Analysis of CGF Biomolecules, Structure and Cell Population: Characterization of the Stemness Features of CGF Cells and Osteogenic Potential
Abstract
:1. Introduction
2. Results
2.1. Untargeted and Targeted GC/MS Metabolomic Analysis of CGF
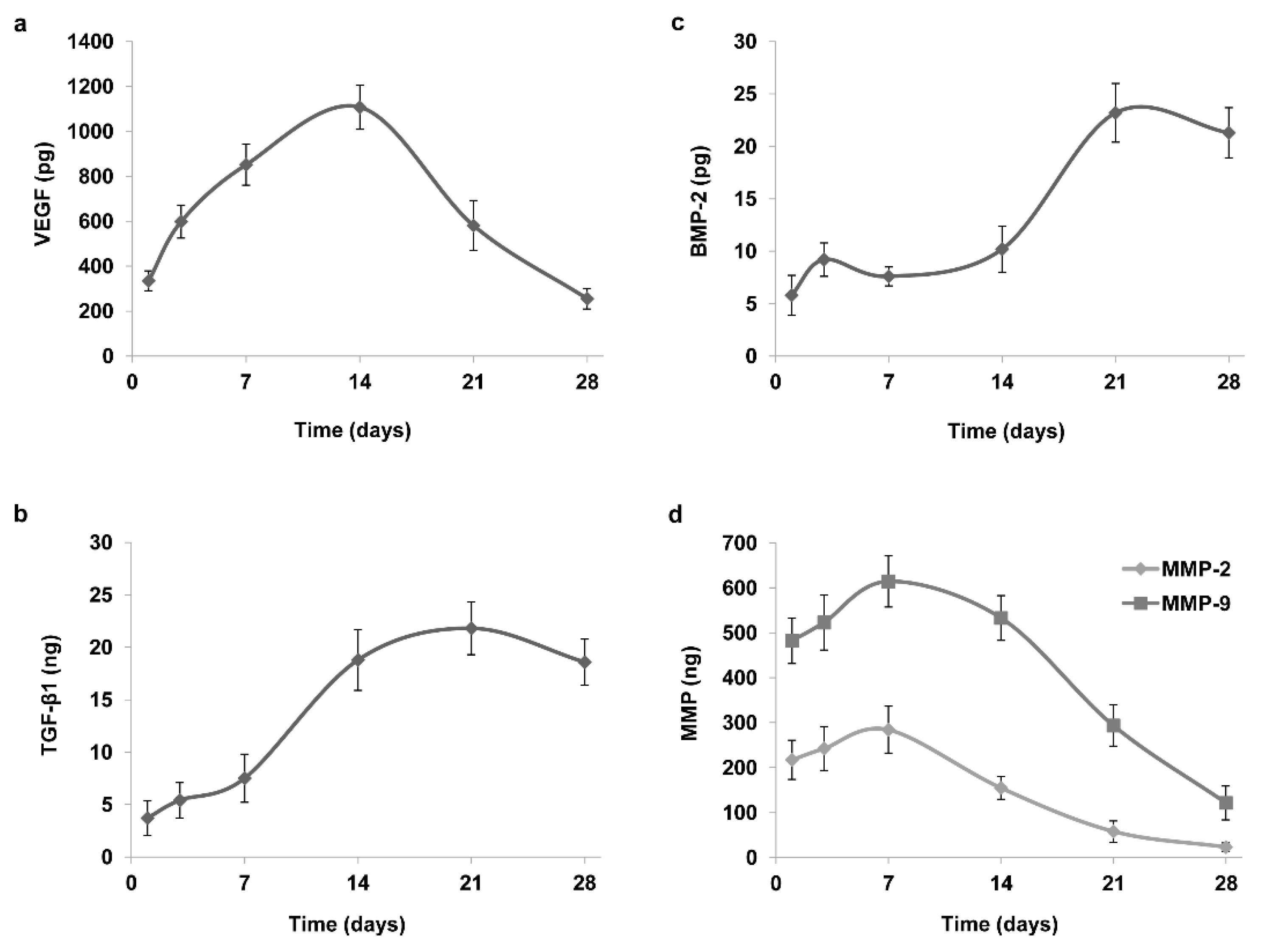
2.2. Analysis of CGF Content and Release of Growth Factors and Matrix Metalloproteinases
2.3. CGF: Fibrin and Cellular Components
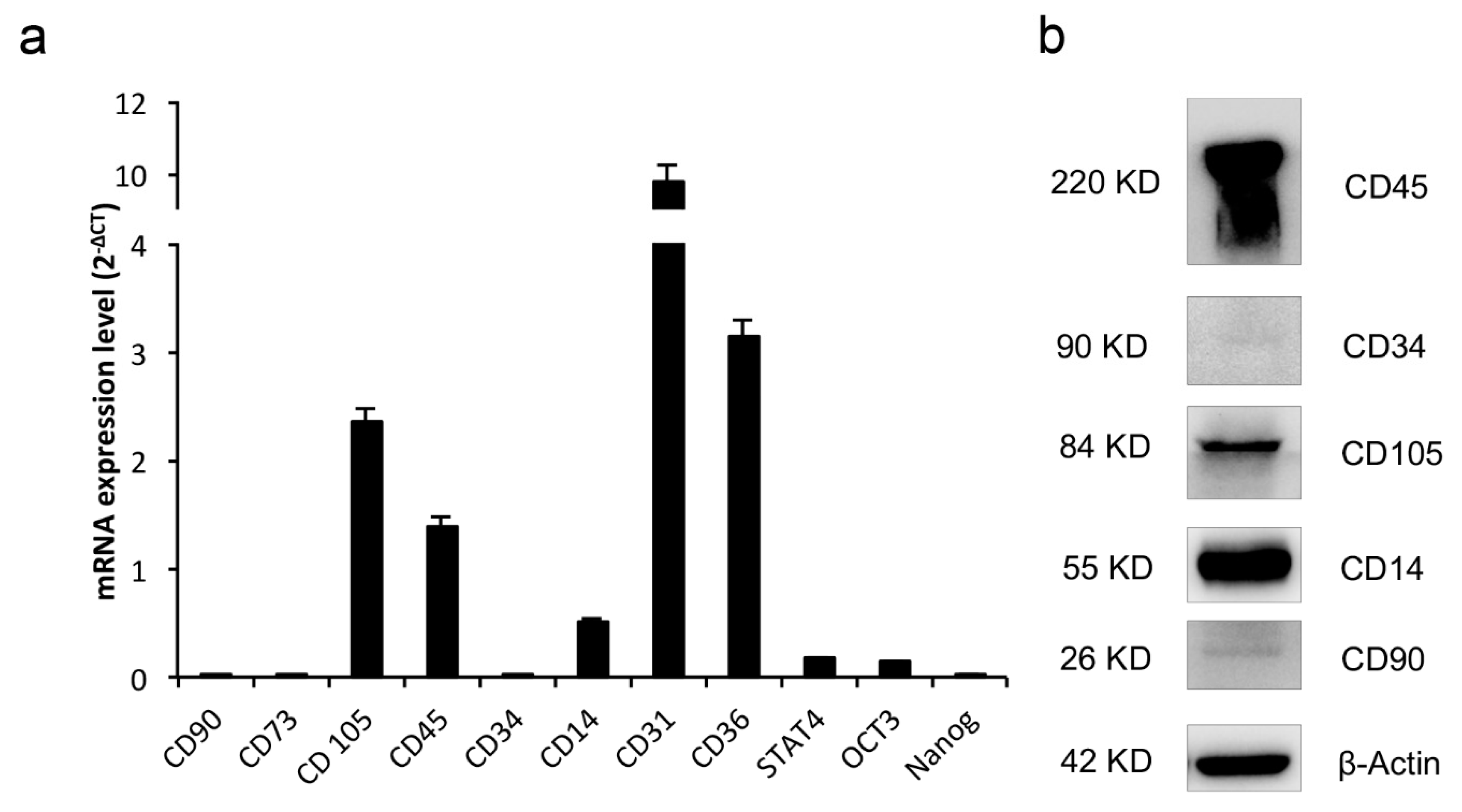
2.4. CGF Cells Display Pluripotency and Stem Cell Markers
2.5. Osteogenic Differentiation of CGF Primary Cells
3. Discussion
4. Materials and Methods
4.1. Preparation of CGF
4.2. GC/MS Analysis
4.3. Growth Factors and MMPs Content and Release
4.4. SEM Analysis
4.5. Immunohistochemistry and Cell Count
4.6. Isolation and Culture of Primary Cell Populations from CGF
4.7. Cell Surface Marker Analysis by Flow Cytometry
4.8. Proliferation Assay
4.9. Western-Blot Analysis
4.10. Real-Time PCR
4.11. Osteogenic Differentiation Protocol
4.12. Alizarin Red Staining
4.13. Statistical Analysis
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Rodella, L.F.; Favero, G.; Boninsegna, R.; Buffoli, B.; Labanca, M.; Scarì, G.; Sacco, L.; Batani, T.; Rezzani, R. Growth factors, CD34 positive cells, and fibrin network analysis in concentrated growth factors fraction. Microsc. Res. Tech. 2011, 74, 772–777. [Google Scholar] [CrossRef]
- Masoudi, E.; Ribas, J.; Kaushik, G.; Leijten, J.; Khademhosseini, A. Platelet-Rich Blood Derivatives for Stem Cell-Based Tissue Engineering and Regeneration. Curr. Stem. Cell Rep. 2016, 2, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Sun, X.; Yu, J.; Wang, J.; Zhai, P.; Chen, S.; Liu, M.; Zhou, Y. Platelet-Rich Fibrin as a Bone Graft Material in Oral and Maxillofacial Bone Regeneration: Classification and Summary for Better Application. BioMed Res. Int. 2019, 2019, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, E.; Flückiger, L.; Fujioka-Kobayashi, M.; Sawada, K.; Sculean, A.; Schaller, B.; Miron, R.J. Comparative release of growth factors from PRP, PRF, and advanced-PRF. Clin. Oral. Investig. 2016, 20, 2353–2360. [Google Scholar] [CrossRef] [PubMed]
- Masuki, H.; Okudera, T.; Watanebe, T.; Suzuki, M.; Nishiyama, K.; Okudera, H.; Nakata, K.; Uematsu, K.; Su, C.Y.; Kawase, T. Growth factor and pro-inflammatory cytokine contents in platelet-rich plasma (PRP), plasma rich in growth factors (PRGF), advanced platelet-rich fibrin (A-PRF), and concentrated growth factors (CGF). Int. J. Implant Dent. 2016, 2, 19. [Google Scholar] [CrossRef] [Green Version]
