A New Glycosyltransferase Enzyme from Family 91, UGT91P3, Is Responsible for the Final Glucosylation Step of Crocins in Saffron (Crocus sativus L.)
Abstract
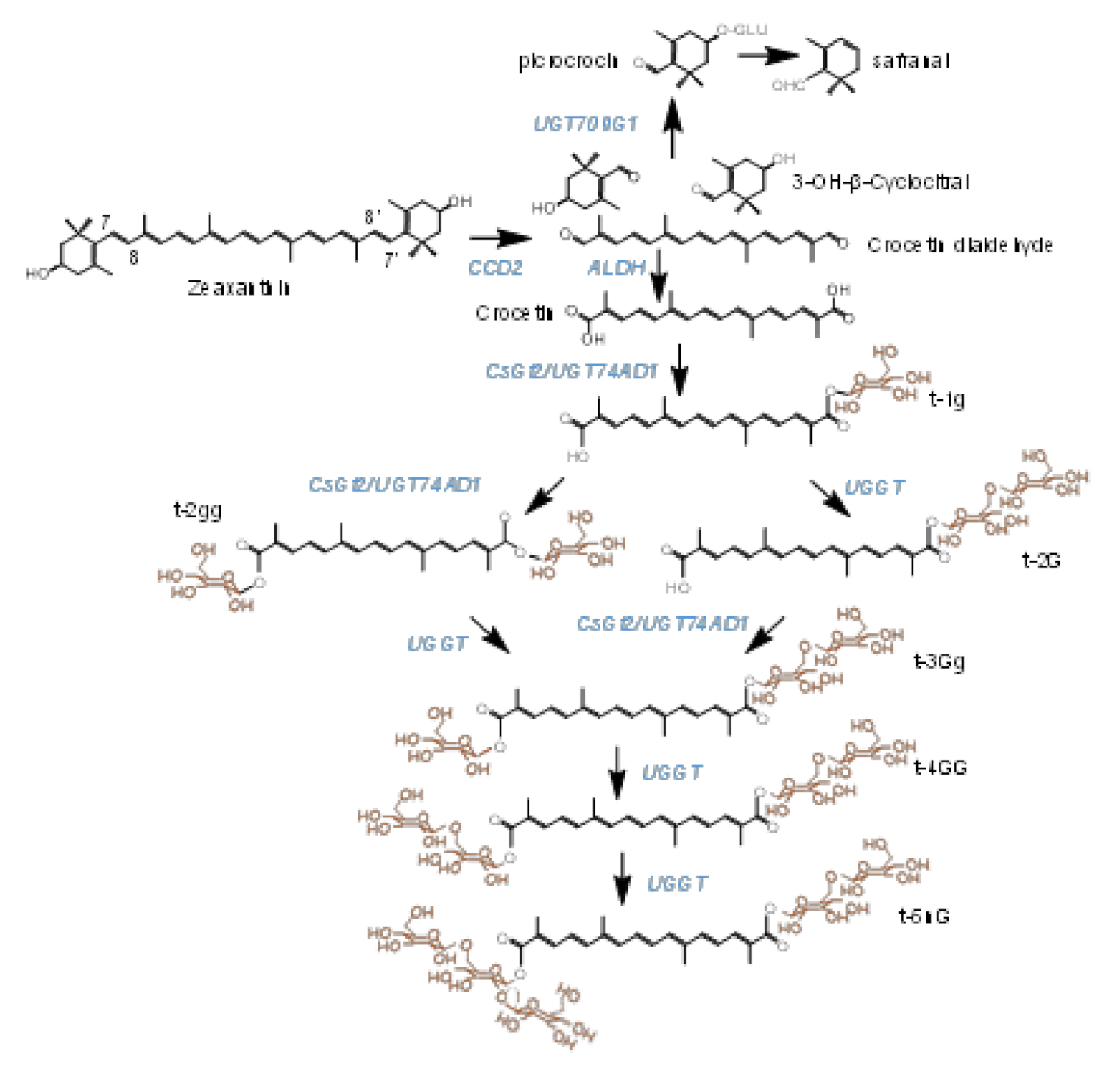
1. Introduction
2. Results
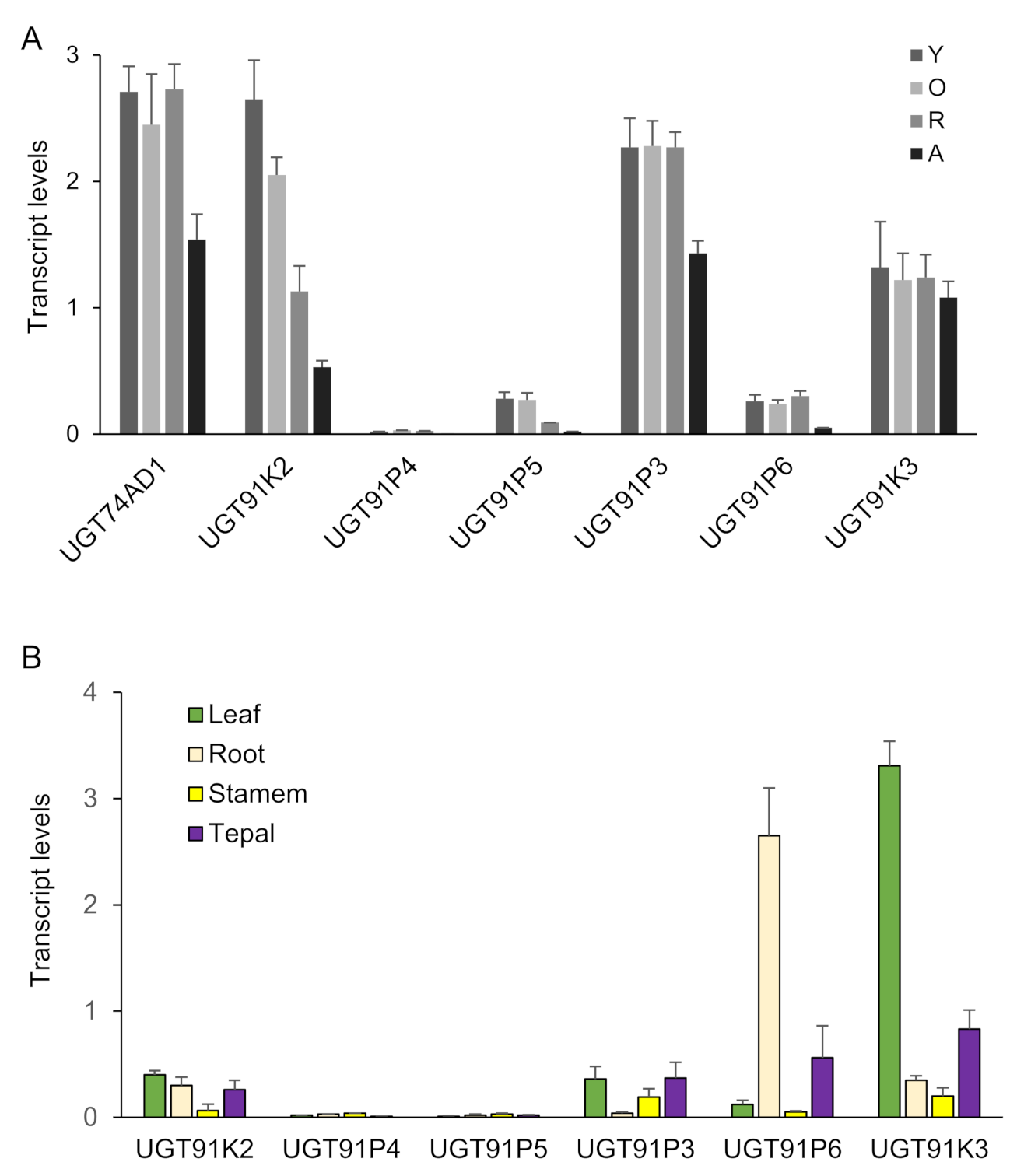
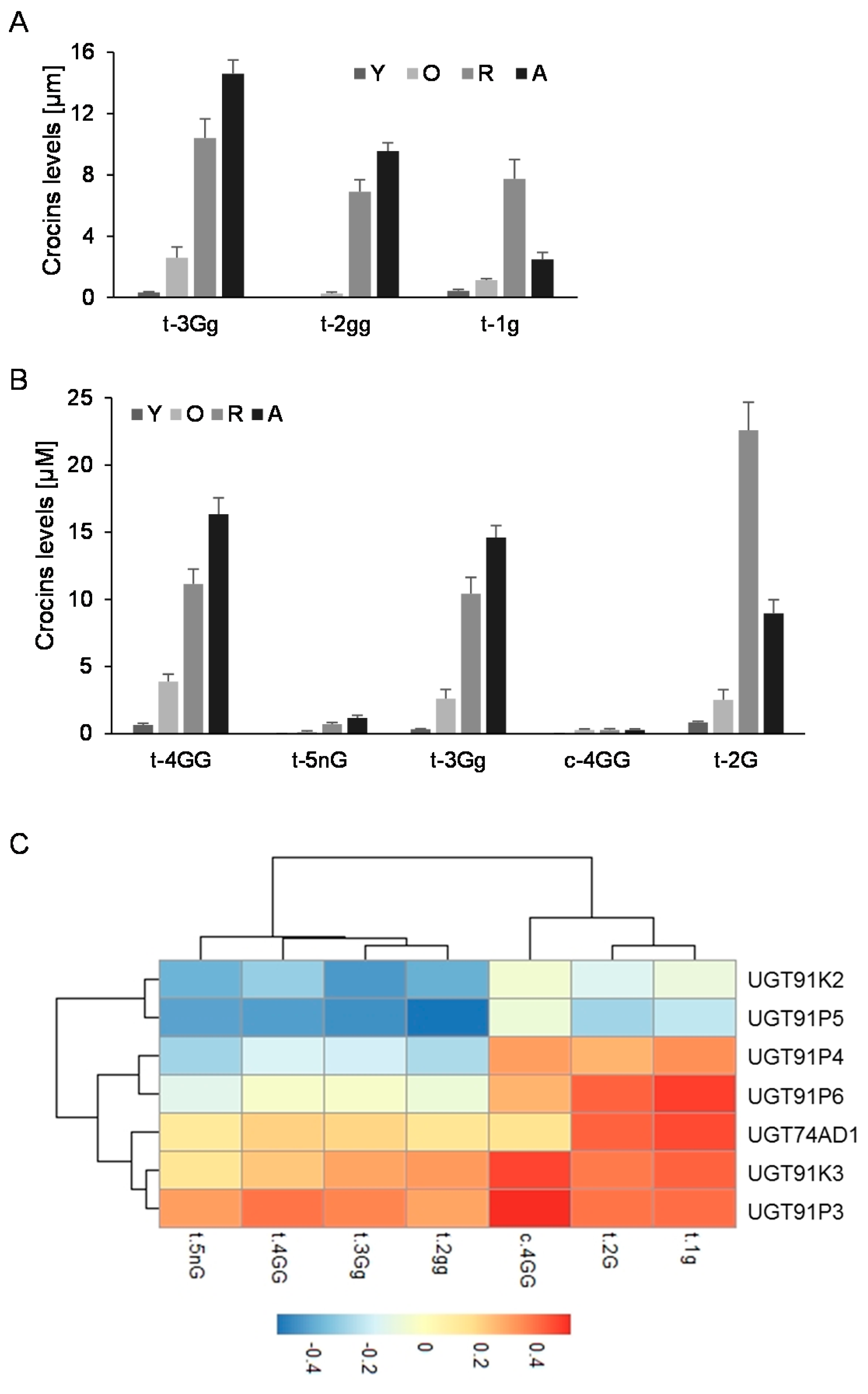
2.1. Dentification of Candidate Glycosyltransferases by Expression Analyses, Crocins Accumulation and Co-Expression with UGT74AD1
2.2. UGT91P3 Catalyzes the Formation of Crocins with Three, Four and Five Glucose Molecules
2.3. Three Alleles of UGT91P3 Are Expressed in Stigmas and Showed Differential Activity
2.4. Interaction between UGT91P3 and UGT74AD1 Revealed by Yeast Two-Hybrid (Y2H) Analysis
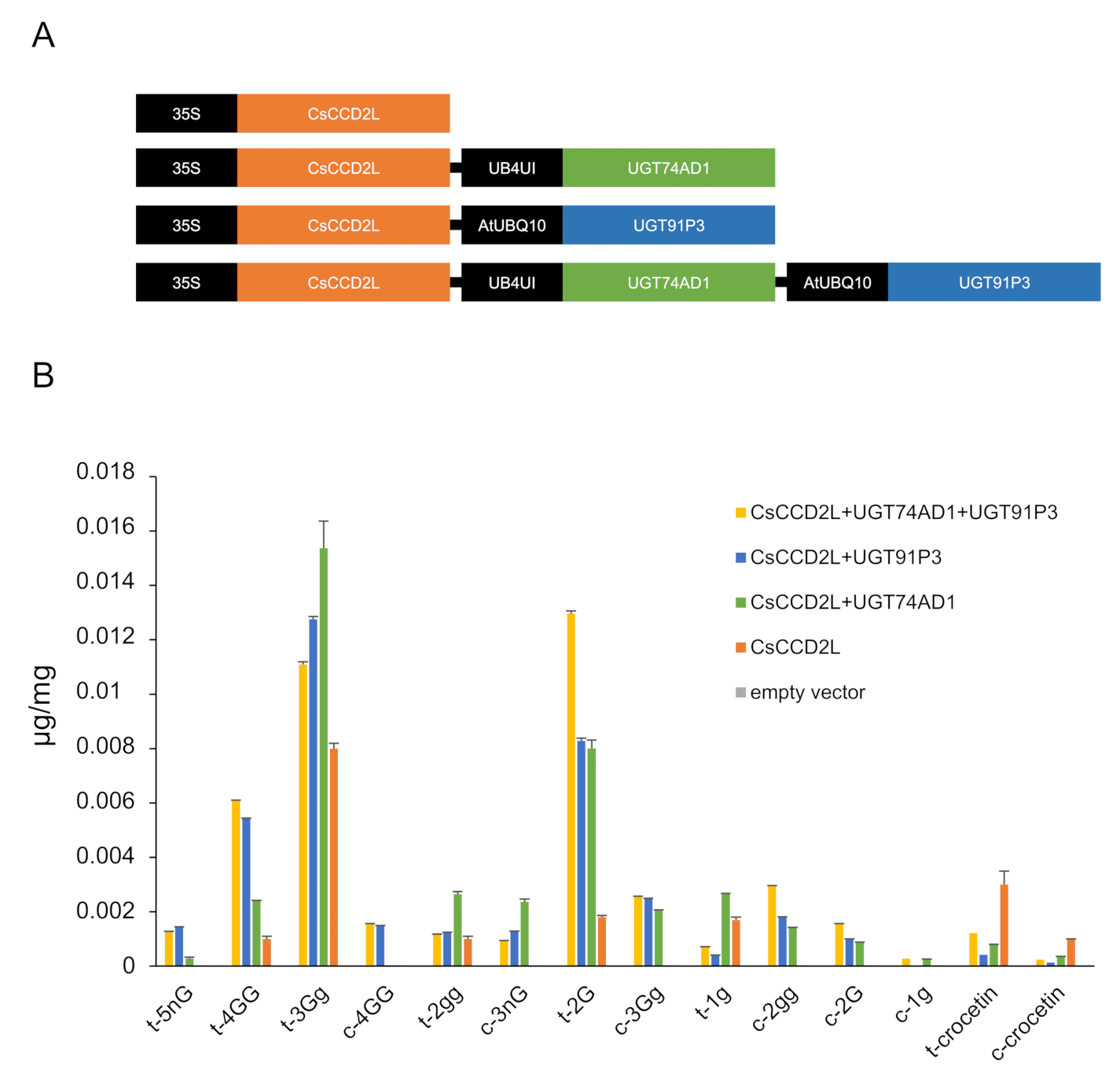
2.5. In Planta Assessment of UGT91P3 and UGT74AD1 Activities
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Chemicals
4.3. Cloning of C. sativus cDNAs
4.4. Protein Sequence and Phylogenetic Analysis
4.5. Topology Alignment
4.6. RNA Isolation and RT-qPCR Analysis
4.7. HPLC-DAD Analysis of Crocins in Saffron Stigmas
4.8. Heterologous Expression of UGT91P3 in E. coli and In Vitro Assays
4.9. Transient Expression in N. benthamiana Leaves
4.10. LC-DAD-HESI-HRMS Analysis of Crocins in UGT Assays
4.11. Yeast Two-Hybrid (Y2H) Analysis
4.12. Data Integration
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahrazem, O.; Diretto, G.; Argandona, J.; Rubio-Moraga, A.; Julve, J.M.; Orzaez, D.; Granell, A.; Gomez-Gomez, L. Evolutionarily distinct carotenoid cleavage dioxygenases are responsible for crocetin production in Buddleja davidii. J. Exp. Bot. 2017, 68, 14. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Nebauer, S.G.; Molina, R.V.; Gomez-Gomez, L. Saffron: Its Phytochemistry, Developmental Processes, and Biotechnological Prospects. J. Agric. Food Chem. 2015, 63, 13. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, E.; Kadoglou, N.P.; Kostomitsopoulos, N.; Valsami, G. Saffron: A natural product with potential pharmaceutical applications. J. Pharm. Pharmacol. 2015, 67, 1634–1649. [Google Scholar] [CrossRef] [PubMed]
