P.; Jemielity, M.; Perek, B.; Bukowska, D.; Kempisty, B. Expression Profile of Genes Encoding Proteins Involved in Regulation of Vasculature Development and Heart Muscle Morphogenesis—A Transcriptomic Approach Based on a Porcine Model. Int. J. Mol. Sci. 2021, 22, 8794. https://doi.org/10.3390/ijms22168794
Nawrocki MJ, Jopek K, Zdun M, Mozdziak P, Jemielity M, Perek B, Bukowska D, Kempisty B. Expression Profile of Genes Encoding Proteins Involved in Regulation of Vasculature Development and Heart Muscle Morphogenesis—A Transcriptomic Approach Based on a Porcine Model. International Journal of Molecular Sciences. 2021; 22(16):8794. https://doi.org/10.3390/ijms22168794
Chicago/Turabian StyleNawrocki, Mariusz J., Karol Jopek, Maciej Zdun, Paul Mozdziak, Marek Jemielity, Bartłomiej Perek, Dorota Bukowska, and Bartosz Kempisty. 2021. "Expression Profile of Genes Encoding Proteins Involved in Regulation of Vasculature Development and Heart Muscle Morphogenesis—A Transcriptomic Approach Based on a Porcine Model" International Journal of Molecular Sciences 22, no. 16: 8794. https://doi.org/10.3390/ijms22168794
APA StyleNawrocki, M. J., Jopek, K., Zdun, M., Mozdziak, P., Jemielity, M., Perek, B., Bukowska, D., & Kempisty, B. (2021). Expression Profile of Genes Encoding Proteins Involved in Regulation of Vasculature Development and Heart Muscle Morphogenesis—A Transcriptomic Approach Based on a Porcine Model. International Journal of Molecular Sciences, 22(16), 8794. https://doi.org/10.3390/ijms22168794