- Sacco, L. Lecture, International Academy of implant prosthesis and osteoconnection. Lecture 2006, 12, 4. [Google Scholar]
- Sohn, D.S.; Heo, J.U.; Kwak, D.H.; Kim, D.E.; Kim, J.M.; Moon, J.W.; Lee, J.H.; Park, I.S. Bone regeneration in the maxillary sinus using an autologous fibrin-rich block with concentrated growth factors alone. Implant Dent. 2011, 20, 389–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schär, M.O.; Diaz-Romero, J.; Kohl, S.; Zumstein, M.A.; Nesic, D. Platelet-rich concentrates differentially release growth factors and induce cell migration in vitro. Clin. Orthop. Relat. Res. 2015, 473, 1635–1643. [Google Scholar] [CrossRef] [Green Version]
- Borsani, E.; Bonazza, V.; Buffoli, B.; Cocchi, M.A.; Castrezzati, S.; Scarì, G.; Baldi, F.; Pandini, S.; Licenziati, S.; Parolini, S.; et al. Biological Characterization and In Vitro Effects of Human Concentrated Growth Factor Preparation: An Innovative Approach to Tissue Regeneration. Biol. Med. 2015, 7, 256. [Google Scholar] [CrossRef] [Green Version]
- Ratiu, C.; Brocks, M.; Costea, T.; Moldovan, L.; Cavalu, S. PRGF-Modified Collagen Membranes for Guided Bone Regeneration: Spectroscopic, Microscopic and Nano-Mechanical Investigations. Appl. Sci. 2019, 9, 1035. [Google Scholar] [CrossRef] [Green Version]
- Rochira, A.; Siculella, L.; Damiano, F.; Palermo, A.; Ferrante, F.; Carluccio, M.A.; Calabriso, N.; Giannotti, L.; Stanca, E. Concentrated Growth Factors (CGF) Induce Osteogenic Differentiation in Human Bone Marrow Stem Cells. Biology 2020, 9, 370. [Google Scholar] [CrossRef]
- Tabatabaei, F.; Aghamohammadi, Z.; Tayebi, L. In vitro and in vivo effects of concentrated growth factor on cells and tissues. J. Biomed. Mater. Res. A 2020, 108, 1338–1350. [Google Scholar] [CrossRef] [PubMed]
- Park, H.C.; Kim, S.G.; Oh, J.S.; You, J.S.; Kim, J.S.; Lim, S.C.; Jeong, M.A.; Kim, J.S.; Jung, C.; Kwon, Y.S.; et al. Early Bone Formation at a Femur Defect Using CGF and PRF Grafts in Adult Dogs: A Comparative Study. Implant Dent. 2016, 25, 387–393. [Google Scholar] [CrossRef]