- Moraga, A.R.; Nohales, P.F.; Perez, J.A.; Gomez-Gomez, L. Glucosylation of the saffron apocarotenoid crocetin by a glucosyltransferase isolated from Crocus sativus stigmas. Planta 2004, 219, 955–966. [Google Scholar] [CrossRef]
- Rubio-Moraga, A.; Trapero, A.; Ahrazem, O.; Gomez-Gomez, L. Crocins transport in Crocus sativus: The long road from a senescent stigma to a newborn corm. Phytochemistry 2010, 71, 1506–1513. [Google Scholar] [CrossRef]
- Ahrazem, O.; Diretto, G.; Argandona Picazo, J.; Fiore, A.; Rubio-Moraga, A.; Rial, C.; Varela, R.M.; Macias, F.A.; Castillo, R.; Romano, E.; et al. The Specialized Roles in Carotenogenesis and Apocarotenogenesis of the Phytoene Synthase Gene Family in Saffron. Front. Plant Sci. 2019, 10, 249. [Google Scholar] [CrossRef]
- Ahrazem, O.; Gomez-Gomez, L.; Rodrigo, M.J.; Avalos, J.; Limon, M.C. Carotenoid Cleavage Oxygenases from Microbes and Photosynthetic Organisms: Features and Functions. Int. J. Mol. Sci. 2016, 17, 1781. [Google Scholar] [CrossRef]
- Frusciante, S.; Diretto, G.; Bruno, M.; Ferrante, P.; Pietrella, M.; Prado-Cabrero, A.; Rubio-Moraga, A.; Beyer, P.; Gomez-Gomez, L.; Al-Babili, S.; et al. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 12246–12251. [Google Scholar] [CrossRef]
- Ahrazem, O.; Rubio-Moraga, A.; Berman, J.; Capell, T.; Christou, P.; Zhu, C.; Gomez-Gomez, L. The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2016, 209, 13. [Google Scholar] [CrossRef]
- Gomez-Gomez, L.; Pacios, L.F.; Diaz-Perales, A.; Garrido-Arandia, M.; Argandona, J.; Rubio-Moraga, A.; Ahrazem, O. Expression and Interaction Analysis among Saffron ALDHs and Crocetin Dialdehyde. Int. J. Mol. Sci. 2018, 19, 1409. [Google Scholar] [CrossRef]