- Anghelina, M.; Krishnan, P.; Moldovan, L.; Moldovan, N.I. Monocytes/macrophages cooperate with progenitor cells during neovascularization and tissue repair. Am. J. Pathol. 2006, 168, 529–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manole, E.; Niculite, C.; Lambrescu, I.M.; Gaina, G.; Ioghen, O.; Ceafalan, L.C.; Hinescu, M.E. Macrophages and Stem Cells-Two to Tango for Tissue Repair? Biomolecules 2021, 11, 697. [Google Scholar] [CrossRef]
- Lin, W.; Xu, L.; Lin, S.; Shi, L.; Wang, B.; Pan, Q.; Lee, W.Y.W.; Li, G. Characterisation of multipotent stem cells from human peripheral blood using an improved protocol. J. Orthop. Transl. 2019, 19, 18–28. [Google Scholar] [CrossRef]
- Cesselli, D.; Beltrami, A.P.; Rigo, S.; Bergamin, N.; D’Aurizio, F.; Verardo, R.; Piazza, S.; Klaric, E.; Fanin, R.; Toffoletto, B.; et al. Multipotent progenitor cells are present in human peripheral blood. Circ. Res. 2009, 104, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.; Pinto, J.; Araújo, A.M.; Jerónimo, C.; Henrique, R.; Bastos, M.L.; Guedes de Pinho, P.; Carvalho, M. GC-MS Metabolomics Reveals Distinct Profiles of Low- and High-Grade Bladder Cancer Cultured Cells. Metabolites 2019, 9, 18. [Google Scholar] [CrossRef] [Green Version]
- Jin, R.; Song, G.; Chai, J.; Gou, X.; Yuan, G.; Chen, Z. Effects of concentrated growth factor on proliferation, migration, and differentiation of human dental pulp stem cells in vitro. J. Tissue Eng. 2018, 9, 2041731418817505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ai, H. Concentrated growth factor promotes proliferation, osteogenic differentiation, and angiogenic potential of rabbit periosteum-derived cells in vitro. J. Orthop. Surg. Res. 2019, 14, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Jiang, H. A Comprehensive Review of Concentrated Growth Factors and Their Novel Applications in Facial Reconstructive and Regenerative Medicine. Aesthetic Plast. Surg. 2020, 44, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Calabriso, N.; Stanca, E.; Rochira, A.; Damiano, F.; Giannotti, L.; Di Chiara Stanca, B.; Massaro, M.; Scoditti, E.; Demitri, C.; Nitti, P.; et al. Angiogenic Properties of Concentrated Growth Factors (CGFs): The Role of Soluble Factors and Cellular Components. Pharmaceutics 2021, 13, 635. [Google Scholar] [CrossRef]
- Everts, P.A.; Knape, J.T.; Weibrich, G.; Schönberger, J.P.; Hoffmann, J.; Overdevest, E.P.; Box, H.A.; van Zundert, A. Platelet-rich plasma and platelet gel: A review. J. Extra Corpor. Technol. 2006, 38, 174–187. [Google Scholar]