- Gomez-Gomez, L.; Parra-Vega, V.; Rivas-Sendra, A.; Segui-Simarro, J.M.; Molina, R.V.; Pallotti, C.; Rubio-Moraga, A.; Diretto, G.; Prieto, A.; Ahrazem, O. Unraveling Massive Crocins Transport and Accumulation through Proteome and Microscopy Tools during the Development of Saffron Stigma. Int. J. Mol. Sci. 2017, 18, 76. [Google Scholar] [CrossRef]
- Ahrazem, O.; Argandona, J.; Fiore, A.; Aguado, C.; Lujan, R.; Rubio-Moraga, A.; Marro, M.; Araujo-Andrade, C.; Loza-Alvarez, P.; Diretto, G.; et al. Transcriptome analysis in tissue sectors with contrasting crocins accumulation provides novel insights into apocarotenoid biosynthesis and regulation during chromoplast biogenesis. Sci. Rep. 2018, 8, 2843. [Google Scholar] [CrossRef]
- Dufresne, C.; Cormier, F.; Dorion, S. In vitro formation of crocetin glucosyl esters by Crocus sativus callus extract. Planta Med. 1997, 63, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Jimeno, M.L.; Gomez-Gomez, L. Structural characterization of highly glucosylated crocins and regulation of their biosynthesis during flower development in Crocus. Front. Plant Sci. 2015, 6, 971. [Google Scholar] [CrossRef] [PubMed]
- Pfister, S.; Steck, A.; Pfander, H. Isolation and structure elucidation of carotenoid glycoslyesters on gardenia fruits (Gardenia jasminoides) and saffron (Crocus sativus). J. Agric. Food Chem. 1996, 44, 1. [Google Scholar] [CrossRef]
- Nagatoshi, M.; Terasaka, K.; Owaki, M.; Sota, M.; Inukai, T.; Nagatsu, A.; Mizukami, H. UGT75L6 and UGT94E5 mediate sequential glucosylation of crocetin to crocin in Gardenia jasminoides. FEBS Lett. 2012, 586, 1055–1061. [Google Scholar] [CrossRef]
- Xu, Z.; Pu, X.; Gao, R.; Demurtas, O.C.; Fleck, S.J.; Richter, M.; He, C.; Ji, A.; Sun, W.; Kong, J.; et al. Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. 2020, 18, 63. [Google Scholar] [CrossRef]
- Diretto, G.; López-Jiménez, A.J.; Ahrazem, O.; Frusciante, S.; Song, J.; Rubio-Moraga, Á.; Gómez-Gómez, L. Identification and characterization of apocarotenoid modifiers and carotenogenic enzymes for biosynthesis of crocins in Buddleja davidii flowers. J. Exp. Bot. 2021, 72, 3200–3218. [Google Scholar] [CrossRef]