- Karaman, O.; Kumar, A.; Moeinzadeh, S.; He, X.; Cui, T.; Jabbari, E. Effect of surface modification of nanofibres with glutamic acid peptide on calcium phosphate nucleation and osteogenic differentiation of marrow stromal cells. J. Tissue Eng. Regen. Med. 2013, 10, E132–E146. [Google Scholar] [CrossRef] [PubMed]
- Onak, G.; Şen, M.; Horzum, N.; Ercan, U.K.; Yaralı, Z.B.; Garipcan, B.; Karaman, O. Aspartic and Glutamic Acid Templated Peptides Conjugation on Plasma Modified Nanofibers for Osteogenic Differentiation of Human Mesenchymal Stem Cells: A Comparative Study. Sci. Rep. 2018, 8, 17620. [Google Scholar] [CrossRef] [Green Version]
- Prideaux, M.; Kitase, Y.; Kimble, M.; O’Connell, T.M.; Bonewald, L.F. Taurine, an osteocyte metabolite, protects against oxidative stress-induced cell death and decreases inhibitors of the Wnt/β-catenin signaling pathway. Bone 2020, 137, 115374. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhang, X.; Xu, L.; Wu, T.; Cui, L.; Xu, D. Taurine promotes human mesenchymal stem cells to differentiate into osteoblast through the ERK pathway. Amino Acids 2014, 46, 1673–1680. [Google Scholar] [CrossRef]
- Bonazza, V.; Hajistilly, C.; Patel, D.; Patel, J.; Woo, R.; Cocchi, M.A.; Buffoli, B.; Lancini, D.; Gheno, E.; Rezzani, R.; et al. Growth Factors Release from Concentrated Growth Factors: Effect of β-Tricalcium Phosphate Addition. J. Craniofac. Surg. 2018, 29, 2291–2295. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zou, R.; He, J.; Ouyang, K.; Piao, Z. Comparing osteogenic effects between concentrated growth factors and the acellular dermal matrix. Braz. Oral. Res. 2018, 32, e29. [Google Scholar] [CrossRef]
- Dohan Ehrenfest, D.M.; Pinto, N.R.; Pereda, A.; Jime’nez, P.; Corso, M.D.; Kang, B.S.; Nally, M.; Lanata, N.; Wang, H.L.; Quirynen, M. The impact of the centrifuge characteristics and centrifugation protocols on the cells, growth factors, and fibrin architecture of a leukocyte- and platelet-rich fibrin (L-PRF) clot and membrane. Platelets 2018, 29, 171–184. [Google Scholar] [CrossRef]
- Qin, J.; Wang, L.; Zheng, L.; Zhou, X.; Zhang, Y.; Yang, T.; Zhou, Y. Concentrated growth factor promotes Schwann cell migration partly through the integrin beta1-mediated activation of the focal adhesion kinase pathway. Int. J. Mol. Med. 2016, 37, 1363–1370. [Google Scholar] [CrossRef] [Green Version]