- De Bruyn, F.; Maertens, J.; Beauprez, J.; Soetaert, W.; De Mey, M. Biotechnological advances in UDP-sugar based glycosylation of small molecules. Biotechnol. Adv. 2015, 33, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.; Lim, E.K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef]
- Tiwari, P.; Sangwan, R.S.; Sangwan, N.S. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 2016, 34, 714–739. [Google Scholar] [CrossRef]
- Lim, E.-K.; Bowles, D.J. A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 2004, 23, 2915–2922. [Google Scholar] [CrossRef]
- Osmani, S.A.; Bak, S.; Moller, B.L. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 2009, 70, 325–347. [Google Scholar] [CrossRef]
- Song, C.; Gu, L.; Liu, J.; Zhao, S.; Hong, X.; Schulenburg, K.; Schwab, W. Functional Characterization and Substrate Promiscuity of UGT71 Glycosyltransferases from Strawberry (Fragaria × ananassa). Plant Cell Physiol. 2015, 56, 2478–2493. [Google Scholar] [CrossRef]
- Meech, R.; Hu, D.G.; McKinnon, R.A.; Mubarokah, S.N.; Haines, A.Z.; Nair, P.C.; Rowland, A.; Mackenzie, P.I. The UDP-Glycosyltransferase (UGT) Superfamily: New Members, New Functions, and Novel Paradigms. Physiol. Rev. 2019, 99, 1153–1222. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.-C.; Giri, A.; Daniilidis, M.; Sun, G.; Härtl, K.; Hoffmann, T.; Schwab, W. Structural and Functional Analysis of UGT92G6 Suggests an Evolutionary Link Between Mono- and Disaccharide Glycoside-Forming Transferases. Plant Cell Physiol. 2018, 59, 862–875. [Google Scholar] [CrossRef]
- Moraga, A.R.; Rambla, J.L.; Ahrazem, O.; Granell, A.; Gomez-Gomez, L. Metabolite and target transcript analyses during Crocus sativus stigma development. Phytochemistry 2009, 70, 1009–1016. [Google Scholar] [CrossRef]