- Honda, H.; Tamai, N.; Naka, N.; Yoshikawa, H.; Myoui, A. Bone tissue engineering with bone marrow-derived stromal cells integrated with concentrated growth factor in Rattus norvegicus calvaria defect model. J. Artif. Organs 2013, 16, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wan, M.; Li, Z.; Zhong, N.; Liang, D.; Ge, L. A comparative study of the effects of concentrated growth factors in two different forms on osteogenesis in vitro. Mol. Med. Rep. 2019, 20, 1039–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isobe, K.; Watanebe, T.; Kawabata, H.; Kitamura, Y.; Kawase, T. Mechanical and degradation properties of advanced platelet-rich fibrin (APRF), concentrated growth factors (CGF), and platelet-poor plasma-derived fibrin (PPTF). Int. J. Implant Dent. 2017, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, X.; Liu, Y.; Qiao, J. Cytokine release kinetics of concentrated growth factors in different scaffolds. Clin. Oral. Investig. 2018, 23, 1663–1671. [Google Scholar] [CrossRef]
- Lei, L.; Yu, Y.; Han, J.; Shi, D.; Sun, W.; Zhang, D.; Chen, L. Quantification of growth factors in advanced platelet-rich fibrin and concentrated growth factors and their clinical efficacy as adjunctive to the GTR procedure in periodontal intrabony defects. J. Periodontol. 2020, 91, 462–472. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.-P. The Role of Vascular Endothelial Growth Factor in Angiogenesis. Acta Haematol. 2001, 106, 148–156. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef]
- Lazarous, D.F.; Shou, M.; Scheinowitz, M.; Hodge, E.; Thirumurti, V.; Kitsiou, A.N.; Stiber, J.A.; Lobo, A.D.; Hunsberger, S.; Guetta, E.; et al. Comparative Effects of Basic Fibroblast Growth Factor and Vascular Endothelial Growth Factor on Coronary Collateral Development and the Arterial Response to Injury. Circulation 1996, 94, 1074–1082. [Google Scholar] [CrossRef]
- Niu, G.; Chen, X. Vascular endothelial growth factor as an anti-angiogenic target for cancer therapy. Curr. Drug Targets 2010, 11, 1000–1017. [Google Scholar] [CrossRef]
- Lee, K.; Silva, E.A.; Mooney, D.J. Growth factor delivery-based tissue engineering: General approaches and a review of recent developments. J. R. Soc. Interface 2011, 8, 153–170. [Google Scholar] [CrossRef] [Green Version]
- Massagué, J.; Xi, Q. TGF-β control of stem cell differentiation genes. FEBS Lett. 2012, 586, 1953–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.O.; Flavell, R.A. TGF-β: A Master of All T Cell Trades. Cell 2008, 134, 392–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, T.; Wei, L.; Qiao, L.; Qiu, J.; Zha, D. Does bone morphogenetic proteins play an important role in chronic rhinosinusitis? Med. Hypotheses 2009, 72, 228. [Google Scholar] [CrossRef] [PubMed]
- Kalén, A.; Wahlström, O.; Linder, C.H.; Magnusson, P. The content of bone morphogenetic proteins in platelets varies greatly between different platelet donors. Biochem. Biophys. Res. Commun. 2008, 375, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lai, Q.; Li, Y.; Xu, C.; Tang, X.; Ci, J.; Sun, S.; Xu, B.; Li, Y. Acidic pH environment induces autophagy in osteoblasts. Sci. Rep. 2017, 7, 46161. [Google Scholar] [CrossRef] [PubMed]
- Caley, M.P.; Martins, V.L.; O’Toole, E.A. Metalloproteinases and Wound Healing. Adv. Wound Care (New Rochelle) 2015, 4, 225–234. [Google Scholar] [CrossRef] [Green Version]
- Di Liddo, R.; Bertalot, T.; Borean, A.; Pirola, I.; Argentoni, A.; Schrenk, S.; Cenzi, C.; Capelli, S.; Conconi, M.T.; Parnigotto, P.P. Leucocyte and Platelet-rich Fibrin: A carrier of autologous multipotent cells for regenerative medicine. J. Cell Mol. Med. 2018, 22, 1840–1854. [Google Scholar] [CrossRef] [Green Version]
- Kuwana, M.; Okazaki, Y.; Kodama, H.; Izumi, K.; Yasuoka, H.; Ogawa, Y.; Kawakami, Y.; Ikeda, Y. Human circulating CD14+ monocytes as a source of progenitors that exhibit mesenchymal cell differentiation. J. Leukoc. Biol. 2003, 74, 833–845. [Google Scholar] [CrossRef]
- Kapellos, T.S.; Bonaguro, L.; Gemünd, I.; Reusch, N.; Saglam, A.; Hinkley, E.R.; Schultze, J.L. Human Monocyte Subsets and Phenotypes in Major Chronic Inflammatory Diseases. Front. Immunol. 2019, 10, 2035. [Google Scholar] [CrossRef] [Green Version]
- Graf, T.; Stadtfeld, M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell 2008, 3, 480–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frucht, D.M.; Aringer, M.; Galon, J.; Danning, C.; Brown, M.; Fan, S.; Centola, M.; Wu, C.Y.; Yamada, N.; El Gabalawy, H.; et al. Stat4 is expressed in activated peripheral blood monocytes, dendritic cells, and macrophages at sites of Th1-mediated inflammation. J. Immunol. 2000, 164, 4659–4664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amarasekara, D.S.; Yun, H.; Kim, S.; Lee, N.; Kim, H.; Rho, J. Regulation of Osteoclast Differentiation by Cytokine Networks. Immune Netw. 2018, 18, e8. [Google Scholar] [CrossRef] [PubMed]