- Tripathi, S.; Jadaun, J.S.; Chandra, M.; Sangwan, N.S. Medicinal plant transcriptomes: The new gateways for accelerated understanding of plant secondary metabolism. Plant Genet. Resour. 2016, 14, 256–269. [Google Scholar] [CrossRef]
- Ahrazem, O.; Rubio-Moraga, A.; Argandona-Picazo, J.; Castillo, R.; Gomez-Gomez, L. Intron retention and rhythmic diel pattern regulation of carotenoid cleavage dioxygenase 2 during crocetin biosynthesis in saffron. Plant Mol. Biol. 2016, 91, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Argandona, J.; Fiore, A.; Rujas, A.; Rubio-Moraga, A.; Castillo, R.; Gomez-Gomez, L. Multi-species transcriptome analyses for the regulation of crocins biosynthesis in Crocus. BMC Genom. 2019, 20, 320. [Google Scholar] [CrossRef] [PubMed]
- Bourne, Y.; Henrissat, B. Glycoside hydrolases and glycosyltransferases: Families and functional modules. Curr. Opin. Struct. Biol. 2001, 11, 593–600. [Google Scholar] [CrossRef]
- Itkin, M.; Davidovich-Rikanati, R.; Cohen, S.; Portnoy, V.; Doron-Faigenboim, A.; Oren, E.; Freilich, S.; Tzuri, G.; Baranes, N.; Shen, S.; et al. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proc. Natl. Acad. Sci. USA 2016, 113, E7619–E7628. [Google Scholar] [CrossRef]
- Diretto, G.; Ahrazem, O.; Rubio-Moraga, A.; Fiore, A.; Sevi, F.; Argandona, J.; Gomez-Gomez, L. UGT709G1: A novel uridine diphosphate glycosyltransferase involved in the biosynthesis of picrocrocin, the precursor of safranal in saffron (Crocus sativus). New Phytol. 2019, 224, 725–740. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.A.; Harborne, J.B.; Goldblatt, P. Correlations between phenolic patterns and tribal classification in the family iridaceae. Phytochemistry 1986, 25, 2135–2154. [Google Scholar] [CrossRef]
- Mykhailenko, O.; Ivanauskas, L.; Bezruk, I.; Sidorenko, L.; Lesyk, R.; Georgiyants, V. Characterization of Phytochemical Components of Crocus sativus Leaves: A New Attractive By-Product. Sci. Pharm. 2021, 89, 28. [Google Scholar] [CrossRef]
- Petit, E.; Berger, M.; Camborde, L.; Vallejo, V.; Daydé, J.; Jacques, A. Development of screening methods for functional characterization of UGTs from Stevia rebaudiana. Sci. Rep. 2020, 10, 15137. [Google Scholar] [CrossRef]
- Martí, M.; Diretto, G.; Aragonés, V.; Frusciante, S.; Ahrazem, O.; Gómez-Gómez, L.; Daròs, J.-A. Efficient production of saffron crocins and picrocrocin in Nicotiana benthamiana using a virus-driven system. Metab. Eng. 2020, 61, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.-K. The evolutionary paths towards complexity: A metabolic perspective. New Phytol. 2014, 201, 1141–1149. [Google Scholar] [CrossRef]
- Fujiwara, R.; Yokoi, T.; Nakajima, M. Structure and Protein-Protein Interactions of Human UDP-Glucuronosyltransferases. Front. Pharmacol. 2016, 7, 388. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.E.; Tian, L. Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 2019, 100, 1273–1288. [Google Scholar] [CrossRef]
- Durek, P.; Walther, D. The integrated analysis of metabolic and protein interaction networks reveals novel molecular organizing principles. BMC Syst. Biol. 2008, 2, 100. [Google Scholar] [CrossRef]
- Ovádi, J.; Srere, P.A. Macromolecular compartmentation and channeling. Int. Rev. Cytol. 2000, 192, 255–280. [Google Scholar] [PubMed]
- Demurtas, O.C.; Frusciante, S.; Ferrante, P.; Diretto, G.; Azad, N.H.; Pietrella, M.; Aprea, G.; Taddei, A.R.; Romano, E.; Mi, J.; et al. Candidate Enzymes for Saffron Crocin Biosynthesis Are Localized in Multiple Cellular Compartments. Plant Physiol. 2018, 177, 990–1006. [Google Scholar] [CrossRef]
- Baba, S.A.; Mohiuddin, T.; Basu, S.; Swarnkar, M.K.; Malik, A.H.; Wani, Z.A.; Abbas, N.; Singh, A.K.; Ashraf, N. Comprehensive transcriptome analysis of Crocus sativus for discovery and expression of genes involved in apocarotenoid biosynthesis. BMC Genom. 2015, 16, 698. [Google Scholar] [CrossRef]