- Parra-Izquierdo, I.; Castaños-Mollor, I.; López, J.; Gómez, C.; San Román, J.A.; Sánchez Crespo, M.; García-Rodríguez, C. Calcification Induced by Type I Interferon in Human Aortic Valve Interstitial Cells Is Larger in Males and Blunted by a Janus Kinase Inhibitor. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2148–2159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiehn, O. Metabolomics by gas chromatography−mass spectrometry: Combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef] [PubMed]






| Compound | R.T. (min) | Qion (m/z) | Concentration (mg/L) | |
|---|---|---|---|---|
| CGF | PPP | |||
| L-Alanine | 10.614 | 158 | 1.44 ± 0.03 | 1.51 ± 0.03 |
| Glycine | 10.843 | 218 | 0.83 ± 0.06 | 0.62 ± 0.04 |
| L-Valine | 11.903 | 186 | 1.08 ± 0.01 | 1.24 ± 0.01 |
| L-Leucine | 12.371 | 200 | 0.49 ± 0.03 | 0.47 ± 0.03 |
| Isoleucine | 12.672 | 200 | 0.23 ± 0.02 | 0.21 ± 0.02 |
| L-Proline | 14.564 | 184 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| L-Threonine | 15.528 | 404 | 0.47 ± 0.02 | 0.62 ± 0.02 |
| L-Phenylalanine | 15.901 | 234 | 0.22 ± 0.00 | 0.22 ± 0.00 |
| L-Aspartic | 16.485 | 418 | 0.09 ± 0.01 | 0.05± 0.01 |
| L-Glutamic | 17.640 | 432 | 0.56 * ± 0.04 | 0.06 ± 0.00 |
| L-Histidine | 19.426 | 196 | 0.13 ± 0.01 | 0.13 ± 0.01 |
| L-Tyrosine | 19.860 | 466 | 0.21 ± 0.02 | 0.21 ± 0.02 |
| Taurine | 14.154 | 296 | 3.82 * ± 0.11 | 0.08 ± 0.02 |
| FA18 | 19.035 | 341 | 47.9 ± 5.08 | 45.84 ± 4.87 |
| FA16 | 17.743 | 313 | 63.95 ± 0.53 | 63.27 ± 0.57 |
| FA14 | 16.065 | 285 | 3.57 ± 0.07 | 3.37 ± 0.06 |
| FA12 | 14.201 | 257 | 1.99 ± 0.05 | 1.61 ± 0.04 |
| FA10 | 12.187 | 229 | 1.3 ± 0.04 | 1.35 ± 0.03 |
| FA8 | 10.039 | 201 | 2.29 ± 0.03 | 2.17 ± 0.04 |
| Molecules | Quantity |
|---|---|
| VEGF | 792.8 ± 71.9 pg |
| TGF-β1 | 26.6 ± 3.1 ng |
| BMP-2 | 45.5 ± 5.7 pg |
| MMP-2 | 321.1 ± 29.5 ng |
| MMP-9 | 396.3 ± 34.3 ng |
| Gene Name | Accession Number | Sequences (5′–3′) | pb |
|---|---|---|---|
| Thy1 (CD90) | NM_006288.5 | F: ccactctggccattccc R: gagcaggagcagcagcag | 124 |
| CD73 | BC015940.1 | F: agcttacgattttgcacacc R: cggatctgctgaaccttgg | 133 |