- Rubio, A.; Rambla, J.L.; Santaella, M.; Gomez, M.D.; Orzaez, D.; Granell, A.; Gomez-Gomez, L. Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in beta-ionone release. J. Biol. Chem. 2008, 283, 24816–24825. [Google Scholar] [CrossRef] [PubMed]
- Rubio Moraga, A.; Ahrazem, O.; Rambla, J.L.; Granell, A.; Gomez Gomez, L. Crocins with high levels of sugar conjugation contribute to the yellow colours of early-spring flowering crocus tepals. PLoS ONE 2013, 8, e71946. [Google Scholar]
- Hu, J.; Liu, Y.; Tang, X.; Rao, H.; Ren, C.; Chen, J.; Wu, Q.; Jiang, Y.; Geng, F.; Pei, J. Transcriptome profiling of the flowering transition in saffron (Crocus sativus L.). Sci. Rep. 2020, 10, 9680. [Google Scholar] [CrossRef] [PubMed]
- Carmona, M.; Zalacain, A.; Sanchez, A.M.; Novella, J.L.; Alonso, G.L. Crocetin esters, picrocrocin and its related compounds present in Crocus sativus stigmas and Gardenia jasminoides fruits. Tentative identification of seven new compounds by LC-ESI-MS. J. Agric. Food Chem. 2006, 54, 973–979. [Google Scholar] [CrossRef]
- Himeno, H.; Sano, K. Synthesis of crocin, picrocrocin and safranal by saffron stigma-like structures proliferated in vitro. Agric. Biol. Chem. 1987, 51, 5. [Google Scholar] [CrossRef]
- Alder, A.; Holdermann, I.; Beyer, P.; Al-Babili, S. Carotenoid oxygenases involved in plant branching catalyse a highly specific conserved apocarotenoid cleavage reaction. Biochem. J. 2008, 416, 289–296. [Google Scholar] [CrossRef][Green Version]
- Sarrion-Perdigones, A.; Vazquez-Vilar, M.; Palaci, J.; Castelijns, B.; Forment, J.; Ziarsolo, P.; Blanca, J.; Granell, A.; Orzaez, D. GoldenBraid 2.0: A comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 2013, 162, 1618–1631. [Google Scholar] [CrossRef] [PubMed]


| Name | RT | M/Z |
|---|---|---|
| t-4GG | 11.17 | 977.38590 [M + H], 994.41250 [M + NH4] |
| t-3Gg | 12.05 | 815.33310 [M + H], 832.35970 [M + NH4] |
| t-2gg | 13.14 | 653.28030 [M + H], 670.30690 [M + NH4] |
| c-5nG | 13.91 | 1156.46530 [M + H] |
| c-4GG | 14.50 | 977.38590 [M + H], 994.41250 [M + NH4] |
| t-2gg | 15.05 | 653.28030 [M + H], 670.30690 [M + NH4] |
| c-3Gg | 15.55 | 815.33310 [M + H], 832.35970 [M + NH4] |
| t-1g | 16.75 | 491.22750 [M + H], 508.25410 [M + NH4] |
| t-1g | 20.48 | 491.22750 [M + H], 508.25410 [M + NH4] |
| t-crocetin | 19.93 | 329.17470 [M + H] |
| c-crocetin | 22.49 | 329.17470 [M + H] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-jimenez, A.J.; Frusciante, S.; Niza, E.; Ahrazem, O.; Rubio-Moraga, Á.; Diretto, G.; Gómez-Gómez, L. A New Glycosyltransferase Enzyme from Family 91, UGT91P3, Is Responsible for the Final Glucosylation Step of Crocins in Saffron (Crocus sativus L.). Int. J. Mol. Sci. 2021, 22, 8815. https://doi.org/10.3390/ijms22168815
López-jimenez AJ, Frusciante S, Niza E, Ahrazem O, Rubio-Moraga Á, Diretto G, Gómez-Gómez L. A New Glycosyltransferase Enzyme from Family 91, UGT91P3, Is Responsible for the Final Glucosylation Step of Crocins in Saffron (Crocus sativus L.). International Journal of Molecular Sciences. 2021; 22(16):8815. https://doi.org/10.3390/ijms22168815
Chicago/Turabian StyleLópez-jimenez, Alberto José, Sarah Frusciante, Enrique Niza, Oussama Ahrazem, Ángela Rubio-Moraga, Gianfranco Diretto, and Lourdes Gómez-Gómez. 2021. "A New Glycosyltransferase Enzyme from Family 91, UGT91P3, Is Responsible for the Final Glucosylation Step of Crocins in Saffron (Crocus sativus L.)" International Journal of Molecular Sciences 22, no. 16: 8815. https://doi.org/10.3390/ijms22168815
APA StyleLópez-jimenez, A. J., Frusciante, S., Niza, E., Ahrazem, O., Rubio-Moraga, Á., Diretto, G., & Gómez-Gómez, L. (2021). A New Glycosyltransferase Enzyme from Family 91, UGT91P3, Is Responsible for the Final Glucosylation Step of Crocins in Saffron (Crocus sativus L.). International Journal of Molecular Sciences, 22(16), 8815. https://doi.org/10.3390/ijms22168815