| Endoglin (CD105) | NM_001278138.1 | F: gccagcattgtctcacttca R: atgcgcaacaagctctttct | 180 |
| CD34 | M81104.1 | F: caatgaggccacaacaaaca R: gtgactggacagaagagttt | 101 |
| PTPRC (CD45) | NM_080921.3 | F: atgaccatgtatttgtggctta R: tgggggaaggtgttgggc | 97 |
| CD31 | NM_000442.5 | F: atgatgcccagtttgaggtc R: acgtcttcagtggggttgtc | 172 |
| CD36 | NM_001001548.3 | F: agatgcagcctcatttccac R: gccttggatggaagaacaaa | 115 |
| CD14 | NM_000591.4 | F: acctaaagataaccggcacc R: ttgggcaatgctcagtacct | 163 |
| STAT4 | NM_003151.3 | F: aggaacggctgttgctaaag R: ttgtagtctcgcaggatgtc | 193 |
| Oct3 | NM_002701.5 | F: tattcagccaaacgaccatc R: gcaggaacaaattctccagg | 219 |
| Nanog | NM_024865.2 | F: agatgcctcacacggagac R: tcttctgtttcttgaccggg | 162 |
| RunX2 | NM_001278478.2 | F:gacaaccgcaccatggtgg R: tctggtacctctccgaggg | 160 |
| Col1a1 | NM_000088.3 | F: agggaatgcctggtgaacg R: gagagccatcagcacctttg | 90 |
| Ocn | NM_199173.6 | F: gctacctgtatcaatggct R: cgatgtggtcagccaactc | 111 |
| Gapdh | AJ005371.1 | F: atggccttccgtgtccccac R: acgcctgcttcaccaccttc | 245 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stanca, E.; Calabriso, N.; Giannotti, L.; Nitti, P.; Damiano, F.; Stanca, B.D.C.; Carluccio, M.A.; De Benedetto, G.E.; Demitri, C.; Palermo, A.; et al. Analysis of CGF Biomolecules, Structure and Cell Population: Characterization of the Stemness Features of CGF Cells and Osteogenic Potential. Int. J. Mol. Sci. 2021, 22, 8867. https://doi.org/10.3390/ijms22168867
Stanca E, Calabriso N, Giannotti L, Nitti P, Damiano F, Stanca BDC, Carluccio MA, De Benedetto GE, Demitri C, Palermo A, et al. Analysis of CGF Biomolecules, Structure and Cell Population: Characterization of the Stemness Features of CGF Cells and Osteogenic Potential. International Journal of Molecular Sciences. 2021; 22(16):8867. https://doi.org/10.3390/ijms22168867
Chicago/Turabian StyleStanca, Eleonora, Nadia Calabriso, Laura Giannotti, Paola Nitti, Fabrizio Damiano, Benedetta Di Chiara Stanca, Maria Annunziata Carluccio, Giuseppe Egidio De Benedetto, Christian Demitri, Andrea Palermo, and et al. 2021. "Analysis of CGF Biomolecules, Structure and Cell Population: Characterization of the Stemness Features of CGF Cells and Osteogenic Potential" International Journal of Molecular Sciences 22, no. 16: 8867. https://doi.org/10.3390/ijms22168867






